Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Defining Substrate Specificities for Lipase and Phospholipase Candidates
W tym Artykule
Podsumowanie
Many predicted (phospho)lipases are poorly characterized with regard to their substrate specificities and physiological functions. Here we provide a protocol to optimize enzyme activities, search for natural substrates, and propose physiological functions for these enzymes.
Streszczenie
Microorganisms produce a wide spectrum of (phospho)lipases that are secreted in order to make external substrates available for the organism. Alternatively, other (phospho)lipases may be physically associated with the producing organism causing a turnover of intrinsic lipids and frequently giving rise to a remodeling of the cellular membranes. Although potential (phospho)lipases can be predicted with a number of algorithms when the gene/protein sequence is available, experimental proof of the enzyme activities, substrate specificities, and potential physiological functions has frequently not been obtained. This manuscript describes the optimization of assay conditions for prospective (phospho)lipases with unknown substrate specificities and how to employ these optimized conditions in the search for the natural substrate of a respective (phospho)lipase. Using artificial chromogenic substrates, such as p-nitrophenyl derivatives, may help to detect a minor enzymatic activity for a predicted (phospho)lipase under standard conditions. Having encountered such a minor enzymatic activity, the distinct parameters of an enzyme assay can be varied in order to obtain a more efficient hydrolysis of the artificial substrate. After having determined the conditions under which an enzyme works well, a variety of potential natural substrates should be assayed for their degradation, a process that can be followed employing distinct chromatographic methods. The definition of substrate specificities for new enzymes, often provides hypotheses for a potential physiological role of these enzymes, which then can be tested experimentally. Following these guidelines, we were able to identify a phospholipase C (SMc00171) that degrades phosphatidylcholine to phosphocholine and diacylglycerol, in a crucial step for the remodeling of membranes in the bacterium Sinorhizobium meliloti upon phosphorus-limiting conditions of growth. For two predicted patatin-like phospholipases (SMc00930 and SMc01003) of the same organism, we could redefine their substrate specificities and clarify that SMc01003 is a diacylglycerol lipase.
Wprowadzenie
Glycerol-based lipids such as triacylglycerols and (glycero)phospholipids constitute important and probably the best-known lipid classes1. Triacylglycerols (TAGs) are fats or oils, which usually function as storage lipids, and therefore as potential energy and carbon sources. TAGs can be degraded by lipases, which are frequently secreted by the producing organism to digest external TAGs and make them available as carbon sources. Also, lipases have been widely studied over the years due to their important biotechnological applications2.
Due to their amphiphilic nature and their near-cylindric shape, (glycero)phospholipids exhibit membrane-forming properties and usually constitute the major lipidic components of a bilayered membrane3. In simple microorganisms, such as the bacterium Escherichia coli, only three major head group variants, phosphatidylglycerol (PG), cardiolipin (CL), and phosphatidylethanolamine (PE) are encountered, although one should be aware that each one of them can be substituted with a considerable number of different fatty acyl chains at the sn-1 or sn-2 position giving rise to a large number of different molecular species4. Other bacteria might have other phospholipids in addition or instead. For example, Sinorhizobium meliloti, a soil bacterium, which is able to form a nitrogen-fixing root nodule symbiosis with the legume alfalfa (Medicago sativa), contains in addition to PE a second zwitterionic phospholipid, phosphatidylcholine (PC)5. Also, lipids not containing phosphorus or glycerol might be amphiphilic and form part of the cellular membrane. For example, upon phosphorus-limiting growth conditions, in S. meliloti, (glycero)phospholipids are largely replaced by membrane lipids that do not contain phosphorus, i.e., sulfolipids, ornithine lipids, and diacylglyceryl trimethylhomoserine (DGTS)6. In bacteria, DGTS is formed from diacylglycerol (DAG) in a two-step pathway7 but the source for DAG generation was not clear. Pulse-chase experiments suggested that PC might be a precursor for DGTS8 and using the methodology described in this manuscript we could identify a phospholipase C (PlcP, SMc00171) that is formed under phosphorus-limiting conditions and which can convert PC into DAG and phosphocholine8.
In a separate study, we discovered that an acyl-CoA synthetase (FadD)-deficient mutant of S. meliloti or of Escherichia coli accumulated free fatty acids when entering stationary phase of growth9. Although these fatty acids seemed to be derived from membrane lipids, the precise source for the free fatty acids or the enzyme(s) liberating them were not known. Again, employing the strategy outlined in this manuscript, two patatin-like10 (phospho)lipases (SMc00930 and SMc01003) that contributed to the formation of free fatty acids in S. meliloti11 were predicted. Surprisingly, SMc01003 used DAG as substrate converting it to monoacylglycerol and finally glycerol and free fatty acids11. Therefore, SMc01003 is a DAG lipase (DglA).
Although a number of algorithms exist for predicting potential (phospho)lipases12,13, their precise function and physiological role is usually not known. Here we outline a protocol, to clone and overexpress predicted or potential (phospho)lipases. This manuscript explains how enzyme assays can be developed and optimized for the overexpressed (phospho)lipase by using artificial chromogenic substrates. We provide examples how with an optimized enzyme assay the real (phospho)lipase substrate can be encountered and how these findings might enrich our understanding of microbial physiology.
Protokół
1. Clone and Overexpress Structural Gene for Predicted Lipase
- Using polymerase chain reaction (PCR)14 and specific oligonucleotides (Table 1)15, amplify the gene of interest (smc01003, smc00930, or smc00171), predicted to code for a lipase or phospholipase, from the genomic DNA of the host organism (i.e., S. meliloti).
- Introduce specific restriction sites (with the designed sequence of the oligonucleotides). Digest the amplified DNA fragment with the corresponding restriction enzymes and clone it into an expression vector such as plasmids of the pET series16.
- After verifying the correct DNA sequence for the cloned gene, transform the vector to an expression strain such as Escherichia coli BL21(DE3) pLysS16.
- Prepare an overnight pre-culture of the expression host E. coli BL21(DE3) pLysS, harboring the respective pET vector with the cloned gene or the empty vector, in 100 ml culture flasks containing 20 ml of Luria Bertani broth (LB)17 plus the required antibiotics. Culture the cells at 30 °C (or at the usual growth temperature of the bacterium from which the lipase originates).
- Using the overnight pre-cultures, inoculate 500 ml of prewarmed LB medium (plus the required antibiotics) in 2 L culture flasks to obtain an initial optical density at 620 nm (OD620) = 0.05. Follow growth of cultures and at an OD620 = 0.3, add isopropyl-β-D-thiogalactoside (IPTG) to a final concentration of 100 µM, and incubate under agitation at 30 °C for a period of 4 hr.
- At the end of the incubation period, transfer each culture to a 500 ml centrifuge tube and centrifuge at 5,000 x g at 4 °C for 30 min. Resuspend bacterial cell pellets in 5 ml of suspension buffer (e.g., SMc00930- and SMc01003-expressing cells in 50 mM Tris-HCl pH 8.0 and SMc00171-expressing cells in 50 mM diethanolamine-HCl pH 9.8). Store the cell suspensions at -80 °C until use.
2. Prepare Cell-free Protein Extracts and Determine Protein Concentration
- Defrost bacterial cell suspensions and store on ice. Pass cell suspensions three times through a cold pressure cell at 20,000 lb per in2. Remove intact cells and cell debris by centrifugation at 5,000 x g for 30 min at 4 °C.
- After centrifugation, prepare aliquots of 100 and 500 µl from the supernatant for subsequent analysis and store them at -80 °C until use.
- Use one of the 100 µl aliquot to determine protein concentration of distinct cell-free extracts by a method of choice or as described18.
3. Use Artificial Substrates for Optimizing Enzyme Activities of (Phospho)lipases
- For an initial coverage of distinct enzyme activities, use artificial substrates that yield a colored product upon hydrolysis, such as p-nitrophenol (p-NP).
- For enzyme assays optimized already with artificial p-nitrophenyl ester substrates (outlined for phospholipase C PlcP (SMc00171), as well as for the predicted patatin-like phospholipases SMc00930 and SMc01003), use pipetting schemes described in Table 2.
- When exploring a new potential (phospho)lipase, prepare a first standard enzyme assay containing 50 mM Tris-HCl, pH 8.5, 100 mM NaCl, 0.05% Triton X-100, 0.5 mM p-nitrophenyl-containing compound (p-nitrophenyl phosphate, bis-p-nitrophenyl phosphate, p-nitrophenyl decanoate, or p-nitrophenyl palmitate), and cell-free protein extract (check 1, 3, 10, 30, 100, 300, and 1,000 µg) in a total volume of 1 ml in 1 ml plastic cuvettes.
NOTE: Use alkaline pH (Figure 1) when following p-nitrophenyl ester hydrolysis in a continuous assay. Alternatively, use single time-point assays for a range of pH values, adding NaOH at the end of the incubation period to terminate the enzyme reaction and to ensure that all p-NP is present in the phenolate form. - Follow the time course for an increase of absorbance at 405 nm, due to the formation of p-NP, in a spectrophotometer at 30 °C over a 5 min period. Quantify the initially linear formation of p-NP by determining the initial slope of increase of absorbance per time.
- Calculate the change of concentration (Δc) for p-NP using the law of Lambert-Beer (ΔA = ε Δc d)1.
NOTE: ΔA is the linear change of absorbance determined, ε is the molar extinction coefficient at the respective wave length (in units of M-1 cm-1), d is the length of the light path (1 cm), and Δc is the change of concentration (in units of M) to be determined.- Considering that the assay volume is 1 ml, calculate the amount of p-NP formed.
NOTE: Amount = concentration x volume. - Calculate the enzyme activity by dividing the amount of p-NP formed by the time in which it is formed. Determine the specific enzyme activity by dividing enzyme activity by the amount of protein (in mg) which was responsible for generating this activity.
- Considering that the assay volume is 1 ml, calculate the amount of p-NP formed.
- Compare absorbance changes provoked by protein extracts in which a candidate gene (smc00171, smc00930, or smc01003) had been expressed with extracts that harbor only an empty vector.
NOTE: In order to continue with the following steps, the specific activities, caused by protein extracts in which a candidate gene had been expressed, should be at least twice or more than the values obtained for the specific activities caused by protein extracts that harbor only an empty vector. - For further experiments, select those conditions in which hydrolysis of the p-nitrophenyl-containing compound is minimal with cell-free extracts (i.e., empty vector) and for which the most pronounced formation of p-NP and the p-nitrophenolate anion (Figure 1) can be observed when protein extracts are employed, in which a candidate gene had been expressed.
- After determining the initial enzyme activity in 3.1, optimize the assay conditions for the respective enzyme by varying pH, type of buffer, buffer strength, concentrations of NaCl, detergents such as Triton X-100, and the absence or presence of different bivalent cations.
- For different concentrations of each variable, determine the specific enzyme activity (see 3.1.4.2) (the highest number obtained defines the condition of maximal enzyme activity). Use the combination of the optimal conditions encountered for each variable to define an optimized enzyme assay in which each variable is present in its optimal concentration.
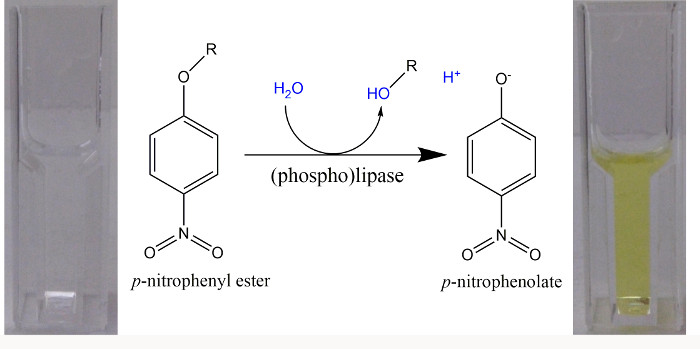
Figure 1. p-Nitrophenyl esters as artificial substrates for (phospho)lipases in a spectrophotometric assay. Upon hydrolysis of p-nitrophenyl esters, an acid (R-OH) and p-nitrophenol (p-NP) are formed. Due to the pKa = 7.2 for the dissociation of the phenolic H+ from p-NP, at a pH > 9.2 more than 99% are in the bright yellow p-nitrophenolate form and a molar extinction coefficient of 18,000 M-1 cm-1 can be used at a wave length of 405 nm for the quantification of free p-nitrophenolate22. When buffers with a pH of 8.5 were used, the absorbance was determined at 400 nm and a molar extinction coefficient of 14,500 M-1 cm-1 was employed23. Please click here to view a larger version of this figure.
NOTE: After having defined the optimal conditions for the activity of the enzyme of interest, embark on the search for the real/physiological substrate of this lipase. In principle, take two, often complementary, approaches to achieve this goal, an in vivo approach or an in vitro approach.
4. In Vivo Identification of the Physiological Substrate of a Lipase
NOTE: In an in vivo approach, express the lipase of interest in a host organism8,11 in order to register over time whether expression of the lipase alters the host´s lipid profile. In another in vivo approach, generate a mutant deficient of the gene of interest8,11 and study whether its lipid profile is distinct from the wild type version6,8,11. In order to obtain a quantitative assessment of an organism's lipid profile, a simple method consists in radiolabeling cellular compounds, extracting the lipids, separating them by chromatography, and quantifying the radioactively labeled separated lipids.
- Radiolabeling of lipids.
- Prepare an overnight pre-culture of an organism of interest (E. coli or S. meliloti) in 5 ml of the desired culture medium (complex medium or defined minimal medium) and grow at 30 °C.
- From the pre-culture, inoculate into 20 ml of the same fresh medium in a 100 ml culture flask to obtain an initial OD620 = 0.3 for the culture.
- Take an aliquot (1 ml) of the culture under sterile conditions and transfer to a 14 ml sterile polystyrene round-bottom tube.
- Add 1 µCi of [1-14C]acetate (60 mCi per mmol) to the 1 ml culture.
- Incubate the liquid culture under agitation at 30 °C for a period of 24 hr.
- At the end of the incubation period, transfer the culture to a 1.5 ml microcentrifuge tube and centrifuge at 12,000 x g at room temperature for 5 min.
- Resuspend the pellet in 100 µl of water. At this point, store the cell suspension at -20 °C or immediately continue with the extraction of polar lipids (section 4.2).
- Extraction of polar lipids.
NOTE: The method described here essentially follows the procedure reported by Bligh and Dyer19.- To the 100 µl of aqueous cell suspension, add 375 µl of methanol:chloroform solution (2:1; vol/vol).
- Vortex for 30 sec and incubate for 5 min at room temperature.
- Centrifuge 5 min at 12,000 x g at room temperature.
- Transfer the supernatant to a new 1.5 ml microcentrifuge tube.
- Add 125 µl of chloroform and 125 µl of water, vortex 30 sec.
- Centrifuge 1 min at 12,000 x g at room temperature.
- Transfer the lower chloroform phase to a fresh tube and dry with a stream of nitrogen gas.
- Dissolve dried lipids in 100 µl of chloroform:methanol solution (1:1; vol/vol).
NOTE: At this point, an aliquot of 5 µl of the lipid solution can be quantified by liquid scintillation counting. - For thin-layer chromatographic (TLC) analysis, dry down the remaining 95 µl with a stream of nitrogen gas and redissolve dried lipids in 20 µl of chloroform:methanol solution (1:1; vol/vol). Use a 3 µl aliquot for TLC analysis.
- Separation of polar lipids by thin-layer chromatography (TLC).
NOTE: Depending on the lipid classes to be analyzed, different combinations of solid and mobile phases might be employed for separation. Here a typical separation for charged polar lipids and another, more suited for neutral polar lipids, using high performance thin-layer chromatography (HPTLC) silica gel aluminum sheets as the solid phase, are outlined.- Separation of charged polar lipids by two-dimensional TLC (2D-TLC).
- Apply a 3 µl aliquot of lipid sample in one corner of a HPTLC silica gel aluminum sheet (10 x 10 cm), 2 cm from the edge of the plate.
- Prepare and mix the mobile phase (140 ml chloroform, 60 ml methanol, and 10 ml water) for separation in the first dimension.
- Coat a TLC developing chamber internally with chromatography paper.
NOTE: This is to ensure that the gas phase of the chamber will be saturated rapidly (within 30 min) after the mobile phase for the first dimension has been added to the chamber and the chamber has been closed with a glass plate. - Prepare and mix the mobile phase (130 ml chloroform, 50 ml methanol, and 20 ml glacial acetic acid) for separation in the second dimension and transfer to a second TLC developing chamber internally coated with chromatography paper and let the chamber saturate.
- Carefully transfer the HPTLC silica gel aluminum sheet with the dried lipid sample to the first chamber and develop (i.e., perform chromatography) the plate for 60 min in the closed chamber in the first dimension5.
- Remove the plate from the chamber and let solvents dry off in a flow hood for 30 min.
- After turning the plate by 90 degrees with regard to the previous chromatography, transfer the HPTLC silica gel aluminum sheet, on which the lipids have been separated in one dimension, to the second chamber and develop the plate for 60 min in the second dimension5.
- Remove the sheet from the chamber and let the solvents dry off in a flow hood for at least 2 hr.
- Separation of neutral polar lipids.
- Apply 3 µl aliquots of lipid samples on a HPTLC silica gel aluminum sheet starting 2 cm from the edges of the plate. If multiple samples are analyzed in a one-dimensional chromatography, keep a distance of at least 1.5 cm between the different sample application spots.
- Prepare and mix the mobile phase (140 ml hexane, 60 ml diethylether, and 8 ml acetic acid) and transfer to a TLC developing chamber internally coated with chromatography paper and covered with a glass plate to let the chamber saturate (30 min).
- Transfer the HPTLC silica gel aluminum sheet with the dried lipid samples to the chamber and develop the plate for 30 min in the closed chamber.
- Remove the plate from the chamber and let the solvents dry off in a flow hood for 2 hr.
- Separation of charged polar lipids by two-dimensional TLC (2D-TLC).
- Quantification and visualization of separated polar lipids.
- Once the developed TLC sheet is dry, incubate it with a photostimulable luminescence (PSL) screen in a closed cassette for 3 days.
- Expose the incubated screen to a PSL scanner and acquire a virtual image of the separated radiolabeled lipids.
- Perform quantification using PSL software20.
- Visualization and isolation of individual polar lipids classes.
- Incubate developed TLC sheet for 10 min in a chromatography chamber in the presence of 1 g of iodine crystals.
NOTE: Separated lipidic compounds will dissolve the iodine and appear as brownish spots. - Circle the spots with a pencil, compare them to the relative mobility (Rf) of standard compounds (i.e., 1,2-dipalmitoyl-sn-glycerol, dipalmitoyl-L-α-phosphatidylcholine, DL-α-monopalmitin, or palmitic acid), and identify to which lipid class they might belong.
- In a fume hood, let the iodine evaporate from the TLC sheet.
- With the help of a spatula, scrape the silica gel containing the compound of interest from the sheet, and extract the compound from the silica gel with a mixture of 100 µl of water and 375 µl of methanol:chloroform solution (2:1; vol/vol).
- Continue with extraction according to Bligh and Dyer as outlined (4.2.2 onwards).
- Store purified lipid class in 100 µl of chloroform:methanol solution (1:1; vol/vol) at -20 °C until use.
- Incubate developed TLC sheet for 10 min in a chromatography chamber in the presence of 1 g of iodine crystals.
5. In Vitro Identification of the Physiological Substrate of a Lipase
NOTE: In an in vitro approach, study whether the lipase of interest can convert a mixture of isolated lipids or individual pure lipids to the corresponding hydrolysis products under the conditions defined as optimal in 3.2.
- Use pipetting schemes for enzyme assays as per Table 3 for PC-specific phospholipase C SMc00171 (see 5.2), phospholipase A (see 5.3), and DAG lipase SMc01003 (see 5.4) activity.
- Determination of PC-specific phospholipase C activity (Table 3).
- To a 1.5 ml microcentrifuge tube, add 5,000 counts per minute (cpm) of total 14C-labeled PC and a solution of Triton X-100.
- Mix and dry under a stream of nitrogen.
- Add diethanolamine-HCl, pH 9.8 buffer, as well as NaCl and MnCl2 solutions and bidistilled water to obtain a final volume of 99.5 µl. Vortex for 5 sec.
- Add 0.5 µl of enzyme (5 µg protein) (i.e., a cell-free extract in which overexpressed SMc00171 is present) to initiate the reaction. Mix briefly.
- Incubate at 30 °C for 4 hr.
- Stop the reaction by the addition of 250 µl of methanol and 125 µl of chloroform.
- Extract lipids as described previously (see 4.2).
- Separate lipids by one-dimensional (1D)-TLC (see 4.3.2 and 4.4), and analyze them by PSL imaging.
- Determination of phospholipase A activity (Table 3).
- To a 1.5 ml microcentrifuge tube, add 5,000 cpm of total 14C-labeled phospholipids and a solution of Triton X-100.
- Mix and dry under a stream of nitrogen.
- For a final 100 µl assay, add Tris-HCl, pH 8.5 buffer, NaCl solution and water. Vortex for 5 sec.
- Add 5 µl of enzyme (50 µg protein) (i.e., a cell-free extract in which overexpressed SMc00930 or SMc01003 is present).
- Incubate at 30 °C for 5 hr.
- Stop the reaction by the addition of 250 µl of methanol and 125 µl of chloroform.
- Extract lipids as described previously (see 4.2), separate them by 1D-TLC using 130 ml chloroform, 50 ml methanol, and 20 ml glacial acetic acid as the mobile phase, and analyze them by PSL imaging.
- Determination of diacylglycerol (DAG) lipase activity.
- Preparation of 14C-labeled DAG.
- Radiolabel S. meliloti cultures (see 4.1) and extract polar lipids (see 4.2) as described. Separate S. meliloti total lipid extracts by 1D-TLC in chloroform:methanol:acetic acid (130:50:20; vol/vol) using conditions described for separation in second dimension in 4.3.1.
- Visualize PC by iodine staining and use a pencil to mark the localization of phosphatidylcholine (PC).
- Isolate radiolabeled PC as described in 4.5.
- Quantify extracted PC by scintillation counting.
NOTE: About 320,000 cpm PC is expected. - Treat PC (250,000 cpm) with 0.1 U of phospholipase C from Clostridium perfringens in 50 mM Tris-HCl, pH 7.2, 0.5% Triton X-100 and 10 mM CaCl2 for 2 hr in a total volume of 100 µl and stop the reaction by adding 250 µl of methanol and 125 µl of chloroform.
- Extract lipids as described previously and separate by them by 1D-TLC (see 4.3.2).
- Isolate diacylglycerol from the silica plate and quantify by scintillation counting (as described in 4.2)
- Diacylglycerol lipase assay (Table 3).
- To a 1.5 ml microcentrifuge tube, add 5,000 cpm of 14C-labeled DAG and a solution of Triton X-100.
- Mix and dry under a stream of nitrogen.
- For a final 100 µl assay, add Tris-HCl (pH 9.0) buffer, a NaCl solution and bidistilled water. Vortex for 5 sec.
- Initiate the reaction by adding 5 µl of enzyme (50 µg protein of cell-free extract).
- Incubate at 30 °C for 4 hr.
- Stop the reaction by the addition of 250 µl of methanol and 125 µl of chloroform and extract lipids as described previously (see 4.2).
- Analyze neutral polar lipids by 1D-TLC (see 4.1.3.2) and subsequent PSL imaging.
- Preparation of 14C-labeled DAG.
Wyniki
Activity of PC-specific Phospholipase C SMc00171 with Bis-p-nitrophenyl Phosphate
Cell-free extracts obtained from E. coli BL21(DE3) x pLysS, which had smc00171 expressed, were studied for their ability to hydrolyze bis-p-nitrophenyl phosphate esters, using a spectrophotometric enzymatic assay, measuring the p-NP formed. No hydrolytic...
Dyskusje
Over the past 20 years, genomes of many organisms have been sequenced and although a wealth of genome sequence data has been generated, functional interpretation is lagging behind and therefore hampers our understanding of genome function. Gene functions in genomes are often assigned based on similarity to genes of known function or occurrence of conserved motifs. However, the precise function of a given gene is often not known. Especially, predicted structural genes for enzymes cannot be easily explored by omic techniqu...
Ujawnienia
The authors have nothing to disclose.
Podziękowania
This work was supported by grants from Consejo Nacional de Ciencias y Tecnología-México (CONACyT-Mexico) (82614, 153998, 253549, and 178359 in Investigación Científica Básica as well as 118 in Investigación en Fronteras de la Ciencia) and from Dirección General de Asuntos de Personal Académico-Universidad Nacional Autónoma de México (DGAPA-UNAM; PAPIIT IN202616, IN203612).
Materiały
| Name | Company | Catalog Number | Comments |
| Chloroform | JT Baker | 9180-03 | TLC analysis & Lipid extraction |
| Methanol | JT Baker | 9070-03 | TLC analysis & Lipid extraction |
| Acetic Acid | JT Baker | 9507-05 | TLC analysis & Lipid extraction |
| Hexanes | JT Baker | 9309-02 | TLC analysis & Lipid extraction |
| Diethylether | Sigma | 32203 | Enzymatic assays |
| bidistilled water | ANY | NA | Enzymatic assays |
| Tris Base | Sigma | T-1503 | Enzymatic assays |
| HCl | Baker | 9535-02 | Enzymatic assays |
| NaCl | Baker | 3624-01 | Enzymatic assays |
| Triton X-100 | Sigma | X-100 | Enzymatic assays |
| LB broth | ANY | NA | Bacterial growth, 10 g tryptone + 5 g yeast extract + 10 g NaCl per liter of bidistilled water |
| tryptone | Becton Dickinson and Company | 211705 | Bacterial growth |
| yeast extract | Becton Dickinson and Company | 212750 | Bacterial growth |
| TY broth | ANY | NA | Bacterial growth, 8 g tryptone + 3 g yeast extract + 66 mg CaCl2·2H2O per liter of bidistilled water |
| CaCl2·2H2O | Baker | 1332-01 | Enzymatic assays |
| isopropyl-β-D-thiogalactoside (IPTG) | Invitrogen | 15529-019 | Bacterial growth |
| Diethanolamine | Sigma | D-8885 | Enzymatic assays |
| MnCl2 | Sigma | 221279 | Enzymatic assays |
| Phospholipase A2 snake venom | Sigma | P0790 | Enzymatic assays |
| Phospholipase C Clostridium perfringens | Sigma | P7633 | Enzymatic assays |
| Bis-p-nitrophenyl phosphate | Sigma | 07422AH | Enzymatic assays |
| p-nitrophenyl stearate | Sigma | N3627 | Enzymatic assays |
| p-nitrophenyl dodecanoate | Sigma | 61716 | Enzymatic assays |
| p-nitrophenyl decanoate | Sigma | N0252 | Enzymatic assays |
| p-nitrophenyl palmitate | Sigma | N2752 | Enzymatic assays |
| p-nitrophenyl butyrate | Sigma | N9876 | Enzymatic assays |
| p-nitrophenyl octanoate | Sigma | 21742 | Enzymatic assays |
| Acetic Acid, sodium salt [1-14C] | Perkin Elmer | NEC084 | Bacterial growth |
| dimethylsulfoxide (DMSO) | JT Baker | 9224-01 | Enzymatic assays |
| Aluminium HPTLC silica gel 60 plates. Silica gel HPTLC plates size 20 x 20 cm, 25 sheets. | Merck | 105547 | TLC analysis & Lipid extraction |
| Spectrometer UV/VIS Lambda 35 | Perkin Elmer | NA | Enzymatic assays |
| Storm 820 Phosphorimager | Molecular Dynamics | NA | Photostimulable Luminescence scanner |
| Multipurpose Scintillation Counter | Beckman Coulter | NA | Radioactivity Quantification |
| French Pressure Cell | ThermoSpectronic | NA | Breakage of cells |
| chromatography paper 3MM Chr | Whatman | 3030917 | TLC analysis |
| Sinorhizobium meliloti 1021our | reference 11 | studied strain | |
| Escherichia coli BL21 (DE3) pLysS Competent cells | Novagen | 69451 | protein expression strain |
| pET9a vector | Novagen | 69431 | protein expression vector |
| pET17b vector | Novagen | 69663 | protein expression vector |
| sterile polystyrene round-bottom tube (14 ml) Falcon | Becton Dickinson | 352057 | radiolabeling of bacterial cultures |
| polypropylene microcentrifuge tubes (1.5 ml) | Eppendorf | 30125.15 | Enzymatic assays |
| 1,2-dipalmitoyl-sn-glycerol | Sigma | D9135 | lipid standard |
| L-α-phosphatidylcholine, dipalmitoyl | Sigma | P6267 | lipid standard |
| DL-α-monopalmitin | Sigma | M1640 | lipid standard |
| palmitic acid | Sigma | P0500 | lipid standard |
Odniesienia
- Nelson, D. L., Cox, M. M. . Lehninger, Principles of Biochemistry. , (2013).
- Jaeger, K. E., Eggert, T. Lipases for biotechnology. Curr. Opin. Biotechnol. 13 (4), 390-397 (2002).
- Dowhan, W., Bogdanov, M., Mileykovskaya, E. Functional roles of lipids in membranes. Biochemistry of Lipids, Lipoproteins and Membranes. , 1-37 (2008).
- Dowhan, W. Molecular basis for membrane phospholipid diversity: why are there so many lipids?. Annu. Rev. Biochem. 66, 199-232 (1997).
- De Rudder, K. E. E., Thomas-Oates, J. E., Geiger, O. Rhizobium meliloti mutants deficient in phospholipid N-methyltransferase still contain phosphatidylcholine. J. Bacteriol. 179, 6921-6928 (1997).
- Geiger, O., Röhrs, V., Weissenmayer, B., Finan, T. M., Thomas-Oates, J. E. The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti. Mol. Microbiol. 32 (1), 63-73 (1999).
- Klug, R. M., Benning, C. Two enzymes of diacylglyceryl-O-4'-(N,N,N,-trimethyl)homoserine biosynthesis are encoded by btaA and btaB in the purple bacterium Rhodobacter sphaeroides. Proc. Natl. Acad. Sci. USA. 98 (10), 5910-5915 (2001).
- Zavaleta-Pastor, M., et al. Sinorhizobium meliloti phospholipase C required for lipid remodeling duringphosphorus limitation. Proc. Natl. Acad. Sci. USA. 107 (1), 302-307 (2010).
- Pech-Canul, A., et al. FadD is required for utilization of endogenous fatty acids released from membrane lipids. J. Bacteriol. 193 (22), 6295-6304 (2011).
- Banerji, S., Flieger, A. Patatin-like proteins: a new family of lipolytic enzymes present in bacteria?. Microbiology. 150 (Pt 3), 522-525 (2004).
- Sahonero-Canavesi, D. X., et al. Fatty acid-releasing activities in Sinorhizobium meliloti include unusual diacylglycerol lipase. Environ. Microbiol. 17 (9), 3391-3406 (2015).
- Fischer, M., Pleiss, J. The Lipase Engineering Database: a navigation and analysis tool for protein families. Nucl. Acid. Res. 31 (1), 319-321 (2003).
- Sigrist, C. J. A., et al. New and continuing developments at PROSITE. Nucleic Acids Res. 41 (Database issue), D344-D347 (2013).
- Lorenz, T. C. Polymerase Chain Reaction: Basic Protocol Plus Troubleshooting and Optimization Strategies. J. Vis. Exp. (63), e3998 (2012).
- Untergasser, A., et al. Primer3 - new capabilities and interfaces. Nucl. Acids Res. 40 (15), e115 (2012).
- Studier, F. W., Rosenberg, A. H., Dunn, J. J., Dubendorff, J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185, 60-89 (1990).
- Miller, J. H. . Experiments in Molecular Genetics. , (1972).
- Desjardins, P., Hansen, J. B., Allen, M. Microvolume Protein Concentration Determination using the NanoDrop 2000c Spectrophotometer. J. Vis. Exp. (33), e1610 (2009).
- Bligh, E. G., Dyer, W. J. A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 37 (8), 911-917 (1959).
- Molecular Dynamics. . Phosphorimager SI User´s Guide. , (1994).
- Dixon, M., Webb, E. . Enzymes: Third Edition. , (1979).
- Rudolph, A. E., et al. Expression, characterization, and mutagenesis of the Yersinia pestis murine toxin, a phospholipase D superfamily member. J. Biol. Chem. 274 (17), 11824-11831 (1999).
- Kato, S., Yoshimura, T., Hemmi, H., Moriyama, R. Biochemical analysis of a novel lipolytic enzyme YvdO from Bacillus subtilis. Biosci. Biotechnol. Biochem. 74 (4), 701-706 (2010).
- Peppelenbosch, M. P. Kinome profiling. Scientifica (Cairo). , (2012).
- Manafi, M., Kneifel, W., Bascomb, S. Fluorogenic and chromogenic substrates used in bacterial diagnostics. Microbiol. Rev. 55 (3), 335-348 (1991).
- Kuznetsova, E., et al. Enzyme genomics: Application of general enzymatic screens to discover new enzymes. FEMS Microbiol. Rev. 29 (2), 263-279 (2005).
- Scopes, R. K. . Protein Purification, Principles and Practice. , (2010).
- Geiger, O., Lopez-Lara, I. M., Sohlenkamp, C. Phosphatidylcholine biosynthesis and function in bacteria. Biochim. Biophys. Acta. 1831 (3), 503-513 (2013).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone