Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Selecting Multiple Biomarker Subsets with Similarly Effective Binary Classification Performances
W tym Artykule
Podsumowanie
Existing algorithms generate one solution for a biomarker detection dataset. This protocol demonstrates the existence of multiple similarly effective solutions and presents a user-friendly software to help biomedical researchers investigate their datasets for the proposed challenge. Computer scientists may also provide this feature in their biomarker detection algorithms.
Streszczenie
Biomarker detection is one of the more important biomedical questions for high-throughput 'omics' researchers, and almost all existing biomarker detection algorithms generate one biomarker subset with the optimized performance measurement for a given dataset. However, a recent study demonstrated the existence of multiple biomarker subsets with similarly effective or even identical classification performances. This protocol presents a simple and straightforward methodology for detecting biomarker subsets with binary classification performances, better than a user-defined cutoff. The protocol consists of data preparation and loading, baseline information summarization, parameter tuning, biomarker screening, result visualization and interpretation, biomarker gene annotations, and result and visualization exportation at publication quality. The proposed biomarker screening strategy is intuitive and demonstrates a general rule for developing biomarker detection algorithms. A user-friendly graphical user interface (GUI) was developed using the programming language Python, allowing biomedical researchers to have direct access to their results. The source code and manual of kSolutionVis can be downloaded from http://www.healthinformaticslab.org/supp/resources.php.
Wprowadzenie
Binary classification, one of the most commonly investigated and challenging data mining problems in the biomedical area, is used to build a classification model trained on two groups of samples with the most accurate discrimination power1,2,3,4,5,6,7. However, the big data generated in the biomedical field has the inherent "large p small n" paradigm, with the number of features usually much larger than the number of samples6,8,9. Therefore, biomedical researchers have to reduce the feature dimension before utilizing the classification algorithms to avoid the overfitting problem8,9. Diagnosis biomarkers are defined as a subset of detected features separating patients of a given disease from healthy control samples10,11. Patients are usually defined as the positive samples, and the healthy controls are defined as the negative samples12.
Recent studies have suggested that there exists more than one solution with identical or similarly effective classification performances for a biomedical dataset5. Almost all the feature selection algorithms are deterministic algorithms, producing only one solution for the same dataset. Genetic algorithms may simultaneously generate multiple solutions with similar performances, but they still try to select one solution with the best fitness function as the output for a given dataset13,14.
Feature selection algorithms can be roughly grouped as either filters or wrappers12. A filter algorithm chooses the top-k features ranked by their significant individual association with the binary class labels based on the assumption that features are independent of each other15,16,17. Although this assumption does not hold true for almost all real-world datasets, the heuristic filter rule performs well in many cases, for instance, the mRMR (Minimum Redundancy and Maximum Relevance) algorithm, the Wilcoxon test based feature filtering (WRank) algorithm, and the ROC (Receiver operating characteristic) plot based filtering (ROCRank) algorithm. mRMR, is an efficient filter algorithm because it approximates the combinatorial estimation problem with a series of much smaller problems, comparing to the maximum-dependency feature selection algorithm, each of which only involves two variables, and therefore uses pairwise joint probabilities which are more robust18,19. However, mRMR may underestimate the usefulness of some features as it does not measure the interactions between features which can increase relevancy, and thus misses some feature combinations that are individually useless but are useful only when combined. The WRank algorithm calculates a non-parametric score of how discriminative a feature is between two classes of samples, and is known for its robustness for outliers20,21. Furthermore, the ROCRank algorithm evaluates how significant the Area Under the ROC Curve (AUC) of a particular feature is for the investigated binary classification performance22,23.
On the other hand, a wrapper evaluates the pre-defined classifier's performance of a given feature subset, iteratively generated by a heuristic rule, and creates the feature subset with the best performance measurement24. A wrapper generally outperforms a filter in the classification performance but runs slower25. For example, the Regularized Random Forest (RRF)26,27 algorithm uses a greedy rule, by evaluating the features on a subset of the training data at each random forest node, whose feature importance scores are evaluated by the Gini index. The choice of a new feature will be penalized if its information gain does not improve that of the chosen features. Additionally, the Prediction Analysis for Microarrays (PAM)28,29 algorithm, also a wrapper algorithm, calculates a centroid for each of the class labels, and then selects features to shrink the gene centroids toward the overall class centroid. PAM is robust for outlying features.
Multiple solutions with the top classification performance may be necessary for any given dataset. Firstly, the optimization goal of a deterministic algorithm is defined by a mathematical formula, e.g., minimum error rate30, which is not necessarily ideal for biological samples. Secondly, a dataset may have multiple, significantly different, solutions with similar effective or even identical performances. Almost all existing feature selection algorithms will randomly select one of these solutions as the output31.
This study will introduce an informatics analytic protocol for generating multiple feature selection solutions with similar performances for any given binary classification dataset. Considering that most biomedical researchers are not familiar with informatic techniques or computer coding, a user-friendly graphical user interface (GUI) was developed to facilitate the rapid analysis of biomedical binary classification datasets. The analytic protocol consists of data loading and summarizing, parameter tuning, pipeline execution, and result interpretations. With a simple click, the researcher is able to generate the biomarker subsets and publication-quality visualization plots. The protocol has been tested using the transcriptomes of two binary classification datasets of Acute Lymphoblastic Leukemia (ALL), i.e., ALL1 and ALL212. The datasets of ALL1 and ALL2 were downloaded from the Broad Institute Genome Data Analysis Center, available at http://www.broadinstitute.org/cgi-bin/cancer/datasets.cgi. ALL1 contains 128 samples with 12,625 features. Of these samples, 95 are B-cell ALL and 33 are T-cell ALL. ALL2 includes 100 samples with 12,625 features as well. Of these samples, there are 65 patients that suffered relapse and 35 patients that did not. ALL1 was an easy binary classification dataset, with a minimum accuracy of four filters and four wrappers being 96.7%, and 6 of the 8 feature selection algorithms achieving 100%12. While ALL2 was a more difficult dataset, with the above 8 feature selection algorithms achieving no better than 83.7% accuracy12. This best accuracy was achieved with 56 features detected by the wrapper algorithm, Correlation-based Feature Selection (CFS).
Protokół
NOTE: The following protocol describes the details of the informatics analytic procedure and pseudo-codes of the major modules. The automatic analysis system was developed using Python version 3.6.0 and the Python modules pandas, abc, numpy, scipy, sklearn, sys, PyQt5, sys, mRMR, math and matplotlib. The materials used in this study are listed in the Table of Materials.
1. Prepare the Data Matrix and Class Labels
- Prepare the data matrix file as a TAB- or comma-delimited matrix file, as illustrated in Figure 1A.
NOTE: Each row has all the values of a feature, and the first item is the feature name. A feature is a probeset ID for the microarray-based transcriptome dataset or may be another value ID like a cysteine residue with its methylation value in a methylomic dataset. Each column gives the feature values of a given sample, with the first item being the sample name. A row is separated into columns by a TAB (Figure 1B) or a comma (Figure 1C). A TAB-delimited matrix file is recognized by the file extension .tsv, and a comma-delimited matrix file has the extension .csv. This file may be generated by saving a matrix as either the .tsv or .csv format from software such as Microsoft Excel. The data matrix may also be generated by computer coding. - Prepare the class label file as a TAB- or comma-delimited matrix file (Figure 1D), similar to the data matrix file.
NOTE: The first column gives the sample names, and the class label of each sample is given in the column titled Class. Maximal compatibility is considered in the coding process, so that additional columns may be added. The class label file may be formatted as a .tsv or .csv file. The names in the column Class may be any terms, and there may be more than two classes of samples. The user may choose any two of the classes for the following analysis.
2. Load the Data Matrix and Class Labels
- Load the data matrix and class labels into the software. Click the button Load data matrix to choose the user-specified data matrix file. Click the button Load class labels to choose the corresponding class label file.
NOTE: After both files are loaded, kSolutionVis will conduct a routine screen of the compatibility between the two files. - Summarize the features and samples from the data matrix file. Estimate the size of the data matrix file.
- Summarize the samples and classes from the class label file. Estimate the size of the class label file.
- Test whether each sample from the data matrix has a class label. Summarize the numbers of the samples with the class labels.
3. Summarize and Display the Baseline Statistics of the Dataset
- Click the button Summarize, without any specified keyword input, and the software will display 20 indexed features and the corresponding features names.
NOTE: Users need to specify the feature name they wish to find to see its baseline statistics and corresponding value distribution among all input samples. - Provide a keyword, e.g. “1000_at”, in the textbox Feature to find a specific feature to be summarized. Click the button Summarize to get the baseline statistics for this given feature.
NOTE: The keyword may appear anywhere in the target feature names, facilitating the search process for users. - Click the button Summarize to find more than one feature with the given keyword, and then specify the unique feature ID to proceed with the above step of summarizing one particular feature.
4. Determine the Class Labels and the Number of Top-ranked Features
- Choose the names of Positive (“P (33)”) and Negative (“N (95)”) classes in the dropdown boxes Class Positive and Class Negative, as shown in Figure 2 (middle).
NOTE: It is suggested to choose a balanced binary classification dataset, i.e., the difference between the numbers of positive and negative samples is minimal. The number of samples is also given in parenthesis after the name of each class label in the two dropdown boxes. - Choose 10 as the number of top-ranked features (parameter pTopX) in the dropdown box Top_X (?) for a comprehensive screen of the feature-subset.
NOTE: The software automatically ranks all the features by the P-value calculated by a t-test of each feature comparing the positive and negative classes. A feature with a smaller P-value has a better discriminating power between the two classes of samples. The comprehensive screening module is computationally intensive. The parameter pTopX is 10 by default. Users can change this parameter in the range of 10 to 50, until they find satisfying feature subsets with good classification performances.
5. Tune System Parameters for Different Performances
- Choose the performance measurement (pMeasurement) Accuracy (Acc) in the dropdown box Acc/bAcc (?) for the selected classifier Extreme Learning Machine (ELM). Another option of this parameter is the measurement Balanced Accuracy (bAcc).
NOTE: Let TP, FN, TN, and FP be the numbers of true positives, false negatives, true negatives and false positives, respectively. The measurement Acc is defined as (TP+TN)/(TP+FN+TN+FP), which works best on a balanced dataset6. But a classifier optimized for Acc tends to assign all the samples to the negative class if the number of negative samples is much larger than that of positive ones. The bAcc is defined as (Sn+Sp)/2, where Sn = TP/(TP+FN) and Sp = TN/(TN+FP) are the correctly predicted rates for positive and negative samples, respectively. Therefore, bAcc normalizes the prediction performances over the two classes, and may lead to a balanced prediction performance over two unbalanced classes. Acc is the default choice of pMeasurement. The software uses the classifier ELM by default to calculate the classification performances. The user may also choose a classifier from SVM (Support Vector Machine), KNN (k Nearest Neighbor), Decision Tree, or Naïve Bayes. - Choose the cutoff value 0.70 (parameter pCutoff) for the specified performance measurement in the input box pCutoff:.
NOTE: Both Acc and bAcc range between 0 and 1, and the user may specify a value pCutoff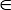 [0, 1] as the cutoff to display the matched solutions. The software carries out a comprehensive feature-subset screening, and an appropriate choice of pCutoff will make the 3D visualization more intuitive and explicit. The default value for pCutoff is 0.70.
[0, 1] as the cutoff to display the matched solutions. The software carries out a comprehensive feature-subset screening, and an appropriate choice of pCutoff will make the 3D visualization more intuitive and explicit. The default value for pCutoff is 0.70.
6. Run the Pipeline and Produce the INTERACTIVE VISUALIZED RESULTS
- Click the button Analyze to run the pipeline and generate the visualization plots, as shown in Figure 2 (bottom).
NOTE: The left table gives all the feature subsets and their pMeasurement calculated by the 10-fold cross validation strategy of the classifier ELM, as described previously5. Two 3D scatter plots and two-line plots are generated for the feature-subset screening procedure with the current parameter settings. - Choose 0.70 as the default value of the pMeasurement cutoff (parameter piCutoff, input box Value), and 10 as the default of the number of best feature subsets (parameter piFSNum).
NOTE: The pipeline is executed using the parameters pTopX, pMeasurement, and pCutoff. The detected feature subsets may be further screened using the cutoff piCutoff, however piCutoff cannot be smaller than pCutoff. Therefore, piCutoff is initialized as pCutoff and only the feature subsets with the performance measurement ≥ piCutoff will be visualized. The default value of piCutoff is pCutoff. Sometimes kSolutionVis detects many solutions, and only the best piFSNum (default: 10) feature subsets will be visualized. If the number of feature subsets detected by the software is smaller than piFSNum, all the feature subsets will be visualized. - Collect and interpret the features detected by the software, as shown in Figure 3.
NOTE: The table in the left box shows the detected feature subsets and their performance measurements. The names of the first three columns are “F1”, “F2”, and “F3”. The three features in each feature subset are given in their ranking order in one row (F1 < F2 < F3). The last column gives the performance measurement (Acc or bAcc) of each feature subset, and its column name (Acc or bAcc) is the value of pMeasurement.
7. Interpret the 3D Scatter Plots-Visualize and Interpret the Feature Subsets with Similarly Effective Binary Classification Performances Using 3D Scatter Plots
- Click the button Analyze to generate the 3D scatter plot of the top 10 feature subsets with the best classification performances (Acc or bAcc) detected by the software, as shown in Figure 3 (middle box). Sort the three features in a feature subset in ascending order of their ranks and use the ranks of the three features as the F1/F2/F3 axes, i.e., F1 < F2 < F3.
NOTE: The color of a dot represents the binary classification performance of the corresponding feature subset. A dataset may have multiple feature subsets with similarly effective performance measurements. Therefore, an interactive and simplified scatter plot is necessary. - Change the value to 0.70 in the input box pCutoff: and click the button Analyze to generate the 3D scatter plot of the feature subsets with the performance measurement ≥ piCutoff, as seen in Figure 3 (right box). Click the button 3D tuning to open a new window to manually tune the viewing angles of the 3D scatter plot.
NOTE: Each feature subset is represented by a dot in the same way as above. The 3D scatter plot was generated in the default angle. To facilitate the 3D visualization and tuning, a separate window will be opened by clicking the button 3D tuning. - Click the button Reduce to reduce the redundancy of the detected feature subsets.
NOTE: If users wish to further select the feature triplets and minimize the redundancy of the feature subsets, the software also provides this function using the mRMR feature selection algorithm. After clicking the Reduce button, kSolutionVis will remove those redundant features in the feature triplets and regenerate the table and the two scatter plots mentioned above. The removed features of the feature triplets will be replaced by the key word in the table. The values of None in the F1/F2/F3 axis will be denoted as the value of piFSNum (the range of the normal value of F1/F2/F3 is [1, top_x]). Therefore, the dots that include a None value may appear to be “outlier” dots in the 3D plots. The manually tunable 3D plots may be found in “Manual tuning of the 3D dot plots” in the supplementary material.
8. Find Gene Annotations and Their Associations with Human Diseases
NOTE: Steps 8 to 10 will illustrate how to annotate a gene from the sequence level of both DNA and protein. Firstly, the gene symbol of each biomarker ID from the above steps will be retrieved from the database DAVID32, and then two representative web servers will be used to analyze this gene symbol from the levels of DNA and protein, respectively. The server GeneCard provides a comprehensive functional annotation of a given gene symbol, and the Online Mendelian Inheritance in Man database (OMIM) provides the most comprehensive curation of disease-gene associations. The server UniProtKB is one of the most comprehensive protein database, and the server Group-based Prediction System (GPS) predicts the signaling phosphorylation’s for a very large list of kinases.
- Copy and paste the web link of the database DAVID into a web browser and open the web page of this database. Click the link Gene ID Conversion seen in Figure 4A and input the feature IDs 38319_at/38147_at/33238_at of the first biomarker subset of the dataset ALL1 (Figure 4B). Click the link Gene List and click Submit List as shown in Figure 4B. Retrieve the annotations of interest and click Show Gene List (Figure 4C). Get the list of gene symbols (Figure 4D).
NOTE: The gene symbols retrieved here will be used for further functional annotations in the next steps. - Copy and paste the web link of the database Gene Cards into a web browser and open the web page of this database. Search a gene’s name CD3D in the database query input box and find the annotations of this gene from Gene Cards33,34, as shown in Table 1 and Figure 5A.
NOTE: Gene Cards is a comprehensive gene knowledgebase, providing nomenclature, genomics, proteomics, subcellular localization, and involved pathways and other functional modules. It also provides external links to various other biomedical databases like PDB/PDB_REDO35, Entrez Gene36, OMIM37, and UniProtKB38. If the feature name is not a standard gene symbol, use the database ENSEMBL to convert it39. CD3D is the name of the gene T-Cell Receptor T3 Delta Chain. - Copy and paste the web link of the database OMIM into a web browser and open the web page of this database. Search a gene’s name CD3D and find the annotations of this gene from the database OMIM37, as shown in Table 1 and Figure 5B.
NOTE: OMIM serves now as one of the most comprehensive and authoritative sources of human gene connections with inheritable diseases. OMIM was initiated by Dr. Victor A. McKusick to catalog the disease-associated genetic mutations40. OMIM now covers over 15,000 human genes and over 8,500 phenotypes, as of December 1st 2017.
9. Annotate the Encoded Proteins and the Post-Translational Modifications
- Copy and paste the web link of the database UniProtKB into a web browser and open the web page of this database. Search a gene’s name CD3D in the query input box of UniProtKB and find the annotations of this gene from the database38, as shown in Table 1 and Figure 5C.
NOTE: UniProtKB collects a rich source of annotations for proteins, including both nomenclature and functional information. This database also provides external links to other widely used databases, including PDB/PDB_REDO35, OMIM37, and Pfam41. - Copy and paste the web link of the web server GPS into a web browser and open the web page of this web server. Retrieve the protein sequence encoded by the biomarker gene CD3D from the UniProtKB database38 and predict the protein’s post-translational modification (PTM) residues using the online tool GPS, as shown in Table 1 and Figure 5D.
NOTE: A biological system is dynamic and complicated, and the existing databases collect only known information. Therefore, biomedical prediction online tools as well as offline programs may provide useful evidence to complement a hypothesized mechanism. GPS has been developed and improved for over 12 years7,42 and may be used to predict a protein’s PTM residues in a given peptide sequence43,44. Tools are also available for various research topics, including the prediction of a protein’s subcellular location45 and transcription factor binding motifs 46 among others .
10. Annotate Protein-Protein Interactions and Their Enriched Functional Modules
- Copy and paste the web link of the web server String into a web browser and open the web page of this web server. Search the list for the genes CD3D and P53, and find their orchestrated properties using the database String47. The same procedure may be carried out using another web server, DAVID32.
NOTE: Besides the aforementioned annotations for individual genes, there are many large-scale informatics tools available to investigate the properties of a group of genes. A recent study demonstrated that individually bad marker genes might constitute a much-improved gene set5. Therefore, it’s worth the computational cost to screen for more complicated biomarkers. The database String may visualize the known or predicted interaction connections, and the David server may detect the functional modules with significant phenotype-associations in the queried genes47,32. Various other large-scale informatics analysis tools are also available.
11. Export the Generated Biomarker Subsets and the Visualization Plots
- Export the detected biomarker subsets as a .tsv or .csv text file for further analysis. Click the button Export the Table under the table of all the detected biomarker subsets and choose which text format to save as.
- Export the visualization plots as an image file. Click the button Save under each plot and choose which image format to save as.
NOTE: The software supports the pixel format .png and the vector format .svg. The pixel images are good for displaying on the computer screen, while the vector images may be converted to any resolution required for journal publication purposes.
Wyniki
The goal of this workflow (Figure 6) is to detect multiple biomarker subsets with similar efficiencies for a binary classification dataset. The whole process is illustrated by two example datasets ALL1 and ALL2 extracted from a recently-published biomarker detection study12,48. A user may install kSolutionVis by following the instructions in the supplementary materials.
Dyskusje
This study presents an easy-to-follow multi-solution biomarker detection and characterization protocol for a user-specified binary classification dataset. The software puts an emphasis on user-friendliness and flexible import/export interfaces for various file formats, allowing a biomedical researcher to investigate their dataset easily using the GUI of the software. This study also highlights the necessity of generating more than one solution with similarly effective modeling performances, previously ignored by many exi...
Ujawnienia
We have no conflicts of interest related to this report.
Podziękowania
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13040400) and the startup grant from Jilin University. Anonymous reviewers and biomedical testing users were appreciated for their constructive comments on improving the usability and functionality of kSolutionVis.
Materiały
| Name | Company | Catalog Number | Comments |
| Hardware | |||
| laptop | Lenovo | X1 carbon | Any computer works. Recommended minimum configuration: 1GB extra hard disk space, 1 GB memory, 2.0MHz CPU |
| Name | Company | Catalog Number | Comments |
| Software | |||
| Python 3.0 | WingWare | Wing Personal | Any python programming and running environments support Python version 3.0 or above |
Odniesienia
- Heckerman, D., et al. Genetic variants associated with physical performance and anthropometry in old age: a genome-wide association study in the ilSIRENTE cohort. Scientific Reports. 7, 15879 (2017).
- Li, Z., et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nature Genetics. 49, 1576-1583 (2017).
- Winkler, T. W., et al. Quality control and conduct of genome-wide association meta-analyses. Nature Protocols. 9, 1192-1212 (2014).
- Harrison, R. N. S., et al. Development of multivariable models to predict change in Body Mass Index within a clinical trial population of psychotic individuals. Scientific Reports. 7, 14738 (2017).
- Liu, J., et al. Multiple similarly-well solutions exist for biomedical feature selection and classification problems. Scientific Reports. 7, 12830 (2017).
- Ye, Y., Zhang, R., Zheng, W., Liu, S., Zhou, F. RIFS: a randomly restarted incremental feature selection algorithm. Scientific Reports. 7, 13013 (2017).
- Zhou, F. F., Xue, Y., Chen, G. L., Yao, X. GPS: a novel group-based phosphorylation predicting and scoring method. Biochemical and Biophysical Research Communications. 325, 1443-1448 (2004).
- Sanchez, B. N., Wu, M., Song, P. X., Wang, W. Study design in high-dimensional classification analysis. Biostatistics. 17, 722-736 (2016).
- Shujie, M. A., Carroll, R. J., Liang, H., Xu, S. Estimation and Inference in Generalized Additive Coefficient Models for Nonlinear Interactions with High-Dimensional Covariates. Annals of Statistics. 43, 2102-2131 (2015).
- Li, J. H., et al. MiR-205 as a promising biomarker in the diagnosis and prognosis of lung cancer. Oncotarget. 8, 91938-91949 (2017).
- Lyskjaer, I., Rasmussen, M. H., Andersen, C. L. Putting a brake on stress signaling: miR-625-3p as a biomarker for choice of therapy in colorectal cancer. Epigenomics. 8, 1449-1452 (2016).
- Ge, R., et al. McTwo: a two-step feature selection algorithm based on maximal information coefficient. BMC Bioinformatics. 17, 142 (2016).
- Tumuluru, J. S., McCulloch, R. Application of Hybrid Genetic Algorithm Routine in Optimizing Food and Bioengineering Processes. Foods. 5, (2016).
- Gen, M., Cheng, R., Lin, L. . Network models and optimization: Multiobjective genetic algorithm approach. , (2008).
- Radovic, M., Ghalwash, M., Filipovic, N., Obradovic, Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinformatics. 18, 9 (2017).
- Ciuculete, D. M., et al. A methylome-wide mQTL analysis reveals associations of methylation sites with GAD1 and HDAC3 SNPs and a general psychiatric risk score. Translational Psychiatry. 7, e1002 (2017).
- Lin, H., et al. Methylome-wide Association Study of Atrial Fibrillation in Framingham Heart Study. Scientific Reports. 7, 40377 (2017).
- Wang, S., Li, J., Yuan, F., Huang, T., Cai, Y. D. Computational method for distinguishing lysine acetylation, sumoylation, and ubiquitination using the random forest algorithm with a feature selection procedure. combinatorial chemistry & high throughput screening. , (2017).
- Zhang, Q., et al. Predicting Citrullination Sites in Protein Sequences Using mRMR Method and Random Forest Algorithm. combinatorial chemistry & high throughput screening. 20, 164-173 (2017).
- Cuena-Lombrana, A., Fois, M., Fenu, G., Cogoni, D., Bacchetta, G. The impact of climatic variations on the reproductive success of Gentiana lutea L. in a Mediterranean mountain area. International journal of biometeorology. , (2018).
- Coghe, G., et al. Fatigue, as measured using the Modified Fatigue Impact Scale, is a predictor of processing speed improvement induced by exercise in patients with multiple sclerosis: data from a randomized controlled trial. Journal of Neurology. , (2018).
- Hong, H., et al. Applying genetic algorithms to set the optimal combination of forest fire related variables and model forest fire susceptibility based on data mining models. The case of Dayu County, China. Science of the Total Environment. 630, 1044-1056 (2018).
- Borges, D. L., et al. Photoanthropometric face iridial proportions for age estimation: An investigation using features selected via a joint mutual information criterion. Forensic Science International. 284, 9-14 (2018).
- Kohavi, R., John, G. H. Wrappers for feature subset selection. Artificial intelligence. 97, 273-324 (1997).
- Yu, L., Liu, H. Efficient feature selection via analysis of relevance and redundancy. Journal of machine learning research. 5, 1205-1224 (2004).
- Wexler, R. B., Martirez, J. M. P., Rappe, A. M. Chemical Pressure-Driven Enhancement of the Hydrogen Evolving Activity of Ni2P from Nonmetal Surface Doping Interpreted via Machine Learning. Journal of American Chemical Society. , (2018).
- Wijaya, S. H., Batubara, I., Nishioka, T., Altaf-Ul-Amin, M., Kanaya, S. Metabolomic Studies of Indonesian Jamu Medicines: Prediction of Jamu Efficacy and Identification of Important Metabolites. Molecular Informatics. 36, (2017).
- Shangkuan, W. C., et al. Risk analysis of colorectal cancer incidence by gene expression analysis. PeerJ. 5, e3003 (2017).
- Chu, C. M., et al. Gene expression profiling of colorectal tumors and normal mucosa by microarrays meta-analysis using prediction analysis of microarray, artificial neural network, classification, and regression trees. Disease Markers. , 634123 (2014).
- Fleuret, F. Fast binary feature selection with conditional mutual information. Journal of Machine Learning Research. 5, 1531-1555 (2004).
- Pacheco, J., Alfaro, E., Casado, S., Gámez, M., García, N. A GRASP method for building classification trees. Expert Systems with Applications. 39, 3241-3248 (2012).
- Jiao, X., et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 28, 1805-1806 (2012).
- Rappaport, N., et al. Rational confederation of genes and diseases: NGS interpretation via GeneCards, MalaCards and VarElect. Biomedical Engineering OnLine. 16, 72 (2017).
- Rebhan, M., Chalifa-Caspi, V., Prilusky, J., Lancet, D. GeneCards: integrating information about genes, proteins and diseases. Trends in Genet. 13, 163 (1997).
- Joosten, R. P., Long, F., Murshudov, G. N., Perrakis, A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 1, 213-220 (2014).
- Maglott, D., Ostell, J., Pruitt, K. D., Tatusova, T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Research. 39, D52-D57 (2011).
- Amberger, J. S., Bocchini, C. A., Schiettecatte, F., Scott, A. F., Hamosh, A. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Research. 43, D789-D798 (2015).
- Boutet, E., et al. the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods in Molecular Biology. 1374, 23-54 (2016).
- Zerbino, D. R., et al. Ensembl 2018. Nucleic Acids Res. , (2017).
- McKusick, V. A., Amberger, J. S. The morbid anatomy of the human genome: chromosomal location of mutations causing disease. Journal of Medical Genetics. 30, 1-26 (1993).
- Finn, R. D., et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research. 44, D279-D285 (2016).
- Xue, Y., et al. GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Research. 33, W184-W187 (2005).
- Deng, W., et al. GPS-PAIL: prediction of lysine acetyltransferase-specific modification sites from protein sequences. Scientific Reports. 6, 39787 (2016).
- Zhao, Q., et al. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Research. 42, W325-W330 (2014).
- Wan, S., Duan, Y., Zou, Q. HPSLPred: An Ensemble Multi-Label Classifier for Human Protein Subcellular Location Prediction with Imbalanced Source. Proteomics. 17, (2017).
- Zhang, H., Zhu, L., Huang, D. S. WSMD: weakly-supervised motif discovery in transcription factor ChIP-seq data. Scientific Reports. 7, 3217 (2017).
- Szklarczyk, D., et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 43, D447-D452 (2015).
- Chiaretti, S., et al. Gene expression profile of adult T-cell acute lymphocytic leukemia identifies distinct subsets of patients with different response to therapy and survival. Blood. 103, 2771-2778 (2004).
- Rowley, J. D., et al. Mapping chromosome band 11q23 in human acute leukemia with biotinylated probes: identification of 11q23 translocation breakpoints with a yeast artificial chromosome. Proceedings of the National Academy of Sciences of the United States of America. 87, 9358-9362 (1990).
- Rabbitts, T. H., et al. The chromosomal location of T-cell receptor genes and a T cell rearranging gene: possible correlation with specific translocations in human T cell leukaemia. Embo Journal. 4, 1461-1465 (1985).
- Yin, L., et al. SH2D1A mutation analysis for diagnosis of XLP in typical and atypical patients. Human Genetics. 105, 501-505 (1999).
- Brandau, O., et al. Epstein-Barr virus-negative boys with non-Hodgkin lymphoma are mutated in the SH2D1A gene, as are patients with X-linked lymphoproliferative disease (XLP). Human Molecular Genetics. 8, 2407-2413 (1999).
- Burnett, R. C., Thirman, M. J., Rowley, J. D., Diaz, M. O. Molecular analysis of the T-cell acute lymphoblastic leukemia-associated t(1;7)(p34;q34) that fuses LCK and TCRB. Blood. 84, 1232-1236 (1994).
- Taylor, G. M., et al. Genetic susceptibility to childhood common acute lymphoblastic leukaemia is associated with polymorphic peptide-binding pocket profiles in HLA-DPB1*0201. Human Molecular Genetics. 11, 1585-1597 (2002).
- Wadia, P. P., et al. Antibodies specifically target AML antigen NuSAP1 after allogeneic bone marrow transplantation. Blood. 115, 2077-2087 (2010).
- Wilson, D. M., et al. 3rd et al. Hex1: a new human Rad2 nuclease family member with homology to yeast exonuclease 1. Nucleic Acids Research. 26, 3762-3768 (1998).
- O'Sullivan, R. J., et al. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nature Structural & Molecular Biology. 21, 167-174 (2014).
- Lee-Sherick, A. B., et al. Aberrant Mer receptor tyrosine kinase expression contributes to leukemogenesis in acute myeloid leukemia. Oncogene. 32, 5359-5368 (2013).
- Guyon, I., Elisseeff, A. An introduction to variable and feature selection. Journal of machine learning research. 3, 1157-1182 (2003).
- John, G. H., Kohavi, R., Pfleger, K. . Machine learning: proceedings of the eleventh international conference. , 121-129 (1994).
- Jain, A., Zongker, D. Feature selection: Evaluation, application, and small sample performance. IEEE transactions on pattern analysis and machine intelligence. 19, 153-158 (1997).
- Taylor, S. L., Kim, K. A jackknife and voting classifier approach to feature selection and classification. Cancer Informatics. 10, 133-147 (2011).
- Andresen, K., et al. Novel target genes and a valid biomarker panel identified for cholangiocarcinoma. Epigenetics. 7, 1249-1257 (2012).
- Guo, P., et al. Gene expression profile based classification models of psoriasis. Genomics. 103, 48-55 (2014).
- Xie, J., Wang, C. Using support vector machines with a novel hybrid feature selection method for diagnosis of erythemato-squamous diseases. Expert Systems with Applications. 38, 5809-5815 (2011).
- Zou, Q., Zeng, J., Cao, L., Ji, R. A novel features ranking metric with application to scalable visual and bioinformatics data classification. Neurocomputing. 173, 346-354 (2016).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone