A subscription to JoVE is required to view this content. Sign in or start your free trial.
Investigating a Zebrafish Larval Brain Injury Using Large-Scale Scanning Transmission Electron Microscopy
In This Article
Overview
This video demonstrates a method to investigate zebrafish larval brain injury using large-scale scanning transmission electron microscopy (STEM), a technique that combines elements of both scanning electron microscopy (SEM) and transmission electron microscopy (TEM) to produce high-resolution images. It outlines the steps involved in preparing samples and the acquisition of high-resolution STEM images to identify brain injury.
Protocol
1. Sample Preparation
- Fixation and Embedding
- Fixation
- Anaesthetize fish larvae in tricaine (0.003%) added to the E3 (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, 10 mM HEPES, pH 7.6) zebrafish water and test for depth of anesthesia by poking gently with a 200 µl pipette tip under dissection microscope until motion is no longer detected (Figure 1A).
- When processing samples for correlative electron microscopy (EM) after acquiring in vivo imaging data using confocal or 2 photon imaging in 1.8% agarose as described, fix larvae in agarose using 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) containing Triton-X-100 (0.05%).
- Cut larvae from agarose and process for large-scale EM.
- Fix larvae in fresh 4% paraformaldehyde in PBS containing Triton-X-100 (0.05%) for 2 hr at room temperature in plastic tubes.
Note: Cells grown in culture (in vitro) can be directly fixed in glutaraldehyde, as described in the next step (1.1.1.5). - Remove the solution and add 100 µl 0.5% PFA, 2% glutaraldehyde (GA), and 0.1 M sodium cacodylate (pH 7.4). Store overnight at 4 °C. Rinse samples twice in 0.1 M sodium cacodylate.
- Cut larval heads rostrally to the hindbrain to facilitate penetration of osmium. Use fine syringes/forceps.
- Postfix larvae in 1% osmium tetroxide (OsO4)/1.5% potassium ferrocyanide (K4[Fe(CN)6]) in 0.1 M sodium cacodylate on ice for 2 hr and rinse 3 x 5 min in double distilled H2O.
- Dehydrate in ethanol by 20 min incubations in increasing ethanol concentrations (50%, 70%, 3 times 100%).
- Embedding
- Prepare epoxy resin according to standard recipes used in EM labs.
Note: We use epoxy resin as follows: mix 19.8 g glycid ether 100, 9.6 g 2-dodecenylsuccinic acid anhydride, and 11.4 g methylnadic anhydride using a magnetic stirrer. Add 0.5 g DMP-30 (tris-(dimethylaminomethyl) phenol) and mix again. - Incubate larvae overnight in 50% epoxy resin in ethanol (v/v) while rotating (RT).
- Replace diluted epoxy-resin with pure resin. Incubate for 30 min, add fresh resin again and incubate for another 30 min. Again, refresh the resin and then incubate 3 hr at RT, followed by 15 min at 58 °C and another 1 hr at RT under low pressure (200 mbar).
- Orient the heads in commercially available silicon flat embedding molds under a dissection microscope using a needle or toothpick with the anterior facing the side of the mold or as needed (Figure 1B).
- Polymerize epoxy resin overnight at 58 °C. If epoxy resin is still too soft allow another day to polymerize and harden at 58 °C. Test stiffness by applying pressure using forceps. If this easily leaves an indentation allow another day to polymerize and harden at 58 °C. When the specimen is fully hardened, proceed to trimming and sectioning.
- Trim away excess resin from the stub using razor blades (Figure 1B).
- Prepare epoxy resin according to standard recipes used in EM labs.
- Fixation
- Sectioning, Contrasting, and Mounting
- Sectioning
- Section epoxy resin block using an ultramicrotome.
- Use a glass knife or a diamond histoknife to cut semithin sections (500 nm) for toluidine blue/basic fuchsin (Figure 1D) staining to detect the right positioning.
- Transfer semi-thin sections on microscopic slides by picking them up with a glass Pasteur pipet whose tip has been closed by melting in a flame. Dry on a hot plate until no water is left. Stain for 10 sec with 1% toluidine blue in water on a hot plate, and after rinsing with water, stain for 10 sec with 0.05% basic fuchsin in 1% sodium tetraborate. Examine with a normal light microscope at 10X to 40X.
- When the proper site/orientation within the brain is reached, continue sectioning the epoxy resin block with a diamond knife to cut ultrathin sections (~ 70 nm; Figure 1E)
- Use easily identifiable anatomic structures in and around the brain, including olfactory pits, eyes, and grey-white matter boundaries, to identify the region of interest during ultrathin sectioning and to adjust the sectioning angle when the sample is tilted.
- Mount the section on single slot L2 x 1 copper grids (Figure 1F), to allow acquisition uninterrupted by grid bars. Use Formvar coated grids to support the sections.
- Contrasting
- Stain ultrathin sections on grids for 2 min in 2% uranyl acetate in methanol followed by Reynolds lead citrate for another 2 min.
- Sectioning
2. Image Acquisition
- Large Scale Scanning Transmission Electron Microscopy (STEM)
Note: We use scanning EM with an external scan generator capable of acquiring multiple large fields of view at high resolution using STEM detection. Typically, one image generated this way is equivalent to the fields of view of ~ 100 Transmission Electron Microscope (TEM) images, significantly reducing the amount of stitching.- Mounting Sample in Microscope
- Place grid with section in the multiple grid sample holder and transfer into the chamber of the SEM.
- Alignment of the STEM Detector
- Put the grid with the sample under the beam and move it up so it will be approx. 5 mm from the lens pole. Acquire an image at 20 kV using the in-lens detector.
- Focus on grid edge and adjust working distance to 4.0 mm.
- Move the sample holder to a safe position and bring in the STEM detector.
- Center the hole of the STEM detector in the image-field by turning the adjustments screws while imaging at maximum speed with the in-lens detector.
- Carefully bring back the sample holder which will just fit between the lens pole and the detector.
- Acquire an image with the in-lens detector to ensure it is on the section area.
- Switch to STEM detection (gain medium and all quadrants set on 'minus'), acquire an image, and select the area to be scanned.
- Raise the acceleration voltage to 21 kV and wait for stabilization of the beam (stable image again), then go to 29 kV in steps of 2 kV.
- Acquiring Images
- Pre-irradiate the sample (to prevent brightness changes caused by the e-beam) by zooming out so the complete area to be scanned fits the image window.
- Change the aperture to 120 µm (to keep the proper dynamic range, the detector gains have to be reduced by one step).
- De-focus so the image is blurred.
- Make the scanned area as tight as possible using the reduced area scan option.
- Set scan speed such that a frame is scanned in approx. 1 - 2 sec.
Note: Depending on the size and tissue, this typically takes ½ hr (100 x 100 µm field of view (FOV)) to up to 3 hr (1,000 x 500 µm). - Zoom in at least 100X and scan a small area for 10 sec.
Note: When this area does not change brightness compared to its surrounding, pre-irradiation is sufficient. - Bring back the 30 µm aperture (and STEM gain on medium)
- Focus on the sample.
- Align the aperture using the wobbler (at ~ 40,000X magnification).
- While in focus, select the lightest area/feature and set the scan speed such that details are visible.
- In the microscope software, adjust brightness and contrast by carefully watching the histogram to keep all pixels in the dynamic range. Do the same for the darkest area/features.
- Go back to the bright area and check again. Be on the safe side so there is some space on both sides of the histogram.
- Zoom out so the complete area to be scanned fits the imaging window and start the large area acquisition program.
- Use the wizard option to set up a mosaic by selecting an area from the screen. Use pixel size 2 - 5 nm depending on what details are necessary. Use dwell time 3 µs for STEM.
- Select the option "autofocus on previous tile", use the large scale acquisition software autofocus with default company settings, check the box "verify setting prior to execution" and define where to save the data.
- Press "optimize" to check microscope settings.
Note: Now, the time needed will be shown. This might be up to 72 hr for 1 x 0.5 mm areas. - In the window, "maximum tile resolution" type 20,000 so individual tiles will not be bigger than 20k x 20k, preventing blunt edge effects. Press "execute."
- When asked for checking focus and Brightness/Contrast (B/C), switch off external scan generator and carefully adjust focus and astigmatism in the microscope software. Do not change B/C.
Note: The stage can be moved and magnification changed, but magnification has to be set back to the desired pixel size. - Switch on the external scan generator again and press "continue."
- Mounting Sample in Microscope
תוצאות
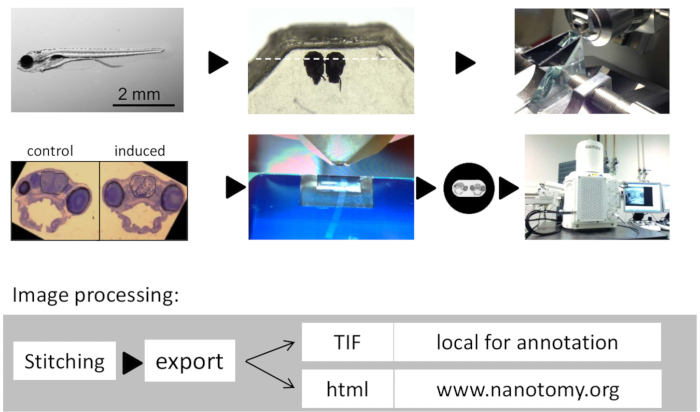
Figure 1: Workflow of Large-scale Tissue Electron Microscopy on Zebrafish Brains. (A) 7 day-old zebrafish larva. (B) Zebrafish larval heads embedded side by side in epoxy resin. (C) Glass knife for semi -thin sectioning and orienting the specimen. (D) Representative semithin toluidine blue stained section showin...
Disclosures
Materials
| Name | Company | Catalog Number | Comments |
| Chemicals | |||
| Low melting point agarose | VWR | 444152G | |
| tricaine | Sigma | E10521 | |
| Triton- X-100 | Sigma | X100 | |
| glutaraldehyde | Polysciences | 1909 | |
| Sodium cacodylate | Sigma | C0250 | |
| osmiumtetroxide | Electron Microscopy Sciences | 19114 | |
| potassiumferrocyanide (K4[Fe(CN)]6) | Merck | 4984 | |
| ethanol | VWR | 20821.365 | |
| uranyl acetate | Merck | 8473 | |
| sodiumtetraborate | Merck | 1063808 | |
| Tolduidene blue | Merck | 1273 | |
| Basic Fuchsin | BDH | 340324 | |
| Lead citrate | BDH | discontinued | |
| EPON | |||
| 2-dodecenylsuccinicacid anhydride | Serva | 20755 | |
| methylnadic anhydride | Serva | 29452 | |
| glycid ether 100 | Serva | 21045 | |
| DMP-30 | Polysciences | 553 | |
| Standard flat embedding mold | Electron Microscopy Sciences | 70901 | |
| diamond knife | Diatome Inc. | ||
| copper grids | Electron Microscopy Sciences | ||
| double sided carbon tape | Electron Microscopy Sciences | ||
| Scanning EM Zeiss Supra55 | Zeiss | ||
| ultramicrotome Leica EM UC7 | Leica | ||
| Atlas external scan generator | Fibics |
References
This article has been published
Video Coming Soon
Source: Kuipers, J. et al., Large-scale Scanning Transmission Electron Microscopy (Nanotomy) of Healthy and Injured Zebrafish Brain. J. Vis. Exp. (2016).
Copyright © 2025 MyJoVE Corporation. All rights reserved