Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Gramicidin-based Fluorescence Assay; for Determining Small Molecules Potential for Modifying Lipid Bilayer Properties
W tym Artykule
Podsumowanie
We introduce a fast fluorescence-based assay that monitors the rate of fluorescence quenching as a measure of gramicidin channel activity. The gramicidin channels are used as molecular force transducers to monitor changes in lipid bilayer properties as sensed by bilayer spanning proteins.
Streszczenie
Many drugs and other small molecules used to modulate biological function are amphiphiles that adsorb at the bilayer/solution interface and thereby alter lipid bilayer properties. This is important because membrane proteins are energetically coupled to their host bilayer by hydrophobic interactions. Changes in bilayer properties thus alter membrane protein function, which provides an indirect way for amphiphiles to modulate protein function and a possible mechanism for "off-target" drug effects. We have previously developed an electrophysiological assay for detecting changes in lipid bilayer properties using linear gramicidin channels as probes 3,12. Gramicidin channels are mini-proteins formed by the transbilayer dimerization of two non-conducting subunits. They are sensitive to changes in their membrane environment, which makes them powerful probes for monitoring changes in lipid bilayer properties as sensed by bilayer spanning proteins. We now demonstrate a fluorescence assay for detecting changes in bilayer properties using the same channels as probes. The assay is based on measuring the time-course of fluorescence quenching from fluorophore-loaded large unilamellar vesicles due to the entry of a quencher through the gramicidin channels. We use the fluorescence indicator/quencher pair 8-aminonaphthalene-1,3,6-trisulfonate (ANTS)/Tl+ that has been successfully used in other fluorescence quenching assays 5,13. Tl+ permeates the lipid bilayer slowly 8 but passes readily through conducting gramicidin channels 1,14. The method is scalable and suitable for both mechanistic studies and high-throughput screening of small molecules for bilayer-perturbing, and potential "off-target", effects. We find that results using this method are in good agreement with previous electrophysiological results 12.
Protokół
1. Generate ANTS-filled Liposomes
- On day 1, remove organic solvent from lipids.
- Remove lipid from freezer and let it equilibrate to room temperature.
- Add 0.6 mL of 25 mg/mL (1,2-dierucoyl-sn-glycero-3-phosphocholine) lipid in chloroform solution to a 25 mL round bottom flask.
- Continually rotate the flask while drying under nitrogen, until all the chloroform has evaporated and a thin white film of lipid coats the entire lower half of the flask.
- Dry in a dessicator under vacuum overnight.
- On day 2, prepare the sample with the fluorophore that will be incorporated into the vesicles.
- Rehydrate in 100 mM NaNO3, 25 mM ANTS (Na salt), 10 mM HEPES. Use HNO3 or NaOH to adjust pH to 7.0. Each ANTS molecule has 2 Na+, for a total of 150 mM [Na+]. Add 1.671 mL of electrolyte to get a 10 mM lipid suspension.
- Parafilm and vortex suspension thoroughly.
- Protect sample from light with foil and let it age at room temperature overnight.
- On Day 3, make large unilamellar vesicles and remove all the fluorophore from outside the vesicles.
- Sonicate for 1 min in a low-power sonicator.
- Freeze-thaw sample 5-6 times, each time with 5 minutes on dry ice and 5 minutes in warm (~50 °C) water.
- Extrude the lipid suspension using an Avanti mini-extruder. Set up the mini-extruder with a 0.1 μm polycarbonate filter and filter supports (see extruder manual for details). Extrude the suspension back and forth 21 times such that the suspension ends up on the opposite syringe. When multiple batches are extruded, always remember to load samples with the same syringe. During extrusion the cloudy initial lipid suspension should become almost translucent (it will still be yellow due to the ANTS fluorophore). If the extrusion suddenly becomes easier and less pressure is needed to move through filter, the filter may have ruptured and will need to be replaced.
- Remove external ANTS by running the extruded suspension over a PD-10 desalting column using a gravity protocol. Equilibrate column with 20-30 mL of Na-buffer (140 mM NaNO3, 10 mM HEPES, pH 7.0, remember to use HNO3 or NaOH to adjust pH). Add 1.5 mL of the extruded lipid suspension to the column and let it seep in. Bring the total volume of sample to 2.5 mL by adding 1 mL Na-buffer. After the buffer is fully incorporated into the column and nothing is leaking out, elute the liposomes with 3 mL of Na-buffer and collect the resulting sample. The resulting ANTS filled LUV stock solution should contain about 4-5 mM lipids and appear translucent milky white. Protect the sample from light with foil.
- If the stock solution is not used immediately, store at 13 °C for a maximum of 7 days. Warning: do not freeze or cool the lipid solution below its liquid- to crystal-phase transition temperature (~12 °C), as this will destroy the vesicles' integrity and the fluorophore will leak out.
2. Mix Fluorescence Solution
- 24 hours prior to use, dilute liposomes and incubate with gramicidin (at 13 °C). Equilibration of gramicidin between the lipid vesicles' inner and outer monolayers is a slow process, requiring a long incubation.
- Thoroughly vortex the ANTS-filled LUV stock solution as the LUVs have a density >1 g/mL and will therefore sediment in the solutions.
- Mix the ANTS-filled LUV stock 1:20 in Na-buffer. Mix all liposomes for use in one day of experiments together in a single flask to enhance sample uniformity.
- Separate out 3/4 of the liposomes and add 260 nM gramicidin pre-diluted in DMSO (500 μg/mL).
- To the remaining 1/4 add the same volume of DMSO (without gramicidin) to keep the solvent concentration constant in all samples.
3. Setting up the Fluorescence Instrument
- We use an Applied Photophysics SX.20 stopped-flow spectrofluorometer with temperature control to measure the rate of fluorescence quenching.
- Turn the instruments on an hour ahead of time to allow for warm up. The components are: computer, electronic control unit, water-bath (both cooler and heater), lamp power supply and nitrogen tank.
- Set up the recording conditions using the Pro-Data SX software.
- Set the monochromator excitation wavelength to 352 nm and slit width apertures for excitation to 1/1.
- Use a λ=455 nm high-pass filter to record the emission.
- Use the "Pressure Hold" setting, which enables the instrument to maintain the nitrogen pressure during the recording and thus prevents pressure oscillations (and recording artifacts) during the first few milliseconds.
- Set "Time" to 1 s and "Points" to 5000.
- Use the repeat box and adjust the number of repeats, normally we use 9 and 13 repeats for buffer and quencher runs, respectively.
- Water-bath temperature should be set to 25°C; the temperature should be monitored in the SX software.
- Set the drive volume to 120 μL, meaning that each time the instrument runs a sample, it mixes 60 μL from each of the two syringes. With this instrumental setup, a 120 μL volume gives a dead time of ≈ 1.2 ms.
- Adjust the high voltage (gain) on the fluorescence detector for the ANTS-liposome's output to be around 8. For a standard experimental mixture the gain should be 400-420 V. Some drugs are fluorescent, such that they can saturate the fluorescence signal. In these cases, gain/voltage should be decreased.
- Always load the fluorescent sample into the left syringe and buffer/quencher in the right syringe. Make sure to thoroughly vortex all samples containing liposomes before loading, and be aware that the LUVs with time will settle in the syringe.
4. Doing an Experiment
- Prepare a sample of ANTS-loaded LUVs with either solvent or compound, and load into the spectrofluorometer together with either Na-buffer or Tl-quencher (50 mM TlNO3, 94 mM NaNO3, 10 mM HEPES at pH 7.0).
- Use 1.5 mL diluted ANTS-LUV solution, with or without gramicidin, prepared the previous day.
- Add the compound at the desired concentration or the solvent for controls. Keep the solvent concentration to a minimum and constant across all the samples. The concentrations will need to be adjusted with any new (unknown) compound to achieve the desired dose-response range. Incubate for 10 min at 25°C in the dark.
- Vortex the LUV sample and load into the left syringe. Load the Na-buffer or Tl-quencher into the right syringe.
- Thoroughly remove air bubbles from both samples by pushing the syringes back and forth.
- Make sure both syringes are loaded equally before closing the valves. Unequal loading will result in pre-mixing of solutions before they reach the recording chamber.
- For the very first sample, record the fluorescence with Na-buffer, repeat 4 times, adjust the high voltage (gain) as necessary, and repeat 5 times.
- For the remaining samples, record 9 repeats with Na-buffer.
- Replace the Na-buffer with the Tl-quencher and record 13 repeats.
- Rinse with water and continue with next sample. As controls we use samples with: no gramicidin and with solvent, no gramicidin and with the maximum compound concentration, with both gramicidin and solvent.
5. Analyzing Data
- Transfer the results to the analysis computer. Read all the data into MATLAB for analyzing. For each sample, read in all the buffer and quencher repeats, excluding the first four in each case, as those contain mixing artifacts. Assuming a drive volume of 120 μL it takes a little less than four shots to completely clear out the previous mixture from the stopped-flow tubing.
- The buffer repeats for all the samples should be very similar. If there is a significant shift in the fluorescence signal, which depends on the compound concentration, then the compound itself may be fluorescent and it may be necessary to first subtract this extra fluorescence signal before normalizing the samples.
- Manually go through the traces and remove "bad" repeats: repeats containing spikes or deviations from multi-exponentiality due to mixing artifacts and/or bubbles.
- Normalize the quencher repeats to the average starting value of the buffer repeats for each sample. Combine averages of all samples into one graph for visualization.
- Traces recorded with no gramicidin should all be similar and show almost no fluorescence quenching; there may be a slow reduction in the fluorescence signal due to slow leakage of Tl+ through the vesicles bilayer 8. If the sample with no gramicidin and no modifier shows significant quenching, then the vesicles have deteriorated and should not be used further. If the samples with no gramicidin but with the modifier show significant quenching, then the modifier perturbs the vesicles bilayer to such an extent that it allows the fluorophore or quencher to cross the bilayer.
- If the compound alters lipid bilayer properties, as sensed by gramicidin channels, the fluorescence time course will be visibly altered.
- For each sample, a stretched exponential
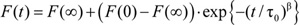 e.g. 4) is fit to the first 2-100 ms of the normalized fluorescence quenching curve of the individual repeats and the rate
e.g. 4) is fit to the first 2-100 ms of the normalized fluorescence quenching curve of the individual repeats and the rate 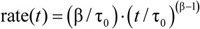 4 calculated at 2 ms. Averages and standard deviations are calculated for a given sample from all the individual repeats.
4 calculated at 2 ms. Averages and standard deviations are calculated for a given sample from all the individual repeats. - Finally, determine the relative change in quench rate by normalizing to the closest control sample in time that has gramicidin and no modifier.
6. Representative Results
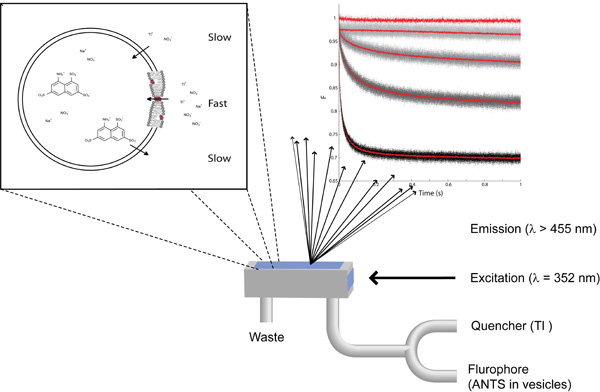
Figure 1: The essentials of the fluorescence quench-based assay to detect changes in lipid bilayer properties. Top left: A zoom-in on a single lipid vesicle with ANTS plus NaNO3 on the inside and NaNO3 plus TlNO3 on the outside. Top right: The fluorescence signal recorded using (form top to bottom) ANTS-filled vesicles without quencher, with quencher, with quencher and pre-doped with 87, 260 and 780 nM gramicidin. Bottom: Schematic representation of the stopped-flow mixing chamber.
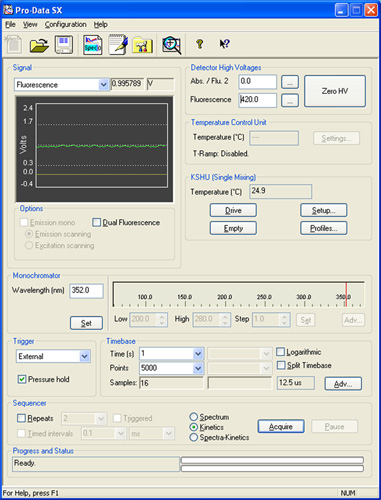
Figure 2: A screen shot from the Pro-Data SX software illustrating the various panels referenced in the description of the experimental setup.
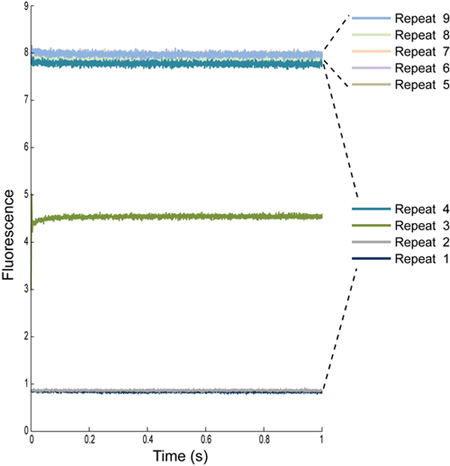
Figure 3: Multiple repeat determinations of the fluorescence signal from ANTS-loaded LUVs with Na-buffer. The first four repeats are always excluded, as they contain mixing artifacts. The tubing connecting the sample syringes to the mixing cell have a defined volume, therefore the first few repeats will give us a reading of what was previously in the tubing: water for repeat 1 and 2, some combination of water and sample for repeat 3, mostly sample for repeat 4, and just sample for the remaining repeats.
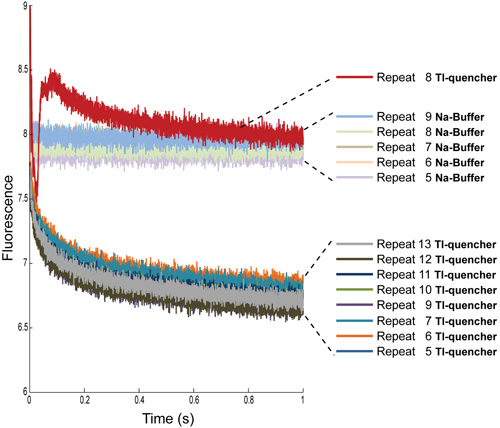
Figure 4: Multiple repeat determinations of the fluorescence signal from ANTS-loaded LUVs with Na-buffer and with Tl-quencher. The first four repeats have been removed from both conditions. Additionally, for the Tl-quencher measurements, repeat 8 needs to be removed due to artifacts, most likely air bubbles.
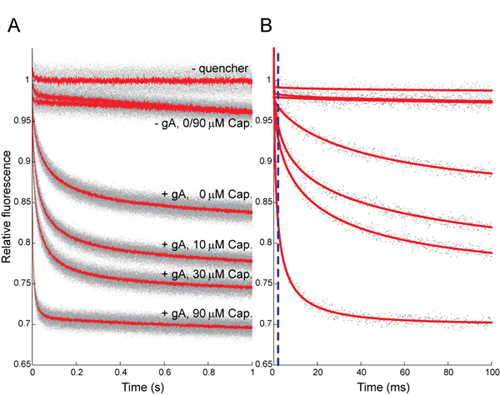
Figure 5: Effect of capsaicin (Cap) on the time course of ANTS fluorescence quenching. (A) Normalized fluorescence signal over 1 s, gray dots denote results from all repeats (n > 5 per condition); red lines denote the average of all repeats. (B) The first 100 ms, gray dots denote results from a single repeat for each condition; red lines are stretched exponential fits (2 - 100 ms) to those repeats. The stippled blue line denotes the 2 ms mark, the time at which the rate of quenching is determined. In both A and B the top trace shows results in the absence of Tl+; the next two traces show results in the absence of gA, with Tl+ ± Cap; the four lower traces show results with 260 nM gA and Tl+, where the numbers denote [Cap] in μM. The rates of quenching for 0, 10, 30 and 90 μM Cap, as determined by the rate of a stretched exponential, are 36±6, 69±6, 85±8 and 247±27 (mean ± s.d., n > 8), respectively.
Dyskusje
We have demonstrated a fast fluorescence-based assay for determining the bilayer modifying potential of drugs and other small amphiphiles. Compounds that modify bilayer properties are likely to alter membrane protein function in an indirect, nonspecific manner, possibly contributing to "off-target" drug effects. The assay exploits the power of gramicidin channels as probes for changes in bilayer properties 12 that are sensed by bilayer-spanning proteins. The results obtained using the fluorescence-based ass...
Ujawnienia
A provisional patent application that covers the methods described in the manuscript has been filed. The co-inventors are HII and OSA.
Podziękowania
We thank Michael J. Bruno, Radda Rusinova and Jon T. Sack for many stimulating discussions. Financial support from NIH, R01GM021342 and ARRA supplement R01GM021342-35S1, and the Josiah Macy, Jr. Foundation to OSA; the Tri-I CMB program for HII; and The Iris L. and Leverett S. Woodworth Medical Scientist Fellowship and NIH MSTP grant GM07739 for RK.
Materiały
| Name | Company | Catalog Number | Comments |
| ANTS | Invitrogen | A-350 | |
| gramicidin | Sigma-Aldrich | G-5002 | |
| 1,2-dierucoyl-sn-glycero-3-phosphocholine | Avanti Polar Lipid, Inc | 850398C | |
| Mini-Extruder kit | Avanti Polar Lipid, Inc | 610000 | |
| PD-10 Desalting column | Sigma-Aldrich | 54805 |
Odniesienia
- Andersen, O. S., Giebisch, G. H., Purcel, E. F. Ion transport through simple membranes. Renal Function. , (1978).
- Andersen, O. S., Koeppe, R. E. Bilayer thickness and membrane protein function: An energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 36, 107-130 (2007).
- Andersen, O. S., Koeppe, R. E., Roux, B., Chung, S. -. H., Andersen, O. S., Krishnamurthy, V. Gramicidin channels. Versatile tools. Biological Membrane Ion Channels: Dynamics, Structure, and Applications. , (2007).
- Berberan-Santos, M. N., Bodunov, E. N., Valeur, B. Mathematical functions for the analysis of luminescence decays with underlying distributions 1. Kohlrausch decay function (stretched exponential. Chem. Phys. 315, 171-182 (2005).
- Bruggemann, E. P., Kayalar, C. Determination of the molecularity of the colicin E1 channel by stopped-flow ion flux kinetics. Proc. Natl. Acad. Sci. USA. 83, 4273-4276 (1986).
- Bruno, M. J., Koeppe, R. E., Andersen, O. S. Docosahexaenoic acid alters bilayer elastic properties. Proc. Natl. Acad. Sci. USA. 104, 9638-9643 (2007).
- Buboltz, J. T., Feigenson, G. W. A novel strategy for the preparation of liposomes: rapid solvent exchange. Biochim. Biophys. Acta. 1417, 232-245 (1999).
- Gutknecht, J. Cadmium & thallous ion permeabilities through lipid bilayer membranes. Biochim. Biophys. Acta. 735, 185-188 (1983).
- Ingólfsson, H. I., Koeppe, R. E., Andersen, O. S. Curcumin is a modulator of bilayer material properties. Biochemistry. 46, 10384-10391 (2007).
- Keserü, G. M., Makara, G. M. The influence of lead discovery strategies on the properties of drug candidates. Nat. Rev. Drug Discov. 8, 203-212 (2009).
- Leeson, P. D., Springthorpe, B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discov. 6, 881-890 (2007).
- Lundb k, J. A., Collingwood, S. A., Ingólfsson, H. I., Kapoor, R., Andersen, O. S., S, O. Lipid bilayer regulation of membrane protein function: gramicidin channels as molecular force probes. J. R. Soc. Interface. 7, 373-395 (2010).
- Moore, H. P. &. a. m. p. ;. a. m. p., Raftery, M. A. Direct spectroscopic studies of cation translocation by Torpedo acetylcholine receptor on a time scale of physiological relevance. Proc. Natl. Acad. Sci. USA. 77, 4509-4513 (1980).
- Neher, E. Ionic specificity of the gramicidin channel and the thallous ion. Biochim. Biophys. Acta. 401, 540-544 (1975).
- O'Connell, A. M., Koeppe, R. E., Andersen, O. S. Kinetics of gramicidin channel formation in lipid bilayers: transmembrane monomer association. Science. 250, 1256-1259 (1990).
- Søgaard, R. GABAA receptor function is regulated by lipid bilayer elasticity. Biochemistry. 45, 13118-13129 (2006).
- Waring, M. J. Defining optimum lipophilicity and molecular weight ranges for drug candidates-Molecular weight dependent lower logD limits based on permeability. Bioorg. Med. Chem. Lett. 19, 2844-2851 (2009).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaPrzeglądaj więcej artyków
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone