Development of a Polymicrobial Colony Biofilm Model to Test Antimicrobials in Cystic Fibrosis
In This Article
Summary
This methodology allows for the establishment of a polymicrobial biofilm model in cystic fibrosis for antimicrobial sensitivity testing in research and clinical laboratories. This model provides accurate and reliable results over a range of outputs.
Abstract
A range of bacteria biofilm models exist for the testing of antibiotics. However, many of these are limited to a single experimental output, such as colony-forming units or metabolic activity. Furthermore, many biofilm models do not reflect the biological and physiochemical properties of the human host environment. This is an important issue in many conditions, but most noticeably in cystic fibrosis (CF). A large proportion of people with CF suffer from both chronic and intermittent infections, and in vitro, antibiotic susceptibility testing poorly correlates with patient treatment outcomes. Some biofilm models incorporate CF lung-relevant media, including synthetic sputum mimics, but do not consider the polymicrobial nature of the environment, which alters biofilm architecture, physiology, and the way microbes respond to treatment. The solid-air interface colony biofilm model described here is highly adaptable and incorporates both CF-relevant media and a polymicrobial context. This model can also be used for mid-throughput screening of antimicrobials and to study their effect on polymicrobial dynamics. Output measurements from the model can be colony-forming units, metabolic activity, and confocal microscopy analysis. The model can easily be adapted to different microorganisms, media, temperatures, and variable oxygen conditions and can be used to test a wide range of chemical, biological, and physical treatments.
Introduction
Cystic fibrosis (CF) is a genetic condition that affects over 11,000 people in the UK and 162,000 people globally1,2. Although CF is a multi-organ disease, a key symptom experienced by people with cystic fibrosis (pwCF) is the formation of abnormally thick, dehydrated mucus within their respiratory tract3. This, along with reduced cilia beating, can enhance the colonization of the lungs by a wide range of bacteria, fungi, viruses, and archaea4,5. Despite the CF lung providing conditions and selective pressures to limit the growth and survival of microorganisms, bacteria such as Pseudomonas aeruginosa are highly adapted to these harsh environments6. This allows for both colonization and survival, resulting in the formation of persistent chronic infections7.
Many of the microbes that cause these chronic infections do so using a phenotypic shift from an initial planktonic to a biofilm style of growth, either surface attached or in aggregates7. These biofilms are characterized by tightly packed bacterial communities encased by an exopolysaccharide (EPS) matrix consisting of a variety of components, including polysaccharides, proteins, lipids, and environmental DNA (eDNA)8. This matrix is a common feature between microorganisms; however, its composition can differ. For example, the main polysaccharide components of the bacterial P. aeruginosa matrix are Psl, Pel, and alginate, unlike in the fungi Candida albicans, whose main polysaccharide matrix components are mannans and glucans9. In mono-species biofilms, this matrix can greatly affect antibiotic tolerance compared to planktonic by reducing the penetration of antibiotics into the biofilm, decreasing their effectiveness10. The biofilm style of growth also induces the formation of persister cells with reduced metabolic activity compared to their planktonic counterparts and, therefore, further decreased susceptibility to antibiotics11. This style of growth is also characterized by the upregulation of some antibiotic resistance mechanisms, such as efflux pumps and those required for horizontal gene transfer, enabling the exchange of resistance genes11,12,13. In addition to these, the disease-related environment of the host very much influences the microbial physiology and the way they respond to antibiotics. This includes the increased micro-aerobiosis within the thick mucus as well as the availability of non-standard carbon sources such as amino acids and eDNA, either host-derived or from microbial degradation of lung products14,15,16.
These specific interactions are further complicated by the polymicrobial nature of biofilms, which introduces an additional layer of complexity with complex interactions not just between bacteria but also between bacteria and fungi. Compared to bacterial biofilms, there is less known regarding some of the interactions between bacterial and fungal biofilms, despite C. albicans being isolated from over 75% of people with CF15. Generally, the interactions between C. albicans and bacteria like P. aeruginosa are antagonistic but can result in more chronic and severe infections18. The combination of microbial interactions, both pathogenic and commensal, alongside a range of CF-related environmental factors can ultimately result in increased antibiotic tolerance19,20. Many of these factors are not often considered in preclinical antibiotic testing despite being attributed to increased antibiotic tolerance in existing models21.
These conditions are also difficult to recapitulate in vitro, and as a result, many models lack the presence of specific tolerance-inducing factors present in pwCF, such as those increasing beta-lactamase production in P. aeruginosa22, the induction of small colony variants in Staphylococcus aureus and the inhibition of hyphenation in C. albicans; all of which have been shown to occur in CF sputum23,24.
There is, therefore, a large disparity between the conditions used in current antibiotic sensitivity testing methods, based on planktonic or agar-plate grown cultures in standardized media such as Mueller-Hinton with disk diffusion, Etest, and CLSI broth microdilution assays, and the conditions encountered in the host environment25. This frequently fails to accurately determine the antibiotic sensitivity26. This problem is further complicated by a lack of standardization in antimicrobial biofilm testing, making it hard to accurately translate antimicrobial efficacy from the laboratory to the clinic27,28.
The polymicrobial model we have developed here demonstrates enhanced tolerance of Pseudomonas aeruginosa to a range of antimicrobials, including meropenem and tobramycin. This illustrates the large variation between current antimicrobial testing using mono-species biofilms and the polymicrobial biofilm model developed in this study to determine minimum inhibitory concentrations. This model also maintains a relatively high throughput and low-cost desirable for antimicrobial testing. The model can also be used to study the impact of antimicrobial therapy on polymicrobial dynamics and establish whether a particular treatment may lead to a particular pathogen becoming dominant, enabling the prediction of further complications. Although this model permits the build-up of complex biofilms, its set-up does not require sophisticated laboratory equipment and provides a platform for a wide range of clinical and research outputs.
Protocol
1. Preparation of Synthetic Cystic Fibrosis media-2 (SCFM2) and plates
- Make SCFM2 according to the recipe described in Palmer et al.16 in a 500 mL borosilicate glass bottle at 2x concentration with a few modifications.
- Weigh out and prepare stocks as indicated in Table 1. Prepare amino acid stocks according to Table 2.
- Weigh out 5 g mucin (from Porcine stomach (Type II)) and 600 mg eDNA (from salmon sperm) in 100 mL of dH2O in a 200 mL borosilicate glass bottle. Stir at 4 °C overnight before sterilization by autoclaving.
- To a 500 mL sterilized bottle, add 0.54 g glucose, 6.06 g NaCl, and 2.228 g KCl. Add 10 mL each of the stocks prepared in step 1.1.1. Adjust pH to 6.9 using 1 M NaOH or HCl.
- Add 10 mL of dH2O followed by 10 mL of 0.175 M CaCl2 and 0.0606 M of MgCl2. Prepare and add 10 mL of 0.93 M L-lactic acid. Immediately before use, add 10 mL of 0.36 mM Fe(III)SO47H2O
- Filter sterilize the SCFM2 using a filter unit and vacuum pump, then add the autoclaved mucin and eDNA.
NOTE: The prepared SCFM2 can be stored at 4 °C for up to 1 month.
- Prepare SCFM2 agar plates as described below.
- Preheat 25 mL of 2x SCFM2 in a 50 mL tube in a 55 °C water bath before mixing with 25 mL of pre-melted 3% (w/v) technical agar, resulting in a mix of 1x SCFM2 and 1.5% (w/v) technical agar.
- Using a serological pipette, transfer 5 mL of the mixture into each well of a 6-well plate.
- Prepare the same number of 13 mm polycarbonate discs with a pore size of 0.2 µm by placing them on the bottom of a Petri dish and applying UV-C for 600 s at a wavelength of 254 nm and a frequency of 60 Hz. Flip the discs over and repeat the UV-C procedure.
- Once the 1x SCFM2-1.5 % (w/v) technical agar has set, use sterile forceps to add a single polycarbonate disc to the surface of each well of the 6-well plate.
2. Preparation of bacteria for infection
- Before setting up the model, prepare agar plate/s for the required bacteria and fungal strains according to the standard agar used in the lab for P. aeruginosa PAO1, S. aureus SH1000 and C. albicans SC5314 (e.g., Lysogeny broth with 1.2% (w/v) agar for bacteria and Sabouraud dextrose agar for C. albicans).
NOTE: To establish the model, specific laboratory reference strains have been used. However, this model has also been used with other strains, including clinical isolates, with some modifications, such as increasing the time for biofilm establishment before treatment for slower-growing strains. - Use a 1 µL inoculation loop to pick a single colony of each microorganism to inoculate 5 mL of standard laboratory liquid media (e.g., Lysogeny broth for P. aeruginosa, S. aureus and Yeast peptone dextrose (YPD) for C. albicans). Incubate these cultures until they reach the exponential growth phase at 200 rpm, 37 °C for the bacteria, and 200 rpm, 30 °C for C. albicans.
NOTE: Aeration will differ depending on the shaking incubator and radius of rotation
3. Setting up the solid air interface model
NOTE: An overall schematic representation of the model can be seen in Supplementary Figure 1.
- Transfer 1 mL of each liquid culture to a separate sterile 1.5 mL tube and centrifuge for 2 min at 8,000 x g to pellet the bacteria/fungi. Perform this and all the following centrifugation steps at room temperature.
- Aspirate the supernatant using a pipette and resuspend the pellets in 1 mL of PBS before centrifugation, once again, for 2 min at 8,000 x g.
- Remove the supernatant as before and resuspend the pellets in 1 mL of PBS. Dilute the resuspended P. aeruginosa and S. aureus cells 1:10 in PBS and measure optical density using a spectrophotometer at a wavelength of 600 nm.
- Adjust washed samples to 1 x 108 colony forming unit (CFU)/mL based on prior validation that an OD600 of 0.05 for P. aeruginosa and 0.1 for S. aureus is equivalent to 1 x 108 CFU/mL.
- For C. albicans, dilute washed samples 1:100 in PBS, add 10 µL of each to two individual chambers of a hemocytometer, and use the fixed dimensions of the chamber and the dilution used to calculate CFU/ mL. Using the improved Neubauer hemocytometer the calculation will be as follow:
Number of cells = average cell count × 10,000 / (dilution factor) - The calculated CFUs represent C. albicans CFU/ mL in the pre-diluted sample. Adjust the C. albicans sample to 1 x 108 CFU/mL in PBS as with the bacterial cultures.
- To prepare the inoculum for addition to the biofilm, adjust the P. aeruginosa inoculum to 1 x 104 CFU/mL (2x 100-fold dilutions), C. albicans to 1 x 105 CFU/mL (100-fold dilution followed by a 10-fold dilution) and finally, S. aureus to 1 x 106 CFU/mL (100-fold dilution) using PBS.
- For mono-species biofilms, add 10 µL of the diluted desired microbe to the center of the polycarbonate disc previously placed on the 1x SCFM2-1.5 % technical agar six-well plates and incubate statically at 37 °C for 24 h before treatment or disruption.
- For polymicrobial biofilms, add 10 µL of each S. aureus and C. albicans on top of one another on the same disc. Incubate the inoculated disk statically for 24 h at 37 °C.
NOTE: The order in which S. aureus and C. albicans inoculums are added does not affect the development of the model and generates the same results. - Following this, transfer the polycarbonate discs to fresh 1x SCFM2-1.5% technical agar plates with sterile forceps and add 10 µL of the 1 x 104 CFU/mL dilution of P. aeruginosa on top of the pre-established S. aureus - C. albicans biofilm. Incubate for another 24 h at 37 °C.
NOTE: P. aeruginosa is added 24 h after S. aureus and C. albicans to reflect the order in which these organisms frequently colonize the CF lung in people with CF. This also prevents P. aeruginosa from outgrowing the other two microorganisms. Once P. aeruginosa has been added, the biofilm can be grown for longer than 24 h; however, the polycarbonate disk with the biofilm needs to be transferred to fresh media every 24 h. Before disruption, biofilms can be used for microscopy using strains that express fluorescent proteins, fluorescent dyes, or strain/microbe-specific antibodies. However, it is worth noting that some dyes stain the polycarbonate disc.
4. Biofilm disruption
- Add ceramic beads with a diameter of 2.8 mm to the plates and UV-crosslink the plates as described in step 1.3. Add 5 ceramic beads to a sterile 2 mL homogenizer tube using flame-sterilized forceps. Then, pipette 1 mL of phosphate-buffered saline (PBS) into the 2 mL homogenization tube.
- Vortex tubes for at least 10 s or until the biofilm has been removed from the polycarbonate disc, determined by visual inspection, at which point remove the polycarbonate disc.
- Place tubes with the beads in the homogenizer and beat for 2x for 10 s at 6 m/s with a 10-s interval between beats.
NOTE: This combination of beads, beating time, and homogenizer use results in sufficient disruption of the biofilm with minimal lysis as determined by microscopy and colony-forming units. This may vary depending on the specific bead beater used. - Pour disrupted biofilm from the homogenizer tube into a 7 mL bijou containing 4 mL of PBS, resulting in a final volume of 5 mL.
- Ensure all liquid from the homogenizer tubes has been transferred to the bijou by centrifuging the tubes at 8,000 x g for 2 min and transferring the remaining liquid to the 7 mL bijou.
NOTE: From this 5 mL of disrupted biofilm, a range of assays can be conducted, but it is key to remember for any calculations the 1 mL biofilm PBS dilution into a final volume of 5 mL.
5. Metabolic activity testing
- Add 200 µL of the disrupted biofilm from step 4 above to a clear-bottomed black 96 well plate. Then, add 10 µL of 0.02% (w/v) resazurin solution.
- Incubate the resazurin-treated biofilm with a plate reader and read the metabolic activity. Incubate the plate at 37 °C, applying orbital shaking every 30 min for 10 s at 200 rpm before taking any fluorescent readings.
- Measure the fluorescence using an excitation of 540 nm and an emission of 590 nm. Repeat this 30 min cycle for 4 h using a plate reader.
NOTE: The 4 h time point is used as it provides more distinct variations between treated versus untreated samples and between samples with varying antibiotic concentrations.
6. Antimicrobial sensitivity testing
- To test the antimicrobial tolerance of standard antibiotics, solubilize the antibiotics of choice in their relevant solvent and add this to the 1x SCFM2-1.5% (w/v) technical agar medium while still liquid before transferring 5 mL to each well of a six-well plate. Once set, transfer the biofilm grown on the polycarbonate disc to the top of the antibiotic-containing media using sterile forceps and add a 10 µL drop of the corresponding antibiotic concentration onto the top of the biofilm.
NOTE: For this study, a 2-log fold serial dilution between 64 µg/mL and 0.125 µg/mL of meropenem and tobramycin was used to reflect clinically relevant concentrations. Administering antibiotics or other antimicrobial/anti-virulence treatments solely on top of the biofilm is sufficient, as similar responses have been observed when added to both the media and onto the biofilm. - After 24 h of incubation at 37 °C with the treatment, image biofilms and disrupt as described in step 4.
- From the 5 mL of disrupted culture, take an aliquot and carry out a 10-fold serial dilution using PBS. Plate out 10 µL spots in triplicate on agar to determine colony-forming units using the Miles and Misra method30.
NOTE: The volume added to agar plates will impact the limit of detection, larger volumes will reduce the limit of detection but require more sample and agar plates. - For mono-species biofilms streak P. aeruginosa and S. aureus onto LB agar plates and C. albicans onto SDA agar.
- For polymicrobial biofilms, place aliquots onto three different selective media containing antibiotics to isolate each of the three species and enable the determination of CFUs. This includes Pseudomonas isolation agar and mannitol salt agar containing nystatin at 10 µg/mL to isolate P. aeruginosa and S. aureus, respectively, and Sabouraud dextrose agar with tetracycline at 125 µg/mL to isolate C. albicans. Incubate S. aureus plates at 37 °C overnight and P. aeruginosa and C. albicans plates at 30 °C.
NOTE: Incubate P. aeruginosa plates at 30 °C to minimize colony overgrowth and reduce swarming.
Representative Results
The simplicity of the solid-air interface biofilm model enables the screening of a large number of antimicrobials under different clinically relevant conditions at one time. This model allows the evaluation of the effectiveness of antibiotics using CFU counts and metabolic assays in both mono and polymicrobial biofilms to be carried out in a week. Due to the nature of the model, it also allows easy manipulation of the environmental conditions, such as changing the media composition and placing the biofilms under reduced oxygen and anaerobic conditions. Using this model, we have shown changes in tolerance to two antibiotics commonly used in pwCF between P. aeruginosa grown in mono-species and polymicrobial biofilms (Figure 1). Alongside this, we have been able to determine how antimicrobial treatment can impact the population dynamics within polymicrobial biofilms (Figure 2).
To carry out these experiments, the initial inoculum was prepared as specified in the above protocol, leading to the formation of stable mono species and polymicrobial biofilms with reproducible CFU/mL and small standard deviations as determined by one or two-way ANOVA. This model also highlighted consistent growth and CFU recovery, as well as allowing for the possibility of combining these classical microbiology techniques with other techniques not performed in this study without the need for sophisticated processing. These include imaging such as live/dead, matrix visualization, and molecular analysis, which other three-dimensional models are unable to offer in such an accessible manner (Figure 1)31. It should be noted that the standard deviation expected for CFU/mL increases when data is displayed as the percentage of survival compared to CFU/mL (Figure 1). When comparing the effectiveness of antibiotic treatment between mono species and polymicrobial biofilms, there was a significant increase in all the antibiotic concentrations required to achieve 50% killing. A 2-log increase in antibiotic concentration was needed to achieve this level of killing for meropenem and tobramycin (Figure 1). There was also an overall increase in P. aeruginosa survival in the presence of S. aureus and C. albicans in the polymicrobial biofilm when treated with 64 µg/mL tobramycin, whereas the opposite was true for meropenem.
The method used to determine the metabolic activity measures the overall activity of the whole polymicrobial biofilm without being able to distinguish the individual contribution from each species. For this reason, only mono-species metabolic changes are shown to demonstrate the use of metabolic activity assays for this model. For mono species biofilms there was a strong relationship between P. aeruginosa survival and metabolic activity for both meropenem and tobramycin (Figure 2). The utilization of both metabolic activity and CFU counts from the same samples enables the easy identification of both bacteriostatic and bactericidal effects. For polymicrobial biofilms, the determination of metabolic activity may indicate how the targeting of one species may induce an overall increase in biofilm metabolic activity and, therefore, potentially increase the microbial growth of other species.
Although CFUs are the gold standard for antimicrobial sensitivity testing, in polymicrobial biofilms they also allow the assessment of species composition within the biofilm. By treating P. aeruginosa in polymicrobial biofilms, we are not only able to show the increased MBEC50 for this organism but also establish the effect this has on the other co-isolated species (Figure 3). This is, for instance, seen with meropenem (antipseudomonal), where the reduction of P. aeruginosa is accompanied by an increase in S. aureus and C. albicans CFU, resulting in S. aureus becoming the most prevalent organism upon exposure to certain antibiotic concentrations (Figure 3A). This highlights the importance of considering the polymicrobial nature of the biofilm due to the potential increase in the population size of other disease-causing species when treating a particular pathogen.
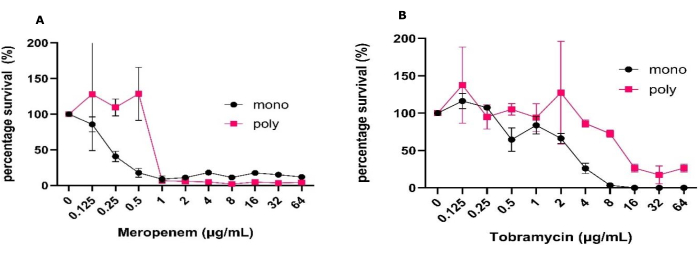
Figure 1: Variation in antimicrobial tolerance of P. aeruginosa to meropenem and tobramycin between mono and polymicrobial biofilms grown in the solid-air interface model. P. aeruginosa PAO1 grown in mono or poly microbial biofilms with S. aureus and C. albicans was established using the solid-air interface models for 24 h on SCFM2. Biofilms were treated with a range of (A) meropenem or (B) tobramycin concentrations from 0.125 µg/mL to 64 µg/mL along with a no antibiotic control. The CFU/mL of each biofilm was determined. P. aeruginosa CFUs were determined in PIA and converted into percentage survival by using the negative control as the 100% survival. Error bars denote standard deviation, and each data point is derived from 3 biological repeats, each with three technical repeats. Please click here to view a larger version of this figure.

Figure 2: Variation in metabolic activity of P. aeruginosa biofilms in the presence of meropenem or tobramycin grown in the solid-air interface model. P. aeruginosa PAO1 biofilms were grown on SCFM2 in the solid-air interface model, and a range of (A) meropenem or (B) tobramycin concentrations from 0.125 µg/mL to 64 µg/mL were added along with a no antibiotic control. The plate was read fluorescently at an excitation of 540 nm and emission of 590 nm every 30 min. Percentage metabolic activity was calculated by determining the percentage of this activity on each antibiotic-treated sample compared to the no antibiotic control. Error bars denote standard deviation, and each data point is derived from 3 biological repeats, each with three technical repeats. Please click here to view a larger version of this figure.
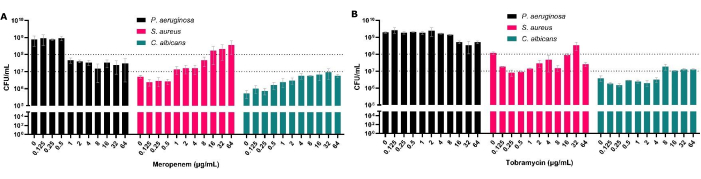
Figure 3: Impact on total CFU counts of P. aeruginosa, S. aureus, and C. albicans recovered from polymicrobial solid-air interface model following antimicrobial treatment. S. aureus and C. albicans were grown on SCFM2 on the solid-air interface model. P. aeruginosa was added, and a range of 0.125 µg/mL to 64 µg/mL concentrations of (A) meropenem or (B) tobramycin were added to the biofilms alongside a no-antibiotic control. The CFU/mL of each biofilm was determined. Error bars denote standard deviation, and each data point was derived from 3 biological repeats, each with three technical repeats. Please click here to view a larger version of this figure.
Supplementary Figure 1: Establishment and workflow of the solid-air interface model. Graphical representation of the solid-air interface model establishment, biofilm disruption, and outputs. Please click here to download this File.
Discussion
The biofilm model described here enables us to mimic some aspects of the CF lung infection environment by incorporating a range of common polymicrobial interactions in a more accessible way than currently used in vitro antimicrobial testing methods such as the CDC reactor and the Lubbock biofilm model31. This is important when a range of factors, including the physical properties of the biofilm, nutrient availability, and molecular interactions between the species within the biofilm, can affect both the activity of the antimicrobial agent and the response of the microbes to the antimicrobial21. By incorporating these factors, we have been able to develop a highly versatile model that may more accurately enable the prediction of antimicrobial treatment outcomes than current models that do not consider the aforementioned factors.
P. aeruginosa, S. aureus, and C. albicans were chosen as representative Gram-positive, Gram-negative, and fungi commonly isolated in CF32. However, we have also been able to show that this model can be used for a range of other CF-relevant microbes, including Burkholderia cenocepacia, Burkholderia multivorans, Aspergillus fumigatus, and is currently being adapted for use with anaerobic bacteria such as Prevotella melaninogenica. This demonstrates the versatility of the model as a wide range of microbes could be incorporated, highlighting this model's adaptability for antimicrobial testing against a wide range of microbial pathogens. The model can also be easily adapted to a range of environmental conditions for more personalized antimicrobial sensitivity testing, including the replacement of SCFM2 with sputum from the patient.
The consideration of polymicrobial biofilm models for the testing of new antimicrobials is also paramount, as they can cover an existing gap in the antimicrobial development pipeline prior to their use in animal or human studies33,34. Alongside this, the model maintains a relatively high throughput and allows for a range of outputs relevant to both industry and research.
Although we have only exemplified CFUs and metabolic activity in this methods paper, we have also demonstrated the utility of the model in the analysis of the biofilm exo-metabolome using liquid extraction surface analysis mass spectrometry (LESA). This allows for probing some of the molecular mechanisms underpinning antimicrobial tolerance35. In addition to these other outputs, we have also demonstrated the use of this model for testing novel antimicrobial delivery systems. These include polymer-ciprofloxacin conjugates to enhance the penetration of this antibiotic within the biofilm while increasing its activity and reducing the development of resistance36.
Critical steps and considerations
It is important to maintain sterility, especially when producing SCFM2 and media, as contamination can result in the introduction of non-naturally occurring species, which can interfere with the results. Different strains of the same microbe and different microbes may require different environmental conditions for growth or may have varying growth characteristics. Therefore, the model must be optimized before carrying out any antibiotic sensitivity testing. We recommend carrying out a small pilot study of 24 h and 48 h for mono species and polymicrobial biofilm to assess growth under standard model conditions. We have found that different P. aeruginosa strains and certain antibiotic concentrations can induce swarming off the polycarbonate disc. This can be easily overcome by using larger-sized discs which are commercially available. Biofilm disruption may also require optimization depending on the bead beater and the microbes used. We optimized the disruption method by determining the CFU/mL of a known sample before and after bead beating. We also used microscopy to visualize the bead beat samples to visualize how well the biofilm was disrupted.
One of the limitations of this CF-mimicking biofilm model is the attachment to a solid surface containing polycarbonate, which is not found in the CF lung, rather than being free-floating aggregates within the CF sputum37 . However, the solid agar model presented here precludes the need to wash the biofilms to remove planktonic microbes and ensures that the results only include microbes derived from the biofilm as opposed to a mixed planktonic biofilm culture. We have also found that the penetration of some antibiotics to the deeper areas of this biofilm model can be reduced, and the use of DNase may be required to alter the rheology of the extracellular matrix and decrease the electrostatic interactions with eDNA.
We believe that the model is highly suitable for antimicrobial testing due to its versatility, adaptability, and inclusion of environmental factors that many existing models fail to take into account. Its use in fundamental research and pre-clinical antimicrobial sensitivity testing can provide a more clinically relevant approach for AST of clinical samples and the development of novel therapeutics.
Acknowledgements
This work has been funded by the National Biofilms Innovation Centre (NBIC) which is an Innovation and Knowledge Centre funded by the Biotechnology and Biological Sciences Research Council, Innovate UK and Hartree Centre [Awards BB/R012415/1 and BB/X002950/1] and by the UK CF Trust and USA CF Foundation Strategic Research Centre: 'An evidence-based preclinical framework for the development of antimicrobial therapeutics in cystic fibrosis' (PIPE-CF) [Award SRC022].
Materials
| Name | Company | Catalog Number | Comments |
| 1 µL inoculation loops | |||
| 13 mm 0.2 µm pore size polycarbonate discs | Isopore | GTTP01300 | Larger discs are also available |
| 2 mL reinforced tubes | Thermofisher | 15545809 | |
| 2.5 mL ceramic beads | Qiagen | 13114-325 | |
| 500 mL borosilicate glass Duran bottle | Sigma Aldrich | Z305197 | larger bottles available in larger volumes are desired |
| 6-well culture plates | Greiner | 657165 | |
| 7 mL Bijou | Thermofisher | 129B | |
| 96-well plates | Thermofisher | 167008 | for serial dilutions in CFU assay |
| Agar plates for preparing plates of P. aerugnisa, S. aureus, and C. albicans | LB miller for P. aeruginosa and S. aureus and Sabouraud dextrose agar for C. albicans | ||
| Bead beater - suitable for 2 mL tubes | Fisherbrand | 15515799 | Thermofisher bead mill 24 |
| bench top centrifuge | must be capable of at least 8000 x g | ||
| Black clear bottom 96 well plates | Costar | 3603 | |
| Bunsen Burner | |||
| Containers for disposing of contaminated equipment and material according to the institutes health and safety regulations. | |||
| deionised water | |||
| eDNA | Sigma Aldrich | 31149 | |
| Filter unit | Fisherbrand | FB12566504 | Interchangeable depending on the vacuum pump used but must have a pore size of 0.2 µm |
| Haemocytometer and cover slip | Hawksley | HC001 | Haemocytometers may differ in size and volume. Double check and adjust CFU calculations accordingly |
| LB broth | oxoid | 1.46813 | |
| Mannitol salt agar | Oxoid | CM0085B | |
| meropenem | abcr | Ab243429 | |
| Mucin from porcine stomach Type II | Sigma Aldrich | M2378 | |
| Nystatin | Millipore | 1003352658 | |
| petri dishes | SLS | SLS2000 | |
| Phosphate buffered saline | |||
| Pseudomonas isolation agar | Millipore | 17208 | |
| Resazurin sodium salt | Sigma Aldrich | 199303 | |
| Sabouraud dextrose agar | Oxoid | CM0041 | |
| selection of forceps | fine tipped and tissue forceps with teeth for transferring ceramic beads | ||
| serological pipette | |||
| shaking and static incubators | must be temperature controlled | ||
| Sparks microtitre plate reader | Tecan | For Resazurin assay the microtitre plate reader must have the appropriate filters or be a monochromator for detecting flourescence. | |
| spectrophotometer | |||
| Technical agar (Agar Technical No.2 ) | Oxoid | LP0012B | |
| tetracycline | Sigma Aldrich | T7660 | |
| UV crosslinker | Spectroline | 11-992-89 | |
| vacuum pump/ flask | Fisherbrand | FB12566504 | |
| water bath | must be capable of maintaining 55 °C | ||
| YPD broth | Millipore | Y1375 | Can be bought pre-made or made using the base ingredients |
References
- . 2021 Cf foundation patient registry highlights report Available from: https://cysticfibrosis.msu.edu/index.php/welcome/news-and-events/39-2021-cf-foundation-patient-registry-highlights-report (2022)
- Guo, J., Garratt, A., Hill, A. Worldwide rates of diagnosis and effective treatment for cystic fibrosis. J Cyst Fibros. 21 (3), 456-462 (2022).
- Mandal, V., Ghosh, N. N., Mitra, P. K., Mandal, S., Mandal, V. Production and characterization of a broad-spectrum antimicrobial 5-butyl-2-pyridine carboxylic acid from Aspergillus fumigatus nhf-01. Sci Rep. 12 (1), 6006 (2022).
- Françoise, A., Héry-Arnaud, G. The microbiome in cystic fibrosis pulmonary disease. Genes. 11 (5), 536 (2020).
- Grasemann, H., Ratjen, F. Cystic fibrosis. New Engl J Med. 389 (18), 1693-1707 (2023).
- Camus, L., Vandenesch, F., Moreau, K. From genotype to phenotype: Adaptations of pseudomonas aeruginosa to the cystic fibrosis environment. Microb Genom. 7 (3), mgen000513 (2021).
- Rossi, E., et al. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat Rev Microbiol. 19 (5), 331-342 (2021).
- Flemming, H. -. C., Wingender, J. The biofilm matrix. Nat Rev Microbiol. 8 (9), 623-633 (2010).
- Grainha, T., Jorge, P., Alves, D., Lopes, S. P., Pereira, M. O. Unraveling pseudomonas aeruginosa and candida albicans communication in coinfection scenarios: Insights through network analysis. Front Cell Infect Microbiol. 10, 550505 (2020).
- Flemming, H. C., et al. Biofilms: An emergent form of bacterial life. Nat Rev Microbiol. 14 (9), 563-575 (2016).
- Høiby, N., Bjarnsholt, T., Givskov, M., Molin, S., Ciofu, O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 35 (4), 322-332 (2010).
- Lorusso, A. B., Carrara, J. A., Barroso, C. D. N., Tuon, F. F., Faoro, H. Role of efflux pumps on antimicrobial resistance in Pseudomonas aeruginosa. Int J Mol Sci. 23 (24), 15779 (2022).
- Michaelis, C., Grohmann, E. Horizontal gene transfer of antibiotic resistance genes in biofilms. Antibiotics .(Basel). 12 (2), 328 (2023).
- Worlitzsch, D., et al. Effects of reduced mucus oxygen concentration in airway pseudomonas infections of cystic fibrosis patients. J Clin Invest. 109 (3), 317-325 (2002).
- König, J., Schreiber, R., Voelcker, T., Mall, M., Kunzelmann, K. The cystic fibrosis transmembrane conductance regulator (CFTR) inhibits ENaC through an increase in the intracellular Cl- concentration. EMBO Reps. 2 (11), 1047-1051 (2001).
- Palmer, K. L., Mashburn, L. M., Singh, P. K., Whiteley, M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol. 187 (15), 5267-5277 (2005).
- Valenza, G., et al. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros. 7 (2), 123-127 (2008).
- Alam, F., Catlow, D., Di Maio, A., Blair, J. M. A., Hall, R. A. Candida albicans enhances meropenem tolerance of Pseudomonas aeruginosa in a dual-species biofilm. J Antimicrob Chemother. 75 (4), 925-935 (2020).
- Burmølle, M., Ren, D., Bjarnsholt, T., Sørensen, S. J. Interactions in multispecies biofilms: Do they actually matter. Trends Microbiol. 22 (2), 84-91 (2014).
- Lories, B., Belpaire, T. E. R., Smeets, B., Steenackers, H. P. Competition quenching strategies reduce antibiotic tolerance in polymicrobial biofilms. NPJ Biofilms Microbiomes. 10 (1), 23 (2024).
- Nabb, D. L., et al. Polymicrobial interactions induce multidrug tolerance in Staphylococcus aureus through energy depletion. Front Microbiol. 10, 2803 (2019).
- Cornforth, D. M., et al. Pseudomonas aeruginosa transcriptome during human infection. Proc Natl Acad Sci U S A. 115 (22), E5125-E5134 (2018).
- Wolter, D. J., et al. Staphylococcus aureus small-colony variants are independently associated with worse lung disease in children with cystic fibrosis. Clin Infect Dis. 57 (3), 384-391 (2013).
- Takagi, J., et al. Mucin o-glycans are natural inhibitors of Candida albicans pathogenicity. Nat Chem Biol. 18 (7), 762-773 (2022).
- Cinical and Laboratory Standard Institute. . Methods for dilution susceptibility tests for bacteria that grow aerobically. 35 (2), (2015).
- Roberts, A. E., Kragh, K. N., Bjarnsholt, T., Diggle, S. P. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J Mol Biol. 427 (23), 3646-3661 (2015).
- Macia, M. D., Rojo-Molinero, E., Oliver, A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin Microbiol Infect. 20 (10), 981-990 (2014).
- Coenye, T., Goeres, D., Van Bambeke, F., Bjarnsholt, T. Should standardized susceptibility testing for microbial biofilms be introduced in clinical practice. Clin Microbiol Infect. 24 (6), 570-572 (2018).
- Turner, K. H., Wessel, A. K., Palmer, G. C., Murray, J. L., Whiteley, M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A. 112 (13), 4110-4115 (2015).
- Miles, A. A., Misra, S. S., Irwin, J. O. The estimation of the bactericidal power of the blood. J Hyg. 38 (6), 732-749 (1938).
- Robertson, S. N., Romero, M., Fenn, S., Kohler Riedi, P. L., Cámara, M. Development, characterization, and evaluation of a simple polymicrobial colony biofilm model for testing of antimicrobial wound dressings. J Appl Microbiol. 135 (3), lxae042 (2024).
- Rumpf, C., Lange, J., Schwartbeck, B., Kahl, B. C. Staphylococcus aureus and cystic fibrosis-a close relationship. What can we learn from sequencing studies. Pathogens. 10 (9), 1177 (2021).
- Tay, W. H., Chong, K. K. L., Kline, K. A. Polymicrobial-host interactions during infection. J Mol Biol. 428 (17), 3355-3371 (2016).
- Orazi, G., O'toole, G. A. "It takes a village": Mechanisms underlying antimicrobial recalcitrance of polymicrobial biofilms. J Bacteriol. 202 (1), e00530-e00519 (2019).
- Arjes, H. A., et al. Three-dimensional biofilm colony growth supports a mutualism involving matrix and nutrient sharing. eLife. 10, e64145 (2021).
- Kasza, K., et al. Ciprofloxacin poly(β-amino ester) conjugates enhance antibiofilm activity and slow the development of resistance. ACS Appl Mater Interf. 16 (5), 5412-5425 (2024).
- Martin, I., Waters, V., Grasemann, H. Approaches to targeting bacterial biofilms in cystic fibrosis airways. Int J Mol Sci. 22 (4), 2155 (2021).
Explore More Articles
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2025 MyJoVE Corporation. All rights reserved