Imaging and Quantifying Mitochondrial Morphology in C. elegans During Aging
In This Article
Summary
This protocol provides a standardized approach for imaging mitochondrial morphology in multiple tissues of C. elegans during aging.
Abstract
Mitochondria, important cellular organelles found in most eukaryotic cells, are major sites of energy production through aerobic respiration. Beyond this well-known role as the 'cellular powerhouse,' mitochondria are also involved in many other essential cellular processes, including the regulation of cellular metabolism, proliferation, immune signaling, and hormonal signaling. Deterioration in mitochondrial function during aging or under mitochondrial stress is often characterized by distinct changes in mitochondrial morphology and volume. The nematode C. elegans is an ideal model for studying these changes due to its transparent body and short lifespan, which facilitate live microscopy throughout its lifetime. However, even within the C. elegans field, numerous transgenic constructs and methods for mitochondrial imaging are available, each with its own limitations. Here, single-copy, matrix-localized GFP constructs are presented as a robust and reliable method for imaging mitochondrial morphology in C. elegans. This study specifically focuses on experimentally controllable factors to minimize errors and reduce variability between replicates and across studies when performing mitochondrial imaging during the aging process. Additionally, mitoMAPR is recommended as a robust method to quantify changes in mitochondrial morphology across tissue types during aging.
Introduction
Mitochondria are cellular organelles enclosed by a double phospholipid membrane and play an important role in the maintenance of cellular bioenergetics as the key site for metabolism and energy production1. A large number of mitochondria continuously generate energy in the form of ATP to meet cellular demands2. In addition to these important roles, mitochondria also participate in complex cellular processes such as signal transduction, autophagy, innate immunity, cell cycle, and cell death pathways2,3. Mitochondria exhibit diverse morphologies, ranging from small individual organelles to extensive interconnected tubular networks, depending on varying energetic requirements and mitochondrial health4.
A key feature of mitochondria is their dynamic nature whereby they can alternate between these various forms through a tightly coordinated and continuous sequence of fission and fusion events4. The precise balance between these opposing processes regulates mitochondrial morphology, number, size, and positioning within the cytoplasm5. In addition, these fusion and fission processes are important for maintaining quality control of mitochondria5. For example, mitochondrial fission is an essential component of removing damaged mitochondria through mitophagy and selective removal of mitochondria through autophagy2. Mitochondrial dynamics also play an important role in cell division, development, resistance to various stressors, and maintenance of cellular metabolism6.
Disruption of mitochondrial dynamics is implicated in numerous diseases, including neurodegenerative disorders, metabolic diseases, cardiovascular diseases, and cancers4. In addition, aging is associated with dramatic remodeling of the mitochondria, largely due to changes in these fusion-fission dynamics7. Therefore, visualizing and monitoring the changes in mitochondrial morphology under various stress or disease conditions and throughout the aging process offers valuable insights into understanding cellular function, disease mechanism, and potential therapeutic strategies.
Like many molecular pathways, mitochondrial dynamics and function are also highly conserved across eukaryotes, including model organisms such as the nematode Caenorhabditis elegans. Similar to human cells, mitochondrial fusion in C. elegans is achieved by the function of dynamin-related guanine triphosphatase (GTPase) proteins, including FZO-1 (ortholog of mammalian Mfn1/2) and EAT-3 (ortholog of mammalian Opa1) which control the fusion of the outer mitochondrial membrane (OMM) and inner mitochondrial membrane (IMM), respectively8. Mitochondrial fission is regulated by the dynamin-related protein (DRP-1, ortholog of human Drp1), which promotes mitochondrial fission by forming ring-like complexes around the mitochondrial outer membrane that constrict and eventually separate the mitochondrial membranes9. Mitochondrial fusion plays a pivotal role in mitochondrial quality control by allowing the mixing of mitochondrial contents, including mitochondrial DNA, proteins, and lipids, allowing complementation of partially damaged mitochondria by contents from healthy mitochondria9. On the other hand, mitochondrial fission allows mitochondria to divide, creating new mitochondria and facilitating their distribution not only within the cytoplasm but also to daughter cells during cell division, ensuring proper mitochondrial inheritance and function9. This event is also essential to segregate damaged or dysfunctional mitochondrial segments, which can then be targeted for degradation through mitophagy9.
C. elegans has long been considered one of the most powerful genetic model systems due to the complete genome, and the availability of a variety of genetic tools, including CRISPR/Cas9 that facilitates genetic modifications10, numerous methods for overexpression of genes, and a bacterial food-based method for RNA interference (RNAi)11. In addition, their transparent anatomy allows microscopic imaging in live organisms12. Finally, their relatively short lifespan, low maintenance costs, and ease of producing a large number of age-matched animals make them an excellent model system for aging biology13. These benefits, mixed with the known conservation of major mitochondrial regulatory pathways, make C. elegans a highly attractive model system for studying mitochondrial dynamics during aging.
Fluorescent markers are widely used in biological research to visualize and study cellular components, including mitochondria. There are specific cell-permeable dyes such as MitoTracker and tetramethylrhodamine ethyl ester (TMRE) that are commonly used14. The former is used to dye the overall negatively charged mitochondrial matrix15, whereas the latter is used to evaluate the relative mitochondrial membrane potential through its positively charged triphenylphosphonium ion16. While they benefit from not requiring transgenics, the thick cuticle of the worms that changes in structure and permeability during aging and the variability of dye infiltration across different tissues make dye-based approaches challenging in C. elegans17. Moreover, the evaluation of mitochondrial morphology is confounded by potential off-target effects of the dyes, such as dye aggregation17. Instead, genetic methods to express mitochondria-localized fluorophores are commonly used in the worm model.
Here, this study focuses on highlighting strains expressing a mitochondrial matrix localized GFP (henceforth referred to as MLS::GFP) under cell-type specific promoters. Importantly, these transgenic lines were made using the MosSCI method to ensure single-copy expression of the reporter in a known genomic locus, which avoids issues with other available strains. For example, high-copy expression of some mitochondria-targeted proteins leads to variability in expression levels due to the integration of extrachromosomal plasmid arrays into a random locus with an unknown copy number18. In addition, they have previously been shown to induce damage to mitochondria since it is a high burden for cells to properly localize the overexpressed mitochondrial proteins19. Therefore, the precise, stable, and low gene expression and ease of crossing to known loci make these MosSCI transgenics the preferred method. Using these strains, this study standardizes methods to image mitochondria in the muscle, intestine, and hypodermis of C. elegans. Furthermore, it highlights the important technical applications and troubleshooting methods that are important to consider and ensure reproducible experimental analysis of mitochondria during aging in C. elegans.
Protocol
Details on the reagents and equipment used in this study are listed in the Table of Materials.
1. Growth and maintenance of C. elegans
- Preparing Nematode Growth Media (NGM) plates
- For the culture of C. elegans, use standard 2% agar plates with NGM containing 1 mM of CaCl2, 12.93 μM (5 µg/mL) of cholesterol, 25 mM of KPO4 (pH 6.0), 1 mM of MgSO4, 2.5 mM (0.25% w/v) of Peptone, and 51.3 mM of NaCl. See the detailed method of pouring NGM plates described in Castro Torres et al.20.
NOTE: For RNA interference (RNAi) experiments, add 1 mL of 1 M IPTG and 1 mL of 100 mg/mL Carbenicillin for 1 L NGM-agar plates. - After the NGM plates have solidified, grow a culture of OP50 in lysogeny broth (LB) for 24-48 h at room temperature or grow a culture of HT115 carrying a pL4440 empty vector plasmid in LB with antibiotics (ampicillin/carbenicillin 100 µg/mL + tetracycline 5 µg/mL) with shaking at 37 °C for 12-16 h.
NOTE: This plasmid is available from commercial sources (see Table of Materials). - Seed 200 µL of OP50 or HT115 culture onto 60 mm plates, or 1 mL onto 100 mm plates.
- Dry the plates until they are no longer wet and store the plates in sealed containers at 4 °C for up to 2 months.
- Optional: Add 100 µL of 10 mg/mL 5-fluoro-2'-deoxyuridine (FUDR) directly onto NGM-agar plates that are seeded with bacteria to chemically sterilize L4/adult worms and prevent the development of younger worms.
NOTE: The strains developed and used in this study are:
(1) RHS191 - uthSi17[myo3p::MLS::GFP::unc-54 3'UTR::cb-unc-119(+)] I
(2) RHS192 - uthSi83[col19p:: MLS::GFP::unc-54 3'UTR::cb-unc-119(+)]I
(3) RHS193 - uthSi80[vha-6p:: MLS::GFP::unc-54 3'UTR::cb-unc-119(+)] IV
- For the culture of C. elegans, use standard 2% agar plates with NGM containing 1 mM of CaCl2, 12.93 μM (5 µg/mL) of cholesterol, 25 mM of KPO4 (pH 6.0), 1 mM of MgSO4, 2.5 mM (0.25% w/v) of Peptone, and 51.3 mM of NaCl. See the detailed method of pouring NGM plates described in Castro Torres et al.20.
- Synchronizing C. elegans by bleaching
- For a large-scale assay that requires a high number of nematodes, cut a 60 mm NGM-agar plate full of animals into small pieces and chunk them onto 100 mm plates that are seeded with bacteria for expansion. Allow 2-3 days for the 100 mm plates to be full of worms when grown at 20 °C after chunking. For more detailed recommendations for expanding animals, refer to Castro Torres et al.20.
- To collect the worms for synchronization, pour 5-10 mL of M9 solution (22 mM of KH2PO4 monobasic, 42.3 mM of Na2HPO4, 85.6 mM of NaCl, 1 mM of MgSO4) onto 100 mm NGM-agar plates with worms, and swirl the M9 solution gently to loosen the worms from the bacterial lawns.
- Draw up the worms with a serological pipette and transfer them into 15 mL conical tubes.
NOTE: Glass serological pipettes are recommended since plastic serological pipettes tend to make the worms stick to the inner wall of the plastic pipettes. - Centrifuge for 30 s at 1,100 x g at room temperature to pellet down the worms and aspirate the supernatant using a vacuum pump.
- Prepare 5 mL of bleaching solution containing 1.5 mL of 6% sodium hypochlorite, 0.75 mL of 5 M NaOH or KOH, and 2.75 mL of dH2O. Pour the 5 mL of bleaching solution into the worm pellet.
CAUTION: Wearing gloves and a lab coat is recommended in this step since sodium hypochlorite and hydroxide solutions are corrosive. - Incubate the worm + bleaching solution mix for ~4-6 min until all bodies of animals are fully dissolved and only the eggs remain. Shake the mix vigorously to help the dissolving process.
NOTE: Check the worms every minute under a dissecting microscope. Shorten or extend the bleaching time based on how much the bodies of adult worms are dissolved. Leaving the eggs inside a bleaching solution for an extended period will result in damage to the eggs and affect the viability of the nematodes. - Centrifuge the eggs/bleaching solution mix for 30 s at 1,100 x g at room temperature to pellet down the eggs and then carefully aspirate the supernatant using a vacuum pump, making sure not to aspirate the egg pellet.
NOTE: It is recommended to perform washes quickly to prevent prolonged exposure of the eggs to the bleaching solution. - Wash the eggs with 5-10 mL of M9 solution 4 times.
- Resuspend the eggs in a small volume of M9 solution and plate them onto NGM plates seeded with bacteria by pipetting up to 50 µL. Calculate a rough estimate of the number of eggs by pipetting 3-5 µL of the egg mixture onto a plate and counting the number of eggs in this volume. Add M9 to dilute the egg mixture to a countable number of eggs in this volume.
NOTE: Plating less than 100 worms per plate is recommended since the worms can starve before day 1 of adulthood when there are too many animals plated. For more detailed recommendations for plating numbers of animals, refer to Castro Torres et al.20. - Alternatively, L1-arrest the nematodes for tighter age synchronization. For L1-arresting, add the appropriate volume of M9 solution to the egg pellet in a 15 mL conical tube to a volume of 10-12 mL. Let the worms spin in a rotator for 16-24 h at 20 °C. Centrifuge (as in step 1.2.7) and aspirate the supernatant. Calculate the concentration of L1 animals and plate them onto NGM plates as described for eggs in step 1.2.9.
- Age out the C. elegans population to the desired age: Transfer the L4/adult animals onto plates containing 5-fluoro-2'-deoxyuridine (FUDR) as described in step 1.1.6 to prevent progeny and eliminate any offspring already produced. For alternative methods to age out worms without FUDR, refer to Castro Torres et al.20.
NOTE: Some important factors need to be considered before choosing bleaching as a method to synchronize worms at their first larval stage. As highlighted earlier, overexposure to bleach can damage embryos, reduce hatch rates, and create selection pressure for bleach-resistant strains, potentially altering the population's genetics. In addition, if embryos are left unfed for extended periods of time, nutrient depletion can impair metabolism and development. Therefore, egg-lay or NemaSync methods can be used as an alternative to bleaching methods. The method for the egg-lay procedure is described stepwise in the referred study by Castro Torres et al.20. NemaSync is a newer and more expensive commercial method, where a manual worm synchronizer is used to synchronize worms without the use of chemicals21.
2. Imaging of mitochondria in C. elegans
- Preparation of slides for imaging of C. elegans
- Add 10-20 μL of M9 solution onto glass slides and transfer a desired number of adult worms at the specific ages to the solution on the slide. Approximately 25 worms are recommended.
NOTE: Using traditional methods to paralyze worms (e.g., tetramisole or sodium azide) may cause mitochondrial fragmentation under certain conditions (see below). Instead, using HistoBond slides is recommended here, which have a surface with a permanent positive surface charge, will reduce the movement of worms without using drugs that may introduce artifacts. It is important to put an appropriate ratio of worms to M9 solution, as low volumes may cause animals to get crushed between the slide and coverslip, whereas high volumes will result in animals moving during image acquisition. Recommendations can be found in Table 1. - Alternatively, for using drugs that paralyze worms, refer to representative results of this manuscript for a range of concentration and time points that can be used with limited fragmentation of mitochondria.
NOTE: Although there was no major fragmentation in healthy, wild-type animals, it is important to remember that certain mutants or conditions may experience mitochondrial fragmentation with the same concentration of these drugs, so some testing is required when using these drugs. - Cover the worms in M9 solution on the slide with a cover glass and use nail polish to seal the sides and prevent evaporation of M9 solution.
- Optional: For muscle imaging, roll worms by gently nudging the cover slip. This allows body-wall muscles on the sides of the worms to be better aligned with the focal plane for higher-quality images.
- Add 10-20 μL of M9 solution onto glass slides and transfer a desired number of adult worms at the specific ages to the solution on the slide. Approximately 25 worms are recommended.
- Imaging of mitochondria using a compound microscope
- Use a standard wide-field microscope for muscle and hypodermal mitochondria (myo-3p::MLS::GFP and col-19p::MLS::GFP).
NOTE: A commercially available imager equipped with a 63x/1.4 Plan Aprochromat objective, GFP filter (11525314), DFC9000 GT camera, and LED5 light source was used, and compatible microscope software was used. - Optimize the imaging settings for each microscope and experimental setup.
NOTE: However, as a starting point, the parameters for imaging with the imager used in this manuscript are as follows: Dimensions are 2048 (x) * 2048(y) pixels at a step size of 0.50 μm for hypodermal mitochondria and "System Optimized" automatically calculated by the software for muscle mitochondria. Exposure time is 50 ms for muscle mitochondria and 50 ms for hypodermal mitochondria. The 475 nm LED was set to 50% power, and the neutral density filter was set to 30% for muscle and 17% for hypodermal mitochondria. Z-range starts from where the GFP signal is visible to where it ends. To minimize the noise in a max-projection, one can decrease the number of z-planes used for the max-projection. - For imaging of intestinal mitochondria (vha-6p::MLS::GFP), use a confocal microscope.
NOTE: Imaging was performed on a commercially available confocal microscope equipped with a 63x/1.4 Plan ApoChromat objective, white light laser (WLL), Acousto-Optical Beam Splitter, HyD S detectors, and run on microscope software. As previously mentioned, one should optimize each individual experimental setup, but as a starting point, the parameters for imaging with the Stellaris used in this study are as follows: The WLL was set to 85.00 % Maximum Power and with a 485 nm laser line at an intensity of 3.00 %. The HyD S detector was set to 490-590 nm with a gain of 25 in the analog setting. We unidirectionally scanned an area of 1024 (x) * 1024(y) or 82.01 μm (x) * 82.01 μm (y) with 5 (z) pixels at a step size of 0.495 μm (dimension z is variable depending on the size of the worms or tissues) with a scanning speed of 1,000 Hz, 2.25 zoom, line average of 2, and pinhole of 1 AU (95.5 μm). The Z-range starts from where the GFP signal is visible to where it ends. To minimize the noise in a max-projection, one can decrease the number of z-planes used for the max-projection.
- Use a standard wide-field microscope for muscle and hypodermal mitochondria (myo-3p::MLS::GFP and col-19p::MLS::GFP).
- Quantification of mitochondrial morphology
NOTE: For details, refer to Schindelin et al.22 and see Supplementary Figure 1A-D.- Download and install FIJI (Fiji is just ImageJ, https://fiji.sc/) software. Then open FIJI.
- To download and install the MitoMAPR macro, download source code 1 from https://doi.org/10.7554/eLife.49158.033. Then copy all the code mentioned under "A.). IJM code for MitoMAPR-1.0." Next, open Fiji, go to Plugins > New > Macro, and paste the previously copied code into the macro window. Go to File > Save as option and save the Macro for future use.
- Open a 3D microscopic image with Z-stacks in FIJI by dragging and dropping files to Fiji toolbar or by going to File > Open.
- Create a max projection of image by going to Image > Stacks > Z Project, then selecting the range of slices with in-focus images to be max projected and selecting Max Intensity as the projection type.
- Save the image as a TIFF by going to File > Save As > Tiff.
- Crop regions of interest (ROIs) from full images by using the "Rectangle" tool, drawing an ROI, and then going to Image > Crop.
- Save the image as a TIFF by going to File > Save As > Tiff.
- Run the MitoMAPR macro previously saved by dragging and dropping the macro file onto the Fiji toolbar, then click on Run. A new window will prompt to select the folder where the Tiff file was saved in 2.3.6.
- A new window will prompt to select an area; create a rectangle in the image using the rectangle tool as mentioned earlier in step 2.3.4 and press Ok.
NOTE: The entire image can be selected if it was cropped previously as described in step 2.3.6. Save the "Data" window with all the values related to mitochondrial morphology as an Excel file by going to File > Save as > Save.
- Batch processing of samples using FIJI macros
- Create a macro for specifying the region of images to be analyzed. Open Fiji and go to Plugins > New > Macro. Paste the following code into the text box and save the macro as described in 2.3.2 (Supplementary Figure 1E-H).
NOTE: Make Rectangle (100, 100, 200, 200); run (Specify..., width = 100, height = 100, x = 100, y =100 scaled). - Create a macro for saving all opened images as TIFF files. Within the macro window, go to File > New and paste the following code into the text box. Save the macro as described in 2.3.2.
NOTE: dir = getDirectory("Select A folder"); ids=newArray(nImages); for (i=0;i<nImages;i++) { selectImage(i+1); title = getTitle; print(title); Stack.setChannel(1); ids[i]=getImageID; saveAs("tiff", dir+title.replace("/", "-"));}. - Create a macro for the batch MitoMAPR. Go to https://doi.org/10.7554/eLife.49158.033 source code 1 and copy all the codes mentioned under B.). IJM code for MitoMAPR-1.0_Batch". Within the macro window, go to File > New, paste the code from the source code 1 into the text box, and save the macro as described in step 2.3.2.
- Create a macro for iterative cropping. Go to https://doi.org/10.7554/eLife.49158.033 source code 1 and copy all the codes mentioned under C.) IJM code for CropR". Within the macro window, go to File > New, paste the code from the source code 1 into the text box, and save the macro as described in step 2.3.2.
- To begin batch processing, first save all images to be processed as TIFF by opening all the images and running the macro saved in step 2.4.2. All images will be saved as TIFF in the specified folder. Close all images once saved.
- If the images saved in step 2.4.5 are 3D images (images containing z-stacks), convert them into 2D images by making them Z-projections. To batch Z-project the images, open the batch processing window by going to Process > Batch > Macro in the main FIJI window.
- For "Input", select the location of the folder containing all the 3D images to be processed from 2.4.5. For "Output", select the desired location for the saved images after processing. Select the output format as TIFF. Paste the following code in the large text box in the window and press the process button: run("Z Project...", "projection=[Max Intensity]").
- Crop ROIs with identical dimensions from all 2D images to be processed by using the iterative crop macro from step 2.4.4 in combination with the specifying region macro from step 2.4.3. Run the iterative crop macro from step 2.4.4. A window will appear asking for a directory.
- Select the folder containing only 2D images from step 2.4.5 or the 2D Z-projected images from step 2.4.6 and press the Select button. The macro will open one of the images in the selected folder, and a window labeled "make a selection" with two buttons will appear.
- Run the specified selection macro from step 2.4.3. Modify the width and height values in that macro text box as required to change the selection dimension and press Run to observe the new selection. Changing the X and Y values in the macro will modify the top-left corner position of the selection.
- Once the selection dimensions are satisfactory, drag the rectangle to the desired area and then press the OK button on the "make a selection" window. The cropped region will be saved to the folder selected in step 2.4.7.
- The macro will iterate through all the remaining images in the folder. For each image, run the macro from step 2.4.8 without changing the dimension values to ensure the same dimensions are cropped for all images. For each image, drag the selection rectangle to the desired location and press the OK button in the "make a selection" window.
- Open the folder containing the cropped images from steps 2.4.8-2.4.9 and move all the cropped images to a new folder.
- Analyze all the cropped images by running the batch MitoMAPR macro from step 2.4.3. A new window will ask you to select a directory. Select the folder with all the cropped images from step 2.4.10. Once complete, a window named "Data" will pop up with all the values related to mitochondrial morphology. Save all data as an Excel file as described in step 2.3.9.
- Create a macro for specifying the region of images to be analyzed. Open Fiji and go to Plugins > New > Macro. Paste the following code into the text box and save the macro as described in 2.3.2 (Supplementary Figure 1E-H).
Representative Results
C. elegans is a great model for mitochondrial imaging due to its transparent body, which allows for easy wholeworm, live animal imaging without excessive sample preparation. In addition, mitochondrial morphology in different tissues can be easily visualized by using tissue-specific promoters to express mitochondria-targeted fluorescent proteins. Here, myo-3p (for muscle mitochondria), vha-6p (for intestinal mitochondria), and col-19p (for hypodermal mitochondria) were utilized to drive expression of a mitochondria-targeted GFP (mitochondrial localization sequence of the ATP-1 protein). The mitochondria of the body wall muscles of C. elegans exhibit tubular morphology, aligning along muscle fibers (myofibrils) (Figure 1A); intestinal mitochondria showed highly interconnected, web-like structures, with less uniform alignment (Figure 1B); and the hypodermis has tubular mitochondria that appear more rounded or oval shaped compared to the intestine or muscle (Figure 1C). Mitochondrial morphology is generally consistent across the length of the worm in the muscle and intestine, but the hypodermis does display some minor differences in the interconnectedness of mitochondria between the proximal and distal ends of the worm. Thus, to achieve reproducible experimental results, it is recommended to focus on a specific region of their bodies. Here, the area between the pharynx and the vulva ~100-200 μm below the pharynx is consistently imaged.
For muscle and hypodermal mitochondria, imaging them under a compound microscope provides sufficient resolution due to the flatness of the cells and low levels of out-of-focus light. However, intestinal mitochondria are hard to observe with a compound microscope, as the large volume of the intestine limits the resolution due to a large amount of out-of-focus light from other sections of the intestine (Figure 2). Therefore, it is recommended to image intestinal mitochondria under a confocal microscope to reduce out-of-focus light and visualize proper mitochondrial morphology.
The shape of mitochondria is highly dynamic and changes based on the metabolic environment of the animals23 or even due to exposure to variable substrate stiffnesses24. Therefore, leaving animals on microscope slides for extended periods of time in the absence of a food source and on the stiff substrate of glass can potentially impact mitochondrial morphology. Here, this study found that the mitochondria of worms fragment after approximately 30 min on a slide in M9 solution, with hypodermal mitochondria displaying the most fragmentation (Figure 3). Therefore, imaging of mitochondria should be performed rapidly after sample preparation.
Chemicals that restrict worm mobility, including sodium azide and tetramisole, are commonly used for live imaging of C. elegans, as stationary worms are required to capture multiple z-sections of animals for 3D imaging. Previous studies have shown that exposure to sodium azide or tetramisole can result in mitochondrial fragmentation25. Surprisingly, it was found that sodium azide - even at high concentrations - had a limited impact on mitochondrial morphology in the muscle or intestine. However, the hypodermis displayed earlier mitochondrial fragmentation than M9 controls (Figure 3). Importantly, high concentrations of tetramisole (100 mM) resulted in significant mitochondrial fragmentation immediately after exposure in all cell types. In comparison, medium concentrations (10 mM) resulted in faster fragmentation compared to an M9 control. Low concentrations (1 mM) had a limited impact on mitochondrial morphology. These data suggest that utilization of sodium azide can be a viable option for rapid imaging of mitochondria, while tetramisole should generally be avoided.
As expected, mitochondria of all tissues of C. elegans display fragmentation during the natural aging process (Figure 4A). The fragmentation can be visualized as mitochondria presenting as more truncated and spherical structures, which are dramatically different from the linear, tubular mitochondria displayed in young animals. It is important to note that the intestine does increase in spherical autofluorescent structures at a later age, so care must be taken not to confuse autofluorescence with actual mitochondrial structures. Because there can be variability of changes in mitochondrial structure across worms, it is important to perform quantification of mitochondrial morphology on a significant sample size rather than just imaging a few worms. Here, mitoMAPR was used, which allows for automated quantification of mitochondrial morphology using a diverse set of metrics, including objects, networks, junctions per network, junction points, object length, mitochondrial footprint, mitochondrial coverage, and object area (Supplementary Table 1 and Supplementary Table 2). The automation removes subjective bias from the user. Here, we report that the object length metric is optimal to quantitively measure the changes in muscle and intestinal mitochondrial morphology during aging, and junction points metrics to measure the changes in hypodermal mitochondrial morphology during aging (Figure 4B).
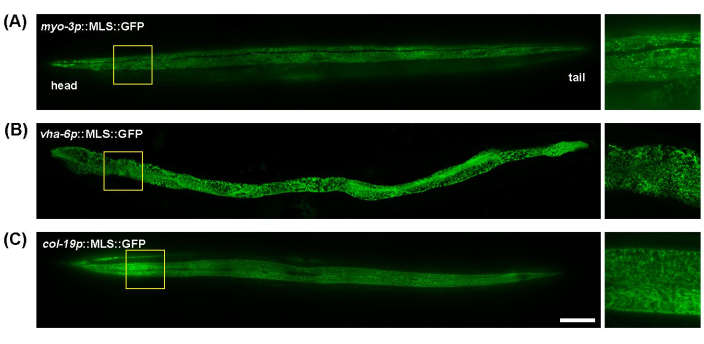
Figure 1: Images of mitochondria throughout the whole body of C. elegans. Mitochondrial imaging was performed throughout the entire length of day 5 adult animals grown on EV from the L1 stage in transgenic animals with MLS::GFP expression in the muscle (A), intestine (B), and hypodermis (C). Scale bar: 100 μm. Please click here to view a larger version of this figure.
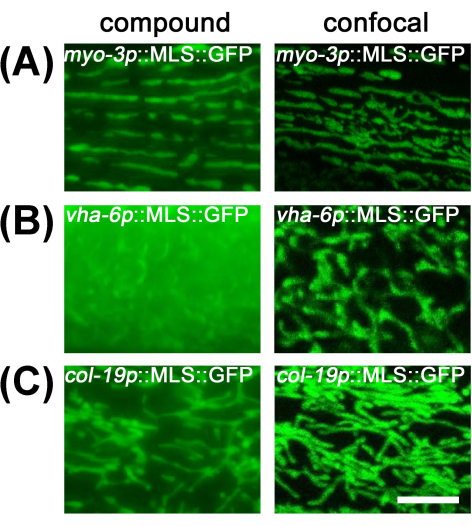
Figure 2: Comparison of mitochondrial imaging between compound microscope and confocal microscope. Mitochondrial imaging was performed in day 1 adult animals grown on EV from the L1 stage in transgenic animals with MLS::GFP expression in the muscle (A), intestine (B), and hypodermis (C). Scale bar: 5 μm. Please click here to view a larger version of this figure.
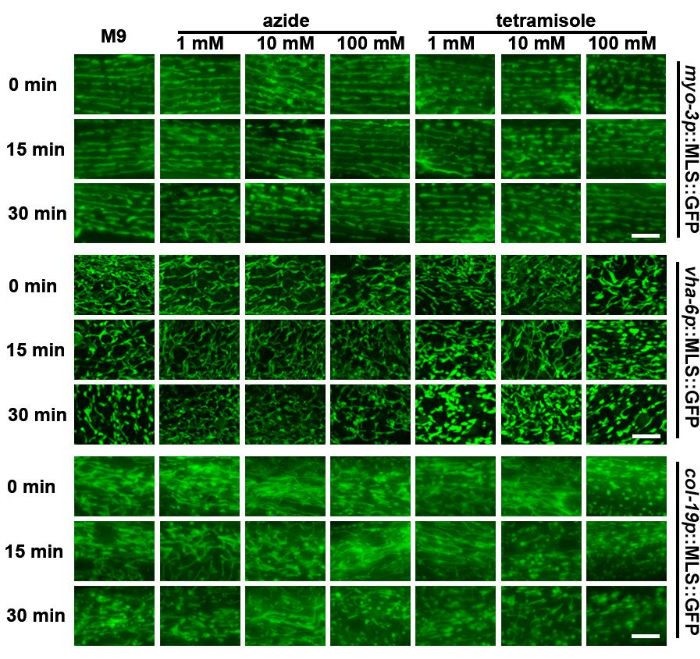
Figure 3: Assessment of mitochondrial fragmentation of different tissues after treatment with M9, sodium azide, and tetramisole. Mitochondrial imaging was performed in day 1 adult animals with expression of MLS::GFP in the muscle, intestine, and hypodermis. Animals were grown on EV from the L1 stage. Animals were placed on slides containing M9, sodium azide (1 mM, 10 mM, and 100 mM), or tetramisole (1 mM, 10 mM, and 100 mM) and imaging was performed immediately after slide preparation (0 min) or 15 min or 30 min after slide preparation. Representative images are shown for n > 5 animals per strain for 2 biological replicates. Scale bars: 5 μm. Please click here to view a larger version of this figure.
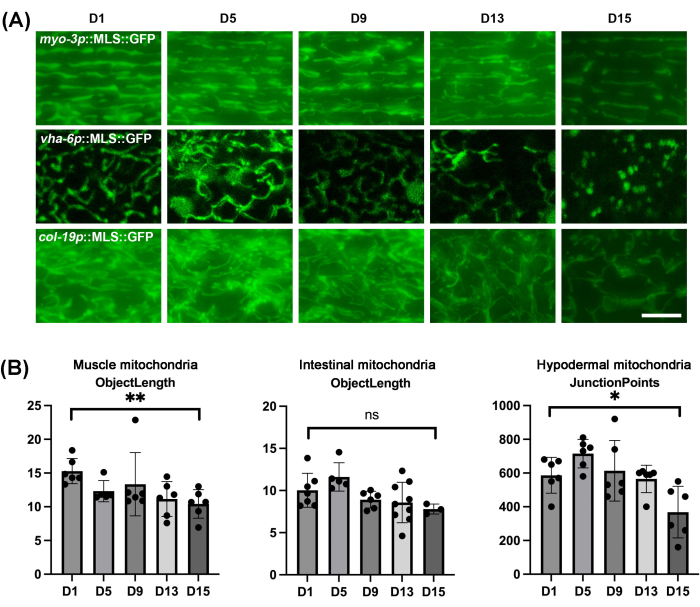
Figure 4: Mitochondrial imaging of C. elegans during aging in different tissues and quantification of mitochondrial morphology using MitoMAPR. (A) Mitochondrial imaging was performed in adult animals on days 1, 5, 9, 13, and 15 of adulthood with expression of MLS::GFP in the muscle, intestine, and hypodermis. Animals were grown on EV from the L1 stage and moved onto EV plates containing FUDR from the day 1 adult stage. Images are representative of n ≥ 5 animals per strain for ≥ 3 biological replicates. Scale bar: 5 μm. (B) The quantification for object length of muscle mitochondria and intestinal mitochondria of worms on days 1, 5, 9, 13, and 15, and the quantification for junction points of hypodermal mitochondria of worms on days 1, 5, 9, 13, and 15. All the individual data points combined with mean ± SD. Graphs were plotted and statistically analyzed using a student t-test. ns = not significant, *p < 0.03; **p < 0.002; ***p < 0.0002; ****p < 0.0001. Please click here to view a larger version of this figure.
| Days of worms | Number of worms | Volume of buffer (mL) |
| 1 | 25 | 13 |
| 5 | 25 | 14 |
| 9 | 20 | 15 |
| 13 | 20 | 15 |
Table 1: Recommended numbers of worms and volumes of buffer when preparing slides.
Supplementary Figure 1: Workflows of the application of a single image quantification using MitoMAPR and batch processing for multiple images. (A) Workflow of quantifying the mitochondrial morphology from a single image using MitoMAPR macro. (B) Z projection of raw 3D image file using Fiji (Max projection). (C) Cropping the region of interest from the Z projected image. (D) Skeletor images acquired from MitoMAPR macro. Scale bars: 5 μm. (E) Workflow of quantifying the mitochondrial morphology from multiple images using batch process using MitoMAPR macro. (F) Screenshot of crop macro. (G) Screenshot of a batch process for Z projection. (H) Screenshot of selection confirmation window of batch cropping macro. Please click here to download this File.
Supplementary Table 1: Quantitative analysis of mitochondrial morphology metrics in C. elegans muscle tissue. Metrics recorded include objects, networks, junctions per network, junction points, object length, mitochondrial footprint, object area, and mitochondrial coverage, measured across different days. Data provide a basis for assessing changes in mitochondrial morphology with aging. Please click here to download this File.
Supplementary Table 2: Effects of immobilizing chemicals on mitochondrial morphology in C. elegans. Data include measurements such as objects, networks, junctions per network, junction points, object length, mitochondrial footprint, object area, and mitochondrial coverage across varying concentrations and time points after sample preparation with M9 buffer or different concentrations of tetramisole and sodium azide. Please click here to download this File.
Discussion
Fluorescent imaging of mitochondrial morphology is the most common way to determine changes in mitochondria. While advanced microscopy techniques such as transmission electron microscopy (TEM), atomic force microscopy, and cryo-electron microscopy offer higher resolution, fluorescence microscopy remains more affordable and accessible to most researchers. In addition, fluorescent microscopy can be performed in live cells, and in clear model organisms like C. elegans, imaging can be performed in whole animals26,27. Imaging of transgenic C. elegans is quite simple, and genetically encoded fluorescent markers allow for more reliable and robust imaging of mitochondria since it does not require complex sample processing or suffer from variable staining across cell types or conditions from conventional mitochondrial dyes like MitoTracker or TMRE28,29,30. Genetically encoded fluorescent markers usually involve direct tagging of mitochondrial proteins with fluorophores or conjugation of fluorophores with a minimal mitochondrial localization sequence. These constructs are often driven by tissue-specific promoters, enabling visualization of mitochondria in different tissues like muscle, intestine, or hypodermis tissues31. Generally, these fluorescently tagged proteins are overexpressed, which can have potentially unwanted physiological effects if full-length mitochondrial proteins are overexpressed; thus, minimal MLS sequences are a better option32. However, even with the overexpression of minimal MLS-fluorescent protein fusions, high overexpression should be avoided, as importing a large amount of protein into the mitochondria can collapse the potential of the mitochondrial membrane and impact animal health33. While an exhaustive characterization of all currently available C. elegans strains is beyond the scope of this manuscript, a detailed comparative analysis of numerous mitochondrial reporters and the pros and cons of each can be found here31.
For live-cell imaging of mitochondria in C. elegans, standard compound or wide-field microscopy may be a preferred option due to the high speed and ease of these methods over confocal microscopy. In this study, it is shown that flat cells like the muscle and hypodermis benefit minimally from confocal microscopy, and compound microscopy allows acquisition with sufficient resolution to visualize proper mitochondrial morphology. Larger cells like the intestine make compound microscopy challenging due to out-of-focus light. Therefore, confocal microscopy is required for reliable imaging of intestinal mitochondrial morphology.
An important consideration for 3D imaging across the entire thickness of tissue in live animals is to prevent the movement of the worms during image acquisition. Researchers often use methods to paralyze worms, such as tetramisole or sodium azide34. Sodium azide inhibits cytochrome c oxidase (complex IV), a critical enzyme in the electron transport chain of mitochondria, leading to general paralysis due to the lack of ATP needed for muscle contractions and other cellular functions34,35. Tetramisole acts by mimicking acetylcholine at the neuromuscular junctions, causing persistent depolarization and muscle contraction36. However, exposure to these drugs can induce mitochondrial fragmentation by causing oxidative stress23. In this study, it was found that tetramisole very rapidly induced mitochondrial fragmentation, but sodium azide had a much more limited effect.
C. elegans offers a very simple and easy way to study the impact of aging on mitochondrial morphology due to its short lifespan and ease of aging out animals. Here, we opted for using exposure to FUDR, which is a robust method to chemically sterilize animals by preventing DNA replication20,37,38. However, FUDR may have unwanted effects on specific aging parameters, and for those concerned about off-target effects of FUDR, other strategies to remove progeny can be utilized39. For example, there are sterile mutants, including the temperature-sensitive germline mutant glp-4, or sperm-deficient mutants, such as CF51240,41,42. As an alternative, animals can also be naturally aged out by manually picking adults from their progeny daily.
Quantification of the changes in mitochondrial morphology is also a very important consideration, as there may be significant variability in mitochondrial morphology across animals. Therefore, performing quantitative analysis and statistics across a sufficient sample size is essential for making significant conclusions on changes to mitochondrial morphology. However, image analysis and quantification can suffer greatly from subjective bias and challenges from imaging highly complex structures like interconnected mitochondria. To this end, here is a summary of an automated method to quantify mitochondrial morphology using numerous metrics developed by the Mair lab. MitoMAPR allows objective, automated quantification of mitochondrial morphology, measuring various aspects of the mitochondria, including the mitochondrial network, object length, distribution, network coverage, and mitochondrial footprint. MitoMAPR is a free macro for ImageJ and thus is available to all labs with a functional computer. One important aspect of using MitoMAPR is to perform quantification across a large sample size to determine which metric of mitochondrial morphology is most robust to determine changes in the experimental conditions being tested43,44. Here, it is found that object length and junction points are the best metrics to determine changes during aging in the muscle, intestine, and hypodermis. An alternative approach for analyzing changes in mitochondrial morphology is the generation of 3D representations of mitochondria from z-stack images, followed by 3D analysis16,45. This can be achieved using commercially available software, such as Image-Pro Plus with the SharpStack Total Deconvolution and 3D Constructor modules. Quantification of 3D mitochondrial representations has been shown to provide more accurate insights into mitochondrial shape and network properties46. However, this method is technically more complex and costly compared to the semi-3D approach, which involves collapsing multiple image sections from a wide-field or confocal z-stack into a single 2D projection and analyzing them with tools like mitoMAPR. This study summarizes the usage of single-copy expression of mitochondrial matrix-targeted GFP and details a few pitfalls to avoid during imaging.
Limitations and time considerations
Although confocal microscopy is recommended for large and thick tissues, such as intestinal tissue, standard line-scanning confocal microscopy techniques may be too slow to perform rapid imaging of mitochondria. This is especially true considering our data that show that keeping worms on slides for long periods of time can result in the fragmentation of mitochondria. Although confocal microscopy technology has advanced significantly, microscopes that can perform rapid imaging, such as spinning disk, Airyscan, or the one used in this study, can be cost-prohibitive for some labs. In these cases, compound microscopy can be combined with computational clearance methods to remove out-of-focus light, such as deconvolution47.
Also, as described, MitoMAPR is a powerful automated macro to quantitatively measure the length of mitochondria and the interconnection of the mitochondrial network. However, it should be used carefully considering the limitations listed here. First, using a matrix-targeted GFP only observes mitochondrial fragmentation of the inner membrane, which may not fully recapitulate mitochondrial morphology of the outer membrane, as inner membrane fission events can occur in the absence of outer membrane fission. Thus, for more refined imaging of both membranes, both a matrix-targeted and mitochondrial outer membrane-targeted fluorophore should be used. Since outer mitochondrial membrane markers can suffer the same consequences as matrix-localized proteins if highly overexpressed, the usage of single-copy markers is recommended, especially those using minimal mitochondrial localization sequences instead of full proteins such as the ones used here31.
As described in Figure 3, mitochondria can rapidly fragment under the microscope, and different methods to prepare sample slides can largely affect morphology depending on the buffer being used. The representative data described in this manuscript show that the mitochondria of the worms undergo minimal fragmentation under M9 and low concentrations of sodium azide. Tetramisole, another widely used chemical to paralyze worms, rapidly fragments the mitochondrial, indicating its use should be generally avoided. Although M9 and sodium azide did not show significant fragmentation of mitochondria, it is important to note that different strains might respond differently than the results shown here. While other studies have shown that sodium azide can fragment mitochondria, it is possible that our strains do not show significant mitochondrial fragmentation with exposure to sodium azide due to the low expression level of our constructs. High expression of mitochondria-localized proteins can collapse membrane potential, and thus, different strains used in other studies may make mitochondria more susceptible to fragmentation from the same concentrations of sodium azide used in this study. Regardless, care should be taken to ensure that chemicals that paralyze worms do not cause mitochondrial fragmentation in specific conditions to be tested prior to use in all studies, as specific mutants or conditions may be more susceptible to drug-induced mitochondrial fragmentation. Furthermore, even keeping the worms in M9 can result in changes to their natural mitochondrial morphology and activity since their swimming activity in the M9 buffer has previously been shown to affect mitochondrial fission and fusion dynamics. In addition, leaving worms in M9 for long periods of time can activate hypoxia responses that significantly affect the mitochondria by causing mitochondrial proteostasis disruptions48,49.
Finally, another important consideration is that while changes to mitochondrial morphology are often correlated with changes to mitochondrial function, there is not always a direct correlation between the two. Thus, a more thorough analysis of mitochondrial function is recommended. For example, oxygen consumption rate can be measured using a Seahorse instrument50, mitochondrial membrane potential can be measured by using membrane potential dyes such as JC9 or TMRE51, the mitochondrial oxidative state can be measured using redox-sensitive dyes such as roGFP52, and mitochondrial stress resilience can be measured using sensitivity to stressors such as rotenone53. Because imaging of mitochondria can be quite rapid, we offer the methods here as an easy first pass to determine whether experimental conditions affect mitochondrial morphology. These methods are amenable even for large-scale screening of drugs or genes, with follow-up of more thorough mitochondrial analysis using additional metrics recommended. Altogether, here we review what is believed to be the simplest methods to image mitochondrial morphology in C. elegans with minimum experimental errors.
Disclosures
The authors have nothing to disclose.
Acknowledgements
J.K. is supported by the USC Provost Fellowship; M.A. and G.G. are supported by T32AG052374; M.V. is supported by 1R25AG076400; and R.H.S. is supported by R01AG079806 from the National Institute on Aging and 2022-A-010-SUP from the Larry L. Hillblom Foundation. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs grant P40 OD010440. Some gene analysis was performed using Wormbase, which is funded on a U41 grant HG002223.
Materials
| Name | Company | Catalog Number | Comments |
| 5-fluoro-2'-deoxyuridine (FUDR) | Spectrum Chemical | F2026-10GMBL | for proliferation inhibition |
| APEX IPTG | Genesee | 18-242 | for RNAi |
| Bacto Agar | VWR | 90000-764 | for NGM plates |
| Bacto Peptone | VWR | 97064-330 | for NGM plates |
| Calcium chloride dihydrate | VWR | 97061-904 | for NGM plates |
| Carbenicillin | VWR | 76345-522 | for RNAi |
| Cholesterol | VWR | 80057-932 | for NGM plates |
| Confocal microscope | Stellaris 5 | ||
| DMSO | VWR | BDH1115-1LP | solvent for drugs |
| HistoBond microscope slides | VWR | 16005-110 | for slides preparation |
| LB Broth | VWR | 95020-778 | for LB |
| LEICA S7E Dissecting Scope | Leica | 10450840 | Standard dissecting microscope |
| LEICA STELLARIS 5 | Leica | 158101100 | Confocal Microscope |
| LEICA THUNDER | Leica | 11525679 | Compound Microscope |
| Magnesium sulfate heptahydrate | VWR | 97062-132 | for NGM plates, M9 |
| pL4440 empty vector plasmid | addgene | plasmid #1654 | for empty vector plasmid |
| Potassium Chloride | VWR | 97061-566 | for bleach soluton |
| Potassium phosphate dibasic | VWR | EM-PX1570-2 | for NGM plates |
| Potassium phosphate monobasic | VWR | EM-PX1565-5 | for M9 |
| Sodium azide | VWR | 97064-646 | for paralyzing worms |
| Sodium Chloride | VWR | EM-SX0420-5 | for NGM plates, M9 |
| Sodium hypochlorite | VWR | RC7495.7-32 | for bleach solution |
| Sodium phosphate dibasic | VWR | 71003-472 | for M9 |
| Tetracycline hydrochloride | VWR | 97061-638 | for RNAi |
| WOB-L® Dry Vacuum Pumps, Standard-Duty, Welch® | VWR | 80077-612 | for aspiration |
References
- Protasoni, M., Zeviani, M. Mitochondrial structure and bioenergetics in normal and disease conditions. Int J Mol Sci. 22 (2), 586 (2021).
- Wang, Y., et al. The role of mitochondrial dynamics in disease. MedComm. 4 (6), e462 (2023).
- Kamer, K. J., Mootha, V. K. The molecular era of the mitochondrial calcium uniporter. Nat Rev Mol Cell Biol. 16 (9), 545-553 (2015).
- Chen, W., Zhao, H., Li, Y. Mitochondrial dynamics in health and disease: mechanisms and potential targets. Signal Transduct Target Ther. 8 (1), 333 (2023).
- Adebayo, M., Singh, S., Singh, A. P., Dasgupta, S. Mitochondrial fusion and fission: The fine-tune balance for cellular homeostasis. FASEB J. 35 (6), e21620 (2021).
- Madan, S., Uttekar, B., Chowdhary, S., Rikhy, R. Mitochondria lead the way: mitochondrial dynamics and function in cellular movements in development and disease. Front Cell Dev Biol. 9, 781933 (2021).
- Sharma, A., Smith, H. J., Yao, P., Mair, W. B. Causal roles of mitochondrial dynamics in longevity and healthy aging. EMBO Rep. 20 (12), e48395 (2019).
- Rolland, S. G., Lu, Y., David, C. N., Conradt, B. The BCL-2-like protein CED-9 of C. elegans promotes FZO-1/Mfn1,2- and EAT-3/Opa1-dependent mitochondrial fusion. J Cell Biol. 186 (4), 525-540 (2009).
- Labrousse, A. M., Zappaterra, M. D., Rube, D. A., van der Bliek, A. M. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 4 (5), 815-826 (1999).
- Dickinson, D. J., Pani, A. M., Heppert, J. K., Higgins, C. D., Goldstein, B. Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 200 (4), 1035-1049 (2015).
- Bosher, J. M., Labouesse, M. RNA interference: Genetic wand and genetic watchdog. Nat Cell Biol. 2 (2), E31-E36 (2000).
- Yu, C. C. J., et al. Expansion microscopy of C. elegans. eLife. 9, e46249 (2020).
- Zhang, S., Li, F., Zhou, T., Wang, G., Li, Z. Caenorhabditis elegans as a useful model for studying aging mutations. Front Endocrinol. 11, 554994 (2020).
- Dingley, S., et al. Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans. Mitochondrion. 10 (2), 125-136 (2010).
- Presley, A. D., Fuller, K. M., Arriaga, E. A. MitoTracker Green labeling of mitochondrial proteins and their subsequent analysis by capillary electrophoresis with laser-induced fluorescence detection. J Chromatogr B Anal Technol Biomed Life Sci. 793 (1), 141-150 (2003).
- Mitra, K., Lippincott-Schwartz, J. Analysis of mitochondrial dynamics and functions using imaging approaches. Curr Protoc Cell Biol. Chapter 4, Unit 4.25.1-Unit 4.25.21 (2010).
- Valera-Alberni, M., Yao, P., Romero-Sanz, S., Lanjuin, A., Mair, W. B. Novel imaging tools to study mitochondrial morphology in Caenorhabditis elegans. Life Sci Alliance. 7 (11), e202402918 (2024).
- Benedetti, C., Haynes, C. M., Yang, Y., Harding, H. P., Ron, D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 174 (1), 229-239 (2006).
- Begelman, D. V., et al. An aco-2::gfp knock-in enables the monitoring of mitochondrial morphology throughout C. elegans lifespan. microPublication Biol. 2022, (2022).
- Castro Torres, T., et al. Surveying low-cost methods to measure lifespan and healthspan in Caenorhabditis elegans. J Vis Exp. 183, e64091 (2022).
- Rasmussen, N. R., Reiner, D. J. Nuclear translocation of the tagged endogenous MAPK MPK-1 denotes a subset of activation events in C. elegans development. J Cell Sci. 134 (17), jcs258456 (2021).
- Schindelin, J., et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 9 (7), 676-682 (2012).
- Luz, A. L., et al. Mitochondrial morphology and fundamental parameters of the mitochondrial respiratory chain are altered in Caenorhabditis elegans strains deficient in mitochondrial dynamics and homeostasis processes. PLoS One. 10 (6), e0130940 (2015).
- Oorloff, M., et al. Mechanical stress through growth on stiffer substrates impacts animal health and longevity in C. elegans. bioRxiv. , (2024).
- Chu, X., Wu, S., Raju, R. NLRX1 regulation following acute mitochondrial injury. Front Immunol. 10, 2431 (2019).
- Shah, P., Bao, Z., Zaidel-Bar, R. Visualizing and quantifying molecular and cellular processes in Caenorhabditis elegans using light microscopy. Genetics. 221 (4), iyac068 (2022).
- Wang, Y., Wang, P., Li, C. Fluorescence microscopic platforms imaging mitochondrial abnormalities in neurodegenerative diseases. Adv Drug Deliv Rev. 197, 114841 (2023).
- Ding, J., et al. An expanded GCaMP reporter toolkit for functional imaging in Caenorhabditis elegans. G3 (Bethesda, Md). 13 (10), jkad183 (2023).
- Sarasija, S., Norman, K. R. Analysis of mitochondrial structure in the body wall muscle of Caenorhabditis elegans. Bio-Protocol. 8 (7), e2801 (2018).
- Chaweeborisuit, P., Suriyonplengsaeng, C., Suphamungmee, W., Sobhon, P., Meemon, K. Nematicidal effect of plumbagin on Caenorhabditis elegans: A model for testing a nematicidal drug. Z Naturforsch C J Biosci. 71 (5-6), 121-131 (2016).
- Valera-Alberni, M., Yao, P., Romero-Sanz, S., Lanjuin, A., Mair, W. B. . Novel imaging tools to study mitochondrial dynamics in Caenorhabditis elegans. 7, 11 (2024).
- Bolognesi, B., Lehner, B. Reaching the limit. eLife. 7, e39804 (2018).
- Jishi, A., Qi, X. Altered mitochondrial protein homeostasis and proteinopathies. Front Mol Neurosci. 15, 867935 (2022).
- Morton, K. S., Wahl, A. K., Meyer, J. N. The effect of common paralytic agents used for fluorescence imaging on redox tone and ATP levels in Caenorhabditis elegans. PLoS One. 19 (4), e0292415 (2024).
- Noumi, T., Maeda, M., Futai, M. Mode of inhibition of sodium azide on H+-ATPase of Escherichia coli. FEBS Lett. 213 (2), 381-384 (1987).
- Siete, C., Xiong, R., Khalid, A., Hsieh, Y. -. W., Chuang, C. -. F. Immobilization of C. elegans with different concentrations of an anesthetic for time-lapse imaging of dynamic protein trafficking in neurons. microPublication Biology. 2024, (2024).
- Mitchell, D. H., Stiles, J. W., Santelli, J., Sanadi, D. R. Synchronous growth and aging of Caenorhabditis elegans in the presence of fluorodeoxyuridine. J Gerontol. 34 (1), 28-36 (1979).
- Santi, D. V., McHenry, C. S. 5-Fluoro-2'-deoxyuridylate: covalent complex with thymidylate synthetase. Proc Natl Acad Sci U S A. 69 (7), 1855-1857 (1972).
- Wang, H., Zhao, Y., Zhang, Z. Age-dependent effects of floxuridine (FUdR) on senescent pathology and mortality in the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 509 (3), 694-699 (2019).
- Beanan, M. J., Strome, S. Characterization of a germ-line proliferation mutation in C. elegans. Development (Cambridge, England). 116 (3), 755-766 (1992).
- Austin, J., Kimble, J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 51 (4), 589-599 (1987).
- Garigan, D., et al. Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics. 161 (3), 1101-1112 (2002).
- Zhang, Y., et al. Neuronal TORC1 modulates longevity via AMPK and cell nonautonomous regulation of mitochondrial dynamics in C. elegans. eLife. 8, e49158 (2019).
- Dutta, N., et al. Investigating impacts of the mycothiazole chemotype as a chemical probe for the study of mitochondrial function and aging. GeroScience. 46 (6), 6009-6028 (2024).
- Tronstad, K., et al. Regulation and quantification of cellular mitochondrial morphology and content. Curr Pharm Des. 20 (35), 5634-5652 (2014).
- Nikolaisen, J., et al. Automated quantification and integrative analysis of 2D and 3D mitochondrial shape and network properties. PLoS One. 9 (7), e101365 (2014).
- Lee, J. S., Wee, T. L. E., Brown, C. M. Calibration of wide-field deconvolution microscopy for quantitative fluorescence imaging. J Biomol Tech. 25 (1), 31-40 (2014).
- Hong, M., Kwon, J. Y., Shim, J., Lee, J. Differential hypoxia response of hsp-16 genes in the nematode. J Mol Biol. 344 (2), 369-381 (2004).
- Yan, J., Sun, C. L., Shin, S., Van Gilst, M., Crowder, C. M. Effect of the mitochondrial unfolded protein response on hypoxic death and mitochondrial protein aggregation. Cell Death Dis. 12 (7), 711 (2021).
- Koopman, M., et al. A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. Nat Protoc. 11 (10), 1798-1816 (2016).
- Shpilka, T., et al. UPRmt scales mitochondrial network expansion with protein synthesis via mitochondrial import in Caenorhabditis elegans. Nat Comm. 12 (1), 479 (2021).
- Braeckman, B. P., Smolders, A., Back, P., De Henau, S. In vivo detection of reactive oxygen species and redox status in Caenorhabditis elegans. antioxidants & redox signaling. 25 (10), 577-592 (2016).
- Bar-Ziv, R., et al. Measurements of physiological stress responses in C. elegans. J Vis Exp. 159, e61001 (2020).
Explore More Articles
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2025 MyJoVE Corporation. All rights reserved