Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Genetic Mapping of Thermotolerance Differences Between Species of Saccharomyces Yeast via Genome-Wide Reciprocal Hemizygosity Analysis
W tym Artykule
Podsumowanie
Reciprocal hemizygosity via sequencing (RH-seq) is a powerful new method to map the genetic basis of a trait difference between species. Pools of hemizygotes are generated by transposon mutagenesis and their fitness is tracked through competitive growth using high-throughout sequencing. Analysis of the resulting data pinpoints genes underlying the trait.
Streszczenie
A central goal of modern genetics is to understand how and why organisms in the wild differ in phenotype. To date, the field has advanced largely on the strength of linkage and association mapping methods, which trace the relationship between DNA sequence variants and phenotype across recombinant progeny from matings between individuals of a species. These approaches, although powerful, are not well suited to trait differences between reproductively isolated species. Here we describe a new method for genome-wide dissection of natural trait variation that can be readily applied to incompatible species. Our strategy, RH-seq, is a genome-wide implementation of the reciprocal hemizygote test. We harnessed it to identify the genes responsible for the striking high temperature growth of the yeast Saccharomyces cerevisiae relative to its sister species S. paradoxus. RH-seq utilizes transposon mutagenesis to create a pool of reciprocal hemizygotes, which are then tracked through a high-temperature competition via high-throughput sequencing. Our RH-seq workflow as laid out here provides a rigorous, unbiased way to dissect ancient, complex traits in the budding yeast clade, with the caveat that resource-intensive deep sequencing is needed to ensure genomic coverage for genetic mapping. As sequencing costs drop, this approach holds great promise for future use across eukaryotes.
Wprowadzenie
Since the dawn of the field, it has been a prime goal in genetics to understand the mechanistic basis of variation across wild individuals. As we map loci underlying a trait of interest, the emergent genes can be of immediate use as targets for diagnostics and drugs, and can shed light on the principles of evolution. The industry standard toward this end is to test for a relationship between genotype and phenotype across a population via linkage or association1. Powerful as these approaches are, they have one key limitation—they rely on large panels of recombinant progeny from crosses between interfertile individuals. They are of no use in the study of species that cannot mate to form progeny in the first place. As such, the field has had little capacity for unbiased dissection of trait differences between reproductively isolated species2.
In this work we report the technical underpinnings of a new method, RH-seq3, for genome-scale surveys of the genetic basis of trait variation between species. This approach is a massively parallel version of the reciprocal hemizygote test4,5, which was first conceived as a way to evaluate the phenotypic effects of allelic differences between two genetically distinct backgrounds at a particular locus (Figure 1A). In this scheme, the two divergent individuals are first mated to form a hybrid, half of whose genome comes from each of the respective parents. In this background, multiple strains are generated, each containing an interrupted or deleted copy of each parent’s allele of the locus. These strains are hemizygous since they remain diploid everywhere in the genome except at the locus of interest, where they are considered haploid, and are referred to as reciprocal since each lacks only one parent’s allele, with its remaining allele derived from the other parent. By comparing the phenotypes of these reciprocal hemizygote strains, one can conclude whether DNA sequence variants at the manipulated locus contribute to the trait of interest, since variants at the locus are the only genetic difference between the reciprocal hemizygote strains. In this way, it is possible to link genetic differences between species to a phenotypic difference between them in a well-controlled experimental setup. To date the applications of this test have been in a candidate-gene framework—that is, cases in which the hypothesis is already in hand that natural variation at a candidate locus might impact a trait.
In what follows, we lay out the protocol for a genome-scale reciprocal hemizygosity screen, using yeast as a model system. Our method creates a genomic complement of hemizygote mutants, by generating viable, sterile F1 hybrids between species and subjecting them to transposon mutagenesis. We pool the hemizygotes, measure their phenotypes in sequencing-based assays, and test for differences in frequency between clones of the pool bearing the two parents’ alleles of a given gene. The result is a catalog of loci at which variants between species influence the trait of interest. We implement the RH-seq workflow to elucidate the genetic basis of thermotolerance differences between two budding yeast species, Saccharomyces cerevisiae and S. paradoxus, which diverged ~5 million years ago6.
Protokół
1. Preparation of the piggyBac-containing plasmid for transformation
- Streak out to single colonies the E. coli strain harboring plasmid pJR487 onto an LB + carbenicillin agar plate. Incubate for 1 night at 37 °C or until single colonies appear.
NOTE: A description of how plasmid pJR487 was cloned can be found in our previous work3. - Inoculate 1 L of LB + carbenicillin at 100 μg/mL with a single colony of E. coli containing pJR487 in a 2 L glass flask. Grow overnight at 37 °C with shaking at 200 rpm until saturated (OD600 ≥ 1.0).
- Purify plasmid DNA from the culture using a large-scale plasmid prep kit as instructed in the manufacturer’s published protocol (see Table of Materials for details). Elute the DNA after a 10 minute incubation with 5 mL of elution buffer warmed to 37 °C.
- Measure the quantity and quality of plasmid DNA with a spectrophotometer (see Table of Materials for details).
- Repeat steps 1.2 – 1.4 until a total of at least 11 mg plasmid DNA at an A260:A280 ratio of at least 1.8 are isolated. This may take a few preps, depending on efficiency.
- Mix all plasmid preps together into a single tube and bring the total volume up to 20 mL with elution buffer or water. Measure the final quantity and quality again with a spectrophotometer. The concentration of plasmid should be at least 538 ng/μL in this final 20 mL volume. If the concentration is higher than 538 ng/μL, dilute the plasmid with elution buffer or water to 538 ng/μL. Plasmid can be stored at 4 °C up to a few weeks until use.
2. Creating a pool of untargeted genome-wide reciprocal hemizygotes
- Preparation of hybrid yeast cells for transformation
- Streak out JR507 from a -80 °C freezer stock strain to single colonies on a YPD agar plate. Incubate at 26 °C for 2 days or until colonies appear.
NOTE: JR507 is a hybrid strain made through single-cell mating of haploid spores of S. cerevisiae DBVPG1373 and S. paradoxus Z1 (using a tetrad-dissection microscope)3. - Inoculate 100 mL of liquid YPD in a 250 mL glass flask with a single colony of JR507 and shake at 28 °C, 200 rpm for 24 hours, or until stationary phase is reached.
- The next day, measure the optical density at 600 nm (OD600) of the overnight culture. Create a new culture by back-diluting some of the overnight culture with fresh liquid YPD into a new 1 L glass flask to an OD600 of 0.2 and a volume of 500 mL.
NOTE: Example calculation of a back-dilution if the overnight culture has an OD600 of 5.0, where C is optical density and V is volume:
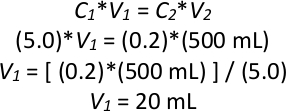
Thus, 20 mL of saturated overnight culture would be added to 480 mL of liquid YPD to make a total of 500 mL of culture at an OD600 of 0.2. - Repeat step 2.1.3 three more times to make a total of four 500 mL cultures at an OD600 of 0.2 in four 1 L glass flasks, using the same overnight culture for all four new cultures. Incubate them all at 28 °C for 6 hours (2-3 generations) shaking at 200 rpm.
- Combine two of the 500 mL cultures to create a 1 L culture. Combine the remaining two 500 mL cultures to create another 1 L culture. At this point, there are two 1 L cultures. Each of these 1 L cultures will be subject to transformation with pJR487 in the following steps.
- Streak out JR507 from a -80 °C freezer stock strain to single colonies on a YPD agar plate. Incubate at 26 °C for 2 days or until colonies appear.
- Transformation of pJR487 into hybrid yeast cells
- Split each of the 1 L cultures into twenty 50 mL aliquots in 20 plastic conical tubes for a total of 40 tubes. Set aside 20 tubes and perform the following steps on 20 tubes at a time.
- Centrifuge each of the twenty tubes for 3 min at 1,000 x g to pellet the yeast cells. Discard the supernatant.
- Resuspend each pellet with 25 mL of sterile H2O by vortexing. Centrifuge for 3 min at 1,000 x g. Discard the supernatant.
- Resuspend each pellet with 5 mL of 1x TE, 0.1 M LiOAc buffer by vortexing. Centrifuge for 3 min at 1,000 x g. Discard the supernatant.
- Repeat step 2.2.4. While the cells are centrifuging, prepare at least 120 mL of a solution of 39.52% polyethylene glycol, 0.12 M LiOAc and 1.2x Tris-EDTA buffer (12 mM Tris-HCl and 1.2 mM EDTA). Store on ice.
- To prepare the plasmid DNA for transformation, first boil 4 mL of salmon sperm DNA at 100 °C for 5 min and immediately cool it on ice for 5 min. Then, mix 20 mL of pJR487 (obtained in section 1) at a concentration of 538 ng/μL with the 4 mL of cooled salmon sperm DNA for a total volume of 24 mL. Keep on ice until use.
- Add 600 μL of plasmid DNA mixed with salmon sperm DNA on top of each cell pellet. Do not resuspend yet.
- Add 3 mL of PEG-LiOAc-TE solution made in step 2.2.5 to each pellet. Resuspend the pellet by pipetting up and down and vortexing.
- Incubate each tube for 10 min at room temperature.
- Heat shock each tube for 26 min in a water bath set to 39 °C.
NOTE: Every few minutes, invert each tube to prevent the cells from settling on the bottom of the tube. - Centrifuge each tube for 3 min at 1,000 x g. Discard the supernatant and resuspend each pellet in 10 mL of YPD by vortexing. Combine all twenty tubes into a new glass flask. The total volume of cells should be ~200 mL.
- Transfer 66.6 mL of cells to a new 1 L glass flask and bring up to a volume of 500 mL with liquid YPD. Repeat two more times to use the entire 200 mL of transformed cells. Measure the OD600 of each new 500 mL culture (expect an OD600 of ~0.35-4).
- Shake all three flasks at 28 °C for 2 hours to recover (<1 generation) at 200 rpm.
- Add 0.5 mL of 300 mg/mL G418 to each of the three flasks, to a final concentration of 300 μg/mL G418 and put back to shake at 28 °C, 200 rpm.
NOTE: Prior to this step, the transformed hybrid cells have been recovering from transformation. Upon the addition of G418, presence of the plasmid pJR487 is selected for. Any cells that did not take up the plasmid during transformation will begin to die. - Repeat steps 2.2.2 – 2.2.14 with the remaining 20 conical tubes of cells. At this point there should be six 1 L glass flasks, each with 500 mL of cells with G418 added.
- Incubate all six flasks of cells at 28 °C, shaking at 200 rpm, for approximately 2 days or until an OD600 of ~2.3 is reached in each flask. Combine all six flasks together to create a single culture.
NOTE: Although all of the cells in this culture will not be used in downstream steps, the goal of using such large volumes has been to create as many unique transformation events as possible and normalize any biases across a single transformation by pooling them all together. - Use the culture created in 2.2.16 to inoculate two new 1 L flasks with 500 mL of YPD + G418 (300 μg/mL) to an OD600 of 0.2. There will be leftover culture that can be discarded.
- Incubate both 1 L flasks at 28 °C overnight, with shaking at 200 rpm, until each reaches an OD600 of ~2.2 (~3.5 generations). Combine both cultures into a single culture and measure the OD600 of the combined culture again.
NOTE: At this point, the culture should be almost entirely comprised of cells harboring plasmid pJR487. In part of the population of cells, the PiggyBac transposon will have been transposed from the plasmid into the genome by the transposase expressed off the plasmid. However, continued expression of the transposase can lead to transposition during the course of a selection, which would obscure the relationship between genotype and phenotype. The goal of the next several steps is to perform a counterselection against the presence of the plasmid, to ensure there is no more expression of the transposase. The resulting pool is a mix of cells with or without the transposon integrated into the genome, but only cells containing the transposon are detected during the subsequent mapping steps. The time in the transformation during which transposase is expressed, before the plasmid encoding is lost, may govern the chance that a given clone after mutagenesis harbors more than one transposon insertion. The frequency of these, which manifest as “secondary” mutations in analysis of any one gene at a time, can be estimated by arraying a defined number of colonies after mutagenesis, then combining their DNA and sequence-confirming the number of independent insertion positions in the pool. - Centrifuge 25 mL of this culture for 3 min at 1,000 x g. Calculate the number of total OD600 units of cells that are in the 25 mL (see example calculation below). Discard the supernatant and resuspend in enough H2O to create a cell suspension of 1.85 OD600/mL by vortexing.
NOTE: Example calculation for resuspension of cells in water if OD600 of combined culture was 2.2:
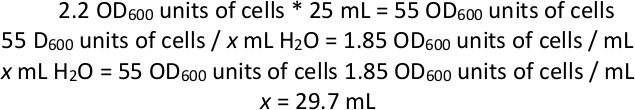
So, after spinning 25 mL of cell culture and discarding the supernatant, add enough H2O to the cells to bring the total volume of cells and water up to ~29.7 mL (since the cell pellet will also have a volume, add less than 29.7 mL of H2O). - Using glass beads, plate 1 mL of resuspended cells in water onto each of 12 large square complete synthetic agar plates with 5-FOA. Incubate each plate at 28 °C for 1-2 days or until a lawn forms on the plate.
- Using small sterile squeegees, scrape the cells off of each of 6 plates and into a tube with 35 mL of sterile water. Repeat with the other 6 plates for a total of two tubes of cells and water. Combine all cell suspensions in a single tube. Measure the OD600 of this suspension, using water as a blank. Bring the OD600/mL concentration of cells to 44.4 OD600 units/mL with water. In our experience, transposition efficiency (the proportion of KAN+ cells that are URA-) is on average 50%.
- Determine the number of -80 °C freezer stocks of cells to store. Each aliquot can be used in the future for a single experiment.
NOTE: Given how time consuming the generation of the pool is, store multiple vials in case of accidental misuse or for performing replicate experiments. 20-30 stocks are a reasonable number. - Each freezer stock will contain 40 OD600 units of cells in 1 mL of 10% DMSO. Add 900 μL of cells to 100 μL of DMSO. Repeat for the total number of freezer stocks created. Store each at -80 °C for future use.
3. Selection of reciprocal hemizygotes in a pooled format
- Thaw from the -80 °C freezer a single aliquot of pooled reciprocal hemizygotes from section 2 at room temperature.
NOTE: Do not let the aliquot sit for long at room temperature once it thaws, use it immediately. - Use the entire 1 mL aliquot to inoculate 150 mL of liquid YPD in a 250 mL glass flask. Measure the OD600 of this culture, and then incubate at 28 °C, shaking at 200 rpm, for ~7 hours, or until the culture has gone through 2-3 population doublings. At this point, the culture is ready to be used to inoculate cultures undergoing selection.
NOTE: Example calculation: If the OD600 of the original flask measures 0.25, incubate the culture until it reaches an OD600 of at least 1.0. If any sample points are desired at “Time-zero” (T-0), as a way to investigate the hemizygote population before selection, cell pellets can be taken now by centrifuging 5-10 mL of culture per pellet at 1,000 x g for 3 min, discarding the supernatant and freezing at -80 °C. - Use the grown hemizygote pool to inoculate cultures for selection in a suitable replicate scheme, at both high temperature (39 °C) and permissive temperature (28 °C). At a minimum, set up three biological replicate selection cultures at each temperature, for a total of six selection cultures.
- Create each selection culture with 500 mL total in a 2 L glass flask with liquid YPD and inoculate to an OD600 of 0.02. Shake each selection culture at 100 rpm at either 28 °C or 39 °C until 6-7 population doublings have occurred (corresponding to an OD600 of ~1.28-2.56). Try to match as closely as possible the final OD600 of all selection cultures.
NOTE: Selection cultures at 28 °C will grow faster than selection cultures at 39 °C. Consequently, selection cultures at 39 °C will spend a longer period of time in the incubator. Proceed with the following steps with each flask as it becomes ready, regardless of the total number of hours spent in the incubator. In our experience, cultures at 28 °C or 39 °C took ~12 or ~18 hours, respectively, to reach an OD of ~2.0. Long selections could have the advantage of amplifying small fitness effects, but also permit de novo background mutations to arise, which would introduce noise into the final distribution of fitnesses across transposon mutants in any one gene/allele. As such it is important to limit selection time in an RH-seq experiment.
- Create each selection culture with 500 mL total in a 2 L glass flask with liquid YPD and inoculate to an OD600 of 0.02. Shake each selection culture at 100 rpm at either 28 °C or 39 °C until 6-7 population doublings have occurred (corresponding to an OD600 of ~1.28-2.56). Try to match as closely as possible the final OD600 of all selection cultures.
- Harvest cell pellets from each selection culture. Calculate the volume required to obtain 7 OD600 units of cells and centrifuge at 1,000 x g for 3 min at least four pellets of this volume from each selection culture as technical replicates for library preparation and sequencing (see sections 4 and 5, below). Discard the supernatant and store at -80 °C.
NOTE: Example if a selection flask has a final OD600 of 2.0:
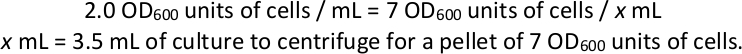
4. Tn-seq library construction and Illumina sequencing to determine abundance of transposon mutant hemizygotes
- Thaw on ice each cell pellet from section 3 that is going to be sequenced.
- Isolate total genomic DNA (gDNA) from each cell pellet using a yeast gDNA purification kit following the manufacturer’s instructions. Resuspend the DNA in 50 μL of elution buffer warmed to 65 °C.
- Quantify the quantity of gDNA from each pellet using a fluorimeter. The minimum total quantity of gDNA required for each cell pellet to create a next-generation sequencing (NGS) library for Tn-seq using the following procedure is 1 μg.
NOTE: Less than 1 μg of gDNA can be used to create a library, but the final quantity and quality of the library will suffer. - Follow an established protocol for creating Tn-seq libraries7. Note the following relevant information that is unique to this protocol:
- After gDNA shearing, end repair and adapter ligation, amplify the gDNA containing the transposon via PCR. For that PCR, use the following forward and reverse primer, which are specific for the PiggyBac transposon and NGS adapters, respectively:
Forward (N – random nucleotide)
5’ ATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACG
CTCTTCCGATCTNNNNNNAGCAATATTTCAAGAATGCATGCGTCAAT 3’
Reverse (the stretch of Ns represents a unique 6-bp index used for multiplexing. See below for further information on indices)
5’ CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCAG
ACGTGTGCTCTTCCGATCT 3’ - Use the included cleanup steps with size-selective beads to minimize the proportion of cloned fragments in the final library that would be too short to include mappable genomic sequence.
NOTE: Having followed the minimum replicate requirements up until now for selection cultures, there will be 24 individual gDNA samples for sequencing. Given the current cost for sequencing, it is unlikely that each sample will be run on its own. To combine samples on the same lane, create multiple reverse primers, each with a unique 6-base pair index. Samples with differing indexes can be combined into the same sequencing lane and separated computationally afterwards.
- After gDNA shearing, end repair and adapter ligation, amplify the gDNA containing the transposon via PCR. For that PCR, use the following forward and reverse primer, which are specific for the PiggyBac transposon and NGS adapters, respectively:
- Sequence single-end 150 bp reads from each library using NGS technologies across eight lanes.
NOTE: The amount of sequencing reads required depends heavily on the quality of the libraries prepared in the previous step (i.e. the proportion of DNA in the library actually containing transposon DNA, representing DNA coming from reciprocal hemizygotes). There are two main factors contributing to this. First, since cells without an integrated transposon are not counterselected against during pool creation, each culture will be a mix of cells with and without the transposon. Secondly, even within the genomes of transposon-containing reciprocal hemizygotes, most of the genome is not transposon containing sequence, and this gDNA will unavoidably be part of the library preparation. The goal of the final PCR amplification of transposon-containing DNA is to increase the ratio of transposon-containing DNA to these two sources of background gDNA. The more efficient this amplification is, the higher proportion of reads will be able to be used in downstream analysis. The lower quality the libraries are, the more sequencing will need to be done, since an increasing proportion of reads will not contain transposon DNA and will not be useful. Given the above constraints, eight lanes of sequencing were capable of tracking reciprocal hemizygote abundances to a reasonable degree. More sequencing would allow a deeper analysis.
5. Mapping the locations of transposon insertions and RH-seq analysis
NOTE: The following data analysis was accomplished with custom Python scripts (found online at https://github.com/weiss19/rh-seq), but could be redone using other scripting languages. Below, the major steps in the process are outlined. Perform the following steps on each individual replicate read file unless it is noted to combine them.
- Trim adapter sequences off of reads and separate out each replicate’s reads according to index.
- Find reads containing transposon-genome junctions. To accomplish this, search within each read for the last 20 base pairs of the transposon, CAGACTATCTTTCTAGGGTTAA. Discard all reads not containing this sequence.
NOTE: In our experience, the proportion of reads mapping to the end of the transposon is 83-95%. - Trim the remaining, transposon-containing reads to contain only the sequence downstream of the 3’ end of the transposon. By mapping this sequence to the yeast genome, determine the genomic context of the transposon insertion for each read (step 5.4 below).
- Use BLAT or an equivalent mapping tool to map the sequence downstream of the transposon to the S. cerevisiae DBVPG1373 x S. paradoxus Z1 hybrid genome (Script name: map_and_pool_BLAT.py).
- Discard any reads for which there are fewer than 50 base pairs of usable sequence downstream of 3’ end of the transposon. Short sequences are difficult to map uniquely.
- If using BLAT, use the following parameters: identity = 95, tile size = 12.
- Create a basic hybrid genome to use for mapping by concatenating the latest versions of reference genomes of S. cerevisiae S288c and S. paradoxus CBS432.
NOTE: A basic annotation file describing the genomic boundaries of individual genes across the hybrid genome can be found at the Github repository listed above (Filename: YS2+CBS432+plasmid_clean). Only use reads which map to a single location in the hybrid genome (i.e. are unique to either S. cerevisiae or S. paradoxus). A uniform frequency of insertion events across the genome is expected; the distribution of insertion positions across the genomes is reported elsewhere3.
- Tally the total number of reads mapping to each unique transposon insertion location, which we infer all originated from cells of a single transposon insertion mutant clone. The sum of all such values from a single library is referred to as the total number of mapped reads for that library.
- In cases where there are multiple insertions mapping within 3 base pairs of one another, combine them all to a single insertion point, assigning all the reads to the single location with the highest read count. This value, ninsert, represents the abundance of that insertion clone in the cell pellet from which gDNA was sequenced. At this point, there will be lists of ninsert, each the abundance of a unique mapped transposon insertion, one list for every cell pellet sequenced.
NOTE: The PiggyBac transposon inserts at TTAA sequences in the genome, a 4 base pair sequence. Thus, we infer that insertions mapping within 3 base pairs of each other must have originated from the same TTAA site. - Since there will be a slightly different number of total reads coming from each sequenced library, normalize the ninsert values across all files if they are to be compared. Do so by tabulating the total number of mapped reads from each individual library, npellet, and take the average of all npellet across all libraries, <npellet>. Multiply each ninsert in an individual library’s data by the ratio of <npellet> / npellet to calculate ainsert, the normalized abundance of a given transposon insertion clone.
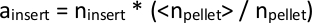
Alternatively, library size can be estimated using available tools like DESeq28 (Script name: total_reads_and_normalize.py). - Tabulate the set of all insertions mapped across all libraries. For insertions found in some libraries but not in others, set ainsert = 1 for downstream calculations.
- Filter the reads to find those insertions that fall within genes according to the annotation file (Script name: remove_NC_and_plasmid_inserts.py).
- For each unique insertion, calculate the average abundance across technical replicates of each selection (each culture at either 28 °C or 39 °C), <ainsert>technical (Script name: combine_tech_reps_V2.py).
- For each unique insertion, calculate the average abundance across biological replicates of each temperature, <ainsert>total, by taking the mean of all <ainsert>technical at each temperature. At the same time, calculate the coefficient of variation for each insertion, CVinsert,total across the <ainsert>technical (Script name: combine_bio_reps.py).
NOTE: At this point, for each temperature, 28 °C and 39 °C, there is a list of unique transposon insertions, their average abundance and the coefficient of variation between biological replicates for each. These data for our experiment are reported elsewhere3. - Filter the list of all insertions for those that have, at either 28 °C or 39 °C, <ainsert>total > 1.1, and CVinsert,total ≤ 1.5 (Script name: filter_inserts.py).
- For each unique insertion, calculate the log2(<ainsert>total,28 °C / <ainsert>total,39 °C). This value represents the “thermotolerance” of a given transposon insertion mutant clone (Script name: fitness_ratios.py).
- Sort all of the unique insertions by gene and by allele (S. cerevisiae or S. paradoxus), and tabulate the number of insertions in each allele. Filter genes so that only genes that have at least 5 insertions in each allele are analyzed (Script name: organize_and_filter_genes.py).
NOTE: Multiple unique insertions across each allele allow for a more accurate measure of that reciprocal hemizygote’s thermotolerance. Lowering the number of insertions required per allele is possible but will compromise the accuracy of this measure and increase the multiple testing burden by allowing more genes to be tested. Additionally, filtering out genes with too few insertions per allele will help reduce the impact on test results of any individual hemizygote clone harboring a secondary site mutation that confers a very disparate phenotype. - For each gene remaining in the data set after the above filtering, compare the thermotolerances (log2 ratios) of all the insertions in the S. cerevisiae allele to those in the S. paradoxus allele using a Mann-Whitney U test. Alternatively, a regression model could be implemented, adapted from DESeq28 (Script name: mann_whitney_u.py).
- Correct p-values for multiple testing using the Benjamini-Hochberg method.
- Genes with significant p-values (say, ≤ 0.01) are candidates for genes important for differences in thermotolerance between the two species.
Wyniki
We mated S. cerevisiae and S. paradoxus to form a sterile hybrid, which we subjected to transposon mutagenesis. Each mutagenized clone was a hemizygote, a diploid hybrid in which one allele of one gene is disrupted (Figure 1A, Figure 2). We competed the hemizygotes against one another by growth at 39 °C and, in a separate experiment as a control, at 28 °C (Figure 1B), and we isolated DNA ...
Dyskusje
The advantages of RH-seq over previous statistical-genetic methods are several-fold. In contrast to linkage and association analysis, RH-seq affords single-gene mapping resolution; as such, it will likely be of significant utility even in studies of trait variation across individuals of a given species, as well as interspecific differences. Also, previous attempts at genome-wide reciprocal hemizygosity analysis used collections of gene deletion mutants, some of which harbor secondary mutations that can lead to false posi...
Ujawnienia
The authors have nothing to disclose.
Podziękowania
We thank J. Roop, R. Hackley, I. Grigoriev, A. Arkin and J. Skerker for their contributions to the original study, F. AlZaben, A. Flury, G. Geiselman, J. Hong, J. Kim, M. Maurer, and L. Oltrogge for technical assistance, D. Savage for his generosity with microscopy resources, and B. Blackman, S. Coradetti, A. Flamholz, V. Guacci, D. Koshland, C. Nelson, and A. Sasikumar for discussions; we also thank J. Dueber (Department of Bioengineering, UC Berkeley) for the PiggyBac plasmid. This work was supported by R01 GM120430-A1 and by Community Sequencing Project 1460 to RBB at the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility. The work conducted by the latter was supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Materiały
| Name | Company | Catalog Number | Comments |
| 1-2 plasmid Gigaprep kits | Zymo Research | D4204 | The number of kits required depends on how efficient your preps are in each kit. This kit comes with 5 individual plasmid prep columns. Run 1 L of saturated E. coli culture through each prep column, as using more than 1 L per column can cause clogging of the prep filter, leading to low yield and poor quality DNA. |
| 10X Tris-EDTA (TE) buffer (100 mM Tris-HCl and 10 mM EDTA) | Any | N/A | Filter sterilize through a 0.22 μm filter before use. |
| 1M LiOAc | Any | N/A | Filter sterilize through a 0.22 μm filter before use. |
| 300 mg/mL Geneticin (G418) | Gibco | 11811023 | |
| 52% polyethylene glycol (PEG) 3350 | Sigma | 1546547 | Dissolve in water and filter sterilize through a 0.22 μm filter before use. 1X trafo mix: 228 uL 52% PEG, 36 uL 1M LiOAc, 36 uL 10X TE buffer |
| Autoclaved LB liquid broth | BD Difco | 244620 | Make LB liquid broth using your powder from any brand, and milliQ water. Autoclave it before use. |
| Carbenicillin stock in water (100 mg/mL) | Any | N/A | Filter sterilize through a 0.22 μm filter before use. |
| Complete synthetic agar plates (24.1cm x 24.1cm) with 5-fluoroorotic acid (5-FOA) [0.2% drop-out amino acid mix without uracil or yeast nitrogen base (YNB), 0.005% uracil , 2% D-glucose, 0.67% YNB without amino acids, 0.075% 5-FOA] | 5-FOA: Zymo Research, Drop-out mix: US Biological, Uracil: Sigma, D-glucose: Sigm), YNB: Difco | 5-FOA: F9001-5, Drop-out mix: D9535, Uracil: U0750, D-glucose: G8270, YNB: DF0919 | |
| DMSO | Any | N/A | |
| E. coli strain carrying pJR487 (CEN-/ARS+ piggyBac-containing plasmid) | N/A | N/A | Request from Brem lab. |
| Hybrid yeast strain JR507 (S. cerevisiae DBVPG1373 x S. paradoxus Z1, URA-/URA-) | N/A | N/A | Request from Brem lab. |
| Illumina Hiseq 2500 | used for SE-150 reads | ||
| Large shaking incubators with variable temperature settings | Any | N/A | |
| LB + carbenicillin agar plates (100 μg/mL) | Agar: BD Difco | Agar: 214010 | Make LB agar plates as normal and add carbenicillin to 100 μg/mL before drying. |
| Nanodrop spectrophotometer | Thermo Scientific | ND-2000 | |
| Qubit Fluorimeter | Thermo Scientific | Q33240 | |
| Salmon sperm DNA | Invitrogen | 15632011 | |
| Water bath at 39°C | Any | N/A | |
| Yeast fungal gDNA prep kit | Zymo Research | D6005 | |
| Yeast peptone dextrose (YPD) liquid media | BD Difco | Peptone: 211677, Yeast Extract: 212750 | Add filter-sterilized D-glucose to 2% after autoclaving. |
| YPD + G418 agar plates (300 μg/mL) | Agar: BD Difco | Agar: 214010 | Make YPD agar plates as normal and add G418 to 300 μg/mL before drying. |
| YPD agar plates | Agar: BD Difco | Agar: 214010 |
Odniesienia
- Flint, J., Mott, R. Finding the molecular basis of quantitative traits: successes and pitfalls. Nature Reviews Genetics. 2, 437-445 (2001).
- Allen Orr, H. The genetics of species differences. Trends in Ecology and Evolution. 16, 343-350 (2001).
- Weiss, C. V., et al. Genetic dissection of interspecific differences in yeast thermotolerance. Nature Genetics. 50, 1501-1504 (2018).
- Stern, D. L. Identification of loci that cause phenotypic variation in diverse species with the reciprocal hemizygosity test. Trends in Genetics. 30, 547-554 (2014).
- Steinmetz, L. M., et al. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 416, 326-330 (2002).
- Scannell, D. R., et al. The Awesome Power of Yeast Evolutionary Genetics: New Genome Sequences and Strain Resources for the Saccharomyces sensu stricto Genus. G3 (Bethesda). 1, 11-25 (2011).
- Wetmore, K. M., et al. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio. 6, e00306-e00315 (2015).
- Love, M. I., Huber, W., Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 15, 550 (2014).
- Wilkening, S., et al. An evaluation of high-throughput approaches to QTL mapping in Saccharomyces cerevisiae. Genetics. 196, 853-865 (2014).
- Kim, H. S., Huh, J., Riles, L., Reyes, A., Fay, J. C. A noncomplementation screen for quantitative trait alleles in saccharomyces cerevisiae. G3 (Bethesda). 2, 753-760 (2012).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone