Aby wyświetlić tę treść, wymagana jest subskrypcja JoVE. Zaloguj się lub rozpocznij bezpłatny okres próbny.
Method Article
Isolation and Selection of Entomopathogenic Fungi from Soil Samples and Evaluation of Fungal Virulence against Insect Pests
W tym Artykule
Podsumowanie
Here we present a protocol based on the mealworm (Tenebrio molitor)-bait system that was used for isolating and selecting entomopathogenic fungi (EPF) from soil samples. An effective conidia number (ECN) formula is used to select high stress tolerant EPF based on physiological characteristics for pest microbial control in the field.
Streszczenie
Entomopathogenic fungi (EPF) are one of the microbial control agents for integrated pest management. To control local or invasive pests, it is important to isolate and select indigenous EPF. Therefore, the soil bait method combined with the insect bait (mealworm, Tenebrio molitor) system was used in this study with some modifications. The isolated EPF were then subjected to the virulence test against the agricultural pest Spodoptera litura. Furthermore, the potential EPF strains were subjected to morphological and molecular identifications. In addition, the conidia production and thermotolerance assay were performed for the promising EPF strains and compared; these data were further substituted into the formula of effective conidia number (ECN) for laboratory ranking. The soil bait-mealworm system and the ECN formula can be improved by replacing insect species and integrating more stress factors for the evaluation of commercialization and field application. This protocol provides a quick and efficient approach for EPF selection and will improve the research on biological control agents.
Wprowadzenie
Currently, entomopathogenic fungi (EPF) are widely used in the microbial control of agricultural, forest, and horticultural pests. The advantages of EPF are its wide host ranges, good environmental adaptability, ecofriendly nature, and that it can be used with other chemicals to show the synergistic effect for integrated pest management1,2. For the application as a pest control agent, it is necessary to isolate a large number of EPF from either diseased insects or the natural environment.
The sampling of these organisms from their hosts helps in understanding the geographic distribution and prevalence rate of EPF in natural hosts3,4,5. However, the collection of fungal infected insects are usually limited by environmental factors and insect populations in the field4. Considering that insect hosts will die after EPF infection and then fall into the soil, isolation of EPF from soil samples might be a stable resource3,6. For example, saprophytes are known to use the dead host as their resource for growth. The soil bait and selective medium systems have been widely used to detect and isolate EPF from the soil3,4,7,8,9,10.
In the selective medium method, the diluted soil solution is plated onto a medium containing broad-spectrum antibiotics (e.g., chloramphenicol, tetracycline, or streptomycin) to inhibit the growth of bacteria2,3,9,11. However, it has been reported that this method may distort the strain's diversity and density and can cause an over- or under-estimation of many microbial communities6. Moreover, the isolated strains are less pathogenic and compete with saprophytes during isolation. It is difficult to isolate EPF from the diluted soil solution3. Instead of using a selective medium, the soil bait method isolates EPF from the infected dead insects, which can be stored for 2-3 weeks, thereby providing a more efficient and standard EPF separation method3,4,7,6. Because the method is easy to operate, one can isolate a variety of pathogenic strains at a low cost4. Therefore, it is widely used by many researchers.
Upon comparing the different types of insect bait systems, Beauveria bassiana and Metarhizium anisopliae are the most common EPF species that are found in insects belonging to the Hemiptera, Lepidoptera, Blattella, and Coleoptera6,12,13,14. Among these insect baits, Galleria mellonella (order Lepidoptera) and Tenebrio molitor (order Coleoptera) show higher recovery rates of Beauveria and Metarhizium spp., when compared with other insects. Therefore, G. mellonella and T. molitor are commonly used for insect baiting. Over the years, the United States Department of Agriculture (USDA) has established an EPF Library (Agricultural Research Service Collection of EPF cultures, ARSEF) that contains a wide variety of species, including 4081 species of Beauveria spp., 18 species of Clonostachys spp., 878 species of Cordyceps spp., 2473 species of Metarhizium spp., 226 species of Purpureocillium spp., and 13 species of Pochonia spp. among others15. Another EPF Library was constructed by the Entomology Research Laboratory (ERL) from the University of Vermont in the United States for c.a. 30 years. It includes 1345 strains of EPF from the United States, Europe, Asia, Africa, and the Middle East16.
To control local or invasion pests in Taiwan, isolation and selection of indigenous EPF is required. Therefore, in this protocol, we have modified and described the procedure of the soil bait method and combined it with the insect bait (mealworm, Tenebrio molitor) system17. Based on this protocol, an EPF library was established. Two rounds of screening (quantification of inoculation) were performed for the preliminary EPF isolates. EPF isolates showed pathogenicity to insects. The potential strains were subjected to morphological and molecular identifications and further analyzed by the thermotolerance and conidial production assay. Further, a concept of effective conidia number (ECN) was also proposed. Using ECN formula and principal component analysis (PCA), the potential strains were analyzed under simulated environmental pressure to complete the process of establishing and screening the EPF library. Subsequently, pathogenicity of promising EPF strains were tested for the target pest (e.g., Spodoptera litura). The current protocol integrates thermotolerance and conidial production data into the ECN formula and PCA analysis, which can be used as a standard ranking system for EPF related research.
Protokół
NOTE: The whole flowchart is shown in Figure 1.
1. Isolation and selection of potential Entomopathogenic fungi (EPF)
- Collect the soil sample
- Remove 1 cm of the surface soil, and then collect the soil within the 5-10 cm depth using a shovel from each sampling site.
NOTE:Sampling sites would be a mountain, forest, or sparsely populated areas to avoid the contamination of artificially sprayed EPF strains. Ensure that areas for the soil samples collection are covered with weed on the surface. Dry soil or damp soil is not suitable for this experiment. - Record the details of each sampling site, including GPS, elevation, type of field, annual temperature, yearly precipitation, collection time (season), soil type, and pH value.
- Collect 100 g of the soil sample into a plastic bag and maintain it at room temperature if subjecting it to the fungal isolation protocol in the laboratory within 3 h.
NOTE: If the sample cannot be used within 3 h, store the soil at room temperature in dark conditions. If the experiment is not performed immediately, the soil sample can be stored at 4 °C for 1 week until the start of the protocol18,19.
- Remove 1 cm of the surface soil, and then collect the soil within the 5-10 cm depth using a shovel from each sampling site.
- Bait and isolate the EPF with mealworm (Tenebrio molitor)
- Place 100 g of the soil sample in a plastic cup (cap diameter = 8.5 cm, height = 12.5 cm), and then place 5 mealworms on the surface of the soil at room temperature in the dark for 2 weeks.
NOTE: Other types of plastic cup containers can also be used. If soils are too dry (cracked or sandy), spray sterilized ddH2O (about 5-10 mL) on the soils. The body length c.a. 2.5 cm (14th instar) of T. molitor larvae helps in fungal isolate screens20. - Observe and record the larvae daily for mortality and mycosis; keep the dead larvae in the cup until 2 weeks for fungal isolation.
NOTE: The fungal conidia spore in the soil samples will attach to the mealworm larvae during the above process. The fungal mycosis will be observed as the hyphae grow from the intersegmental membrane, and then the whole body will be covered with the mycelium. Sporulation will start after 7 days and the color of the fungal infection will change to the color of the conidia mass. - Transfer the dead insects to a clean bench and use a sterile toothpick to collect the conidia. Streak them on a quarter strength Sabouraud dextrose agar medium (¼ SDA) plate (55 mm) in the laboratory21. Incubate the culture plate at 25 °C for 7 days to obtain the primary culture of fungi.
NOTE: The ¼ SDA plate is prepared as follows: Mix 1.5 g of Sabouraud dextrose broth and 3 g of agar in 200 mL of H2O, and then sterilize for 20 min. Aliquot ¼ SDA into each 55 mm Petri dish before solidification. The solidified ¼ SDA plates are stored at 4 °C until use. - Re-streak each primary culture fungi on one 55 mm ¼ SDA plate in a laminar flow and incubate the culture plate at 25 °C for 7 days to obtain single colonies of fungi.
- Repeat this re-isolation ~2-3 times and observe under light microscopy to obtain single and pure morphological fungal colonies.
NOTE: Isolate all EPF using T. molitor-bait and store as described in the following section.The separation, preservation, pure culture, and streaking must be performed in a laminar flow in the subsequent sections.
- Place 100 g of the soil sample in a plastic cup (cap diameter = 8.5 cm, height = 12.5 cm), and then place 5 mealworms on the surface of the soil at room temperature in the dark for 2 weeks.
- Store the EPF isolates
- Cut 3 of the 5 mm agar blocks at the edge of each isolated pure fungal cultured plate with a cork borer and place it into a 1.5 mL micro-centrifuge tube as one replicate.
NOTE: Three replicates for each fungal isolate are recommended for the EPF strains' storage after 2nd virulence test. Molecular identification is also recommended if storage space is limited. - Add 250 µL of 0.03% surfactant solution (Table of Materials) and 250 µL of 60% glycerol into the 1.5 mL micro-centrifuge tube using a micropipette; then, vortex for 10 s.
- Seal the 1.5 mL micro-centrifuge tube with paraffin film and precool it in a -20 °C refrigerator for 24 h. Then, transfer the precooled fungal stocks to a -80 °C refrigerator for cryopreservation.
NOTE: Extra pure fungal culture plates (aside from the cryo-preserved samples) were used in section 1.4.
- Cut 3 of the 5 mm agar blocks at the edge of each isolated pure fungal cultured plate with a cork borer and place it into a 1.5 mL micro-centrifuge tube as one replicate.
- 1st pathogenicity screen for fungal isolates
- Place five T. molitor larvae directly on the surface of each pure fungal culture plate at 25 °C.
- Observe and record the mycosis and mortality for 10 days. Select the fungal isolate for further analysis.
NOTE: Fungal isolates causing 100% mortality are selected for 2nd virulence test to confirm their virulence to T. molitor larvae. Alternatively, the researcher can adjust the criteria as per their own study.
- 2nd virulence test of fungal isolates
NOTE: Based on the 1st pathogenicity screen, recover the selected fungal isolates from -80 °C for the 2nd virulence test. The purpose of the 2nd virulence test is to quantify the pathogenicity of the selected fungal isolates after the 1st round of screening.- Harvest conidia of each fungal isolate by vortexing for 1 min and count the number of conidia using a hemocytometer.
- Adjust the conidia suspension to a concentration of 1 x 107 conidia/mL in a 0.03% surfactant solution (Table of Materials).
- Spread 10 µL of the fungal suspension onto 55 mm ¼ SDA plates and grow for 7 days at 25 °C in the dark.
- Place five T. molitor larvae directly on the surface of each pure fungal culture plate (c.a. 6 x 107 conidia). Seal the plates with paraffin film and incubate at 25 °C in the dark.
- Observe and record the mycosis and mortality for 10 days.
- Repeat the test (from step 1.51 to 1.5.5) in triplicate for each fungal isolate.
NOTE: Fungal isolates causing 100% mortality are selected for 3rd virulence test to confirm their virulence to target the pest.
- 3rd virulence test of fungal isolates for target pest (Spodoptera litura as an example)
- Repeat steps 1.5.2 to 1.5.6 with selected isolates from 2nd virulence to test virulence against target pest.
- Calculate the LT50 of each fungal isolate22.
NOTE: The LT50 of each fungal isolate was calculated through generalized linear models (GLMs) using R studio (version 3.4.1); the quasibinomial error distribution and a log link function can be used to account for overdispersion.
2. Molecular identification of EPF
- Extraction of fungal genomic DNA
- Collect c.a. 1 cm2 EPF from the 7-day ¼ SDA plate.
- Extract the fungal genomic DNA using a fungal genomic DNA extraction kit according to the manufacturer's instructions23 (Table of Materials).
- PCR amplification and DNA sequencing
- Amplify Fungal ITS region by PCR of the DNA sample21 using the PCR Master Mix (2x), ITS1F/ITS4R primer set24 (Table 1) with the following PCR program: 94 °C for 1 min, and then 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, followed by a 7 min final extension at 72 °C.
NOTE: The ITS1F/ITS4R primer set is for the genus level identification. - Sequence the PCR by commercial sequencing service.
- Use NCBI BLAST search for similar fungi in the NCBI database and select the relative fungal type species for phylogenetic analysis.
NOTE: The fungal species belonging to the genus of Metarhizium or Beauveria should be further identified to the species level with tef-983F/tef-2218R primer set25 (repeat steps 2.1.1 to 2.2.3). For fungi that do not belong to the genera Metarhizium or Beauveria, other molecular markers can be used to identify the species, including DNA lyase (APN2), beta tubulin (BTUB), RNA polymerase II largest subunit (RPB1), RNA polymerase II second largest subunit (RPB2), and 3' portion of translation elongation factor 1 alpha (TEF)25,26.
- Amplify Fungal ITS region by PCR of the DNA sample21 using the PCR Master Mix (2x), ITS1F/ITS4R primer set24 (Table 1) with the following PCR program: 94 °C for 1 min, and then 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, followed by a 7 min final extension at 72 °C.
- Phylogenetic analysis
- Use ClustalX 2.1 software27 to align the multiple sequences from steps 2.2.2 and 2.2.3. Trim the conserved sequences region manually with GeneDoc28.
- Perform the phylogenetic analysis by MEGA7 software29 based on the minimum evolution (ME), Neighbor-Joining (NJ), and maximum likelihood (ML) methods.
NOTE: Performing all three methods can help to confirm and accurately conclude the classification status. The fungal isolates screened by the 1st pathogenicity screen are used for molecular identification at the genus level. The fungal isolates screened by the 2nd virulence test are used for the species level molecular and morphological identification.
3. Morphological identification of EPF
- Observation of the fungal colony morphology
- Use a camera to capture the fungal culture colony growth for 7 days, and record the growth, form (fluffy, firm), and color of the colonies.
- Observation of conidia and conidiophores
- Scrape conidia from the pure culture fungal colony with an inoculation loop and transfer the spores to a glass slide with 0.1% Tween 80 solution. Then, cover with a coverslip for light microscopic observation of conidia.
- Use a scalpel to cut a 5 mm2 agar block of the hyphal strand at the edge of the fungal colony, and then transfer the agar block to a glass slide.
- Perform the cleaning as follows: Add the 0.1% Tween 80 solution on the agar block with a plastic dropper and wash off most of the excess conidia using tweezers. Then, cover it with a coverslip for lightmicroscopic observation.
NOTE: 0.1% Tween 80 can be substituted with another more potent surfactant (Table of Materials) depending on fungal species and hydrophobicity. - Measure and record the width and length of the conidia and conidiophores to compare the differences between different fungal isolates.
- Use Welch's ANOVA test and Games-Howell test (post-hoc test) to analyze the conidial width and length of each strain using R studio (version 3.4.1).
NOTE: Data analysis of morphological characters can be adjusted by cases. The fungal isolates screened with the 3rd virulence test are used for the physiological characterization and ECN ranking in sections 4 and 5.
4. Investigation of conidial productivity and thermotolerance
- Conidial production assay
- Culture the selected fungal isolate on ¼ SDA medium at 25 ± 1 °C in dark for 10 days.
- Prepare 1 mL of the conidial suspension of the fungal isolate in 0.03% surfactant solution and adjust to 1 x 107 conidia/mL as described above.
- Drop three droplets of 10 µL of conidial suspension on¼ SDA and incubate at 25 °C in the dark for 7, 10, and 14 days to count the sporulation of fungi.
NOTE: 10 µL is the best volume to collect the 5 mm block with even fungal sporulation after fungal growth for 7-14 days. - Use the cork borer to detach 5 mm agar block from the center of the colony and transfer into 1 mL of 0.03% surfactant solution (Table of Materials) in a 1.5 mL micro-centrifuge tube at each time point.
- Vortex the tube at 3,000 rpm at room temperature for 15 min and use a hemocytometer to count the number of conidia.
NOTE: The formula used for counting is the number of conidia per 25 squares of the smallest cell (size = 0.025 mm2; chamber depth = 0.1 mm):
Total No of conidia in 5 squares ÷ 80 × (4 × 106) - Repeat three times for each isolate.
- Thermotolerance assay
- Culture the selected fungal isolate on ¼ SDA medium at 25 ± 1 °C in dark for 10 days.
- Prepare 1 mL of the conidial suspension of fungal isolate in 0.03% surfactant solution and adjust to 1 x 107 conidia/mL as described above.
- Vortex the conidial suspension and heat it in a 45 °C dry bath for 0, 30, 60, 90, and 120 min. Drop three droplets of 5 µL of the conidial suspension on 55 mm ¼ SDA medium at each time point post heat-exposure and incubate at 25 ± 1 °C for 18 h.
NOTE: Avoid spreading the fungal droplets to be able to better focus on the area. - Count the number of germinated conidia spores with five randomly selected fields under 200x light microscopy to determine germination rate.
- Perform three replicates for each isolate.
5. Effective conidia number (ECN) ranking
- ECN calculation
NOTE: Obtain the conidial production and thermotolerance data of each potential fungal strain before calculating the total ECN21.- Calculate the fold-change (FC) of conidial production at each time point:
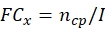
where, x = time point for data collection; ncp = number of conidia after each day of growth; and I = initial number of seeded conidia. - Calculate the conidia number under the stress treatment at each time point using the following formula:

Where, y = the ECN of the time point under treatment; TT0 = the germination rate of conidia not undergoing heat stress (= germination rate of 0 min heat treatment); TTz = stress coefficient is the conidia germination rate at different times of heat treatment (z). - Calculate the total ECN using the following formula:

- Compare the ECN of each fungal strain.
- Calculate the fold-change (FC) of conidial production at each time point:
- Principal component analysis (PCA) of fungal strains
NOTE:The PCA analysis confirms the ranking of ECN and helps in understanding the correlation between the physiological character values. Compare the ECN values and select EPF isolates having higher ECN values. - Use R software to create PCA by coding:
#Input PCA data file
a = read.table("PCA.csv",sep=',',header=T)- # Processing sample data
row.names(a) <- c("NCHU-9","NCHU-11", "NCHU-64", "NCHU-69", "NCHU-95", "NCHU-113")
X=row.names(a)
df<- a[2:11] - #PCA calculation
pca <- prcomp(df, center = TRUE, scale = TRUE)
vars <- (pca$sdev)^2
pc1_percent = vars[1] / sum(vars)
pc2_percent = vars[2] / sum(vars)
value = pca$x - #Output PCA visualization file
png(file = 'pca.png', height = 2000, width = 2000, res = 300)
NOTE: Use 7 to 14 days conidial production and all thermotolerance data to execute principal component analysis (PCA) for confirming the ECN ranking.
- # Processing sample data
- Select the best-performing fungal strains based on ECN or PCA and perform the virulence test of target pests for further research.
Wyniki
Isolation and selectionof potential Entomopathogenic fungi (EPF)
By using the Tenebrio molitor-mediated Entomopathogenic fungi (EPF) library construction method, the number of fungi without insect-killing activity would be excluded; thus, the isolation efficiency and selection of EPF could be largely increased. During the application of this method, the information of sampling sites, soil samples, and the fungal germination rates were recorded (Tab...
Dyskusje
Entomopathogenic fungi (EPF) have been used for insect control. There are several methods to isolate, select, and identify EPF30,31,32. Comparing the different types of insect bait methods, Beauveria bassiana and Metarhizium anisopliae were commonly found in insect baits6,12,13,14. Am...
Ujawnienia
The authors declare there is no conflict of interest involved in this work.
Podziękowania
This research was supported by Grant 109-2313-B-005 -048 -MY3 from the Ministry of Science and Technology (MOST).
Materiały
| Name | Company | Catalog Number | Comments |
| Agar Bacteriological grade | BIOMAN SCIENTIFIC Co., Ltd. | AGR001 | Suitable in most cell culture/molecular, biology applications. |
| AGAROSE, Biotechnology Grade | BIOMAN SCIENTIFIC Co., Ltd. | AGA001 | For DNA electrophoresis. |
| BioGreen Safe DNA Gel Buffer | BIOMAN | SDB001T | |
| Brass cork borer | Dogger | D89A-44001 | |
| Canon kiss x2 | Canon | EOS 450D | For record strain colony morphology |
| Constant temperature incubator | Yihder Co., Ltd. | LE-509RD | Fungal keeping. |
| cubee Mini-Centrifuge | GeneReach | MC-CUBEE | |
| DigiGel 10 Digital Gel Image System | TOPBIO | DGIS-12S | |
| Finnpipette F2 0.2 to 2 µL Pipette | Thermo Scientific | 4642010 | |
| Finnpipette F2 1 to 10 µL Pipette | Thermo Scientific | 4642030 | |
| Finnpipette F2 10 to 100 µL Pipette | Thermo Scientific | 4642070 | |
| Finnpipette F2 100 to 1000 µL Pipette | Thermo Scientific | 4642090 | |
| Finnpipette F2 2 to 20 µL Pipette | Thermo Scientific | 4642060 | |
| Finnpipette F2 20 to 200 µL Pipette | Thermo Scientific | 4642080 | |
| GeneAmp PCR System 9700 | Applied Biosystems | 4342718 | |
| GenepHlow Gel/PCR Kit | Geneaid | DFH100 | |
| Genius Dry Bath Incubator | Major Science | MD-01N | |
| Graduated Cylinder Custom A 100mL | SIBATA | SABP-1195906 | Measure the volume of reagents. |
| Hand tally counter | SDI | NO.1055 | |
| Hemocytometer | bioman | AP-0650010 | Calculate the number of spore |
| Inoculating loop | Dogger | D8GA-23000 | |
| lid | IDEAHOUSE | RS92004 | |
| Micro cover glass | MUTO PURE CHEMICALS CO.,LTD | 24241 | |
| Microscope imaging system | SAGE VISION CO.,LTD | SGHD-3.6C | |
| Microscope Slides | DOGGER | DG75001-07105 | |
| Mupid-2plus DNA Gel Electrophoresis | ADVANCE | AD110 | |
| Nikon optical microscope | SAGE VISION CO.,LTD | Eclipse CI-L | |
| Plastic cup | IDEAHOUSE | CS60016 | |
| Presto Mini gDNA Yeast Kit | Geneaid | GYBY300 | Fungal genomic DNA extraction kit |
| Sabouraud Dextrose Broth (Sabouraud Liquid Medium) | HiMedia Leading BioSciences Company | M033 | Used for cultivation of yeasts, moulds and aciduric microorganisms. |
| Scalpel Blade No.23 | Swann-Morton | 310 | |
| Scalpel Handle No.4 | AGARWAL SURGICALS | SSS -FOR-01-91 | |
| Shovel | Save & Safe | A -1580242 -00 | |
| Silwet L-77 | bioman(phytotech) | S7777 | Surfactant |
| Sorvall Legend Micro 17 Microcentrifuge | Thermo Scientific | 75002403 | |
| Steel Tweezers | SIPEL ELECTRONIC SA | GG-SA | |
| Sterile Petri Dish | BIOMAN SCIENTIFIC Co., Ltd. | 1621 | Shallow cylindrical containers with fitted lids, specifically for microbiology or cell culture use. |
| ThermoCell MixingBlock | BIOER | MB-101 | |
| Tween 80 | FUJIFILM Wako Pure Chemical Corporation | 164-21775 | |
| TwinGuard ULT Freezer | Panasonic Healthcare Holdings Co., Ltd. | MDF-DU302VX | -80°C sample stored. |
| Vertical floor type cabinet | Chih Chin | BSC-3 | Fungal operating culturing. |
| Vortex Genie II | Scientific | SIG560 | |
| Zipper storage bags | Save & Safe | A -1248915 -00 | |
| 100 bp DNA Ladder | Geneaid | DL007 | |
| -20°C Freezer | FRIGIDAIRE | Frigidaire FFFU21M1QW | -20°C sample and experimental reagents stored. |
| 2X SuperRed PCR Master Mix | TOOLS | TE-SR01 | |
| 50X TAE Buffer | BIOMAN | TAE501000 |
Odniesienia
- Wraight, S. P., Carruthers, R. I. . Biopesticides: use and Delivery. , 233-269 (1999).
- Chase, A., Osborne, L., Ferguson, V. Selective isolation of the entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae from an artificial potting medium. Florida Entomologist. , 285-292 (1986).
- Meyling, N. V. Methods for isolation of entomopathogenic fungi from the soil environment. University of Copenhagen. , 1-18 (2007).
- Zimmermann, G. The 'Galleria bait method'for detection of entomopathogenic fungi in soil. Journal of applied Entomology. 102 (1-5), 213-215 (1986).
- Schneider, S., Widmer, F., Jacot, K., Kölliker, R., Enkerli, J. Spatial distribution of Metarhizium clade 1 in agricultural landscapes with arable land and different semi-natural habitats. Applied Soil Ecology. 52, 20-28 (2012).
- Hallouti, A., et al. Diversity of entomopathogenic fungi associated with Mediterranean fruit fly (Ceratitis capitata (Diptera: Tephritidae)) in Moroccan Argan forests and nearby area: impact of soil factors on their distribution. BMC Ecology. 20 (1), 1-13 (2020).
- Meyling, N. V., Eilenberg, J. Occurrence and distribution of soil borne entomopathogenic fungi within a single organic agroecosystem. Agriculture, Ecosystems and Environment. 113 (1-4), 336-341 (2006).
- Skalický, A., Bohatá, A., Šimková, J., Osborne, L. S., Landa, Z. Selection of indigenous isolates of entomopathogenic soil fungus Metarhizium anisopliae under laboratory conditions. Folia Microbiologica. 59 (4), 269-276 (2014).
- Veen, K., Ferron, P. A selective medium for the isolation of Beauveria tenella and of Metarrhizium anisopliae. Journal of Invertebrate Pathology. 8 (2), 268-269 (1966).
- Goettel, M., Inglis, D., Lacy, L. . Manual of Techniques in Insect Pathology. , 213-249 (1997).
- Luz, C., Netto, M. C. B., Rocha, L. F. N. In vitro susceptibility to fungicides by invertebrate-pathogenic and saprobic fungi. Mycopathologia. 164 (1), 39-47 (2007).
- Mantzoukas, S., et al. Trapping entomopathogenic fungi from vine terroir soil samples with insect baits for controlling serious pests. Applied Sciences. 10 (10), 3539 (2020).
- Goble, T., Dames, J., Hill, M., Moore, S. The effects of farming system, habitat type and bait type on the isolation of entomopathogenic fungi from citrus soils in the Eastern Cape Province, South Africa. BioControl. 55 (3), 399-412 (2010).
- Nishi, O., Iiyama, K., Yasunaga-Aoki, C., Shimizu, S. Isolation of entomopathogenic fungi from soil by using bait method with termite, Reticulitermes speratus. Enotomotech. 35, 21-26 (2011).
- Castrillo, L. . ARS Collection of Entomopathogenic Fungal Cultures (ARSEF). , (2014).
- Kim, J. C., et al. Tenebrio molitor-mediated entomopathogenic fungal library construction for pest management. Journal of Asia-Pacific Entomology. 21 (1), 196-204 (2018).
- Keyser, C. A., Henrik, H., Steinwender, B. M., Meyling, N. V. Diversity within the entomopathogenic fungal species Metarhizium flavoviride associated with agricultural crops in Denmark. BMC Microbiology. 15 (1), 1-11 (2015).
- Quesada-Moraga, E., Navas-Cortés, J. A., Maranhao, E. A., Ortiz-Urquiza, A., Santiago-Álvarez, C. Factors affecting the occurrence and distribution of entomopathogenic fungi in natural and cultivated soils. Mycological Research. 111 (8), 947-966 (2007).
- Park, J. B., et al. Developmental characteristics of Tenebrio molitor larvae (Coleoptera: Tenebrionidae) in different instars. International Journal of Industrial Entomology. 28 (1), 5-9 (2014).
- Chang, J. -. C., et al. Construction and selection of an entomopathogenic fungal library from soil samples for controlling Spodoptera litura. Frontiers in Sustainable Food Systems. 5, 15 (2021).
- Podder, D., Ghosh, S. K. A new application of Trichoderma asperellum as an anopheline larvicide for eco friendly management in medical science. Scientific reports. 9 (1), 1-15 (2019).
- . Geneaid Biotech Ltd. Presto Mini gDNA Yeast, Ver. 04.27.17 Available from: https://www.geneaid.com/data/files/1605664221308055331.pdf (2021)
- White, T. J., Bruns, T., Lee, S., Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: A guide to methods and applications. 18 (1), 315-322 (1990).
- Kepler, R. M., Humber, R. A., Bischoff, J. F., Rehner, S. A. Clarification of generic and species boundaries for Metarhizium and related fungi through multigene phylogenetics. Mycologia. 106 (4), 811-829 (2014).
- Kepler, R. M. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus. 8 (2), 335-353 (2017).
- Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 25 (24), 4876-4882 (1997).
- Kumar, S., Stecher, G., Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 33 (7), 1870-1874 (2016).
- Herlinda, S., Mulyati, S. I. Selection of isolates of entomopathogenic fungi and the bioefficacy of their liquid production against Leptocorisa oratorius nymphs. Microbiology Indonesia. 2 (3), 9 (2008).
- Herlinda, S., Irsan, C., Mayasari, R., Septariani, S. Identification and selection of entomopathogenic fungi as biocontrol agents for Aphis gossypii from South Sumatra. Microbiology Indonesia. 4 (3), 137-142 (2010).
- Montes-Bazurto, L. G., Peteche-Yonda, Y., Medina-Cardenas, H. C., Bustillo-Pardey, A. E. Selection of entomopathogenic fungi for the biological control of Demotispa neivai (Coleoptera: Chrysomelidae) in oil palm plantations in Colombia. Journal of Entomological Science. 55 (3), 388-404 (2020).
- Shin, T. -. Y., Choi, J. -. B., Bae, S. -. M., Koo, H. -. N., Woo, S. -. D. Study on selective media for isolation of entomopathogenic fungi. International Journal of Industrial Entomology. 20 (1), 7-12 (2010).
- Sharma, L., Oliveira, I., Torres, L., Marques, G. Entomopathogenic fungi in Portuguese vineyards soils: Suggesting a 'Galleria-Tenebrio-bait method'as bait-insects Galleria and Tenebrio significantly underestimate the respective recoveries of Metarhizium (robertsii) and Beauveria (bassiana). MycoKeys. (38), 1 (2018).
- Rodríguez, M., Gerding, M., France, A. Selección de Hongos Entomopatógenos para el Control de Varroa destructor (Acari: Varroidae). Chilean journal of agricultural research. 69 (4), 534-540 (2009).
- Yang, H., et al. Persistence of Metarhizium (Hypocreales: Clavicipitaceae) and Beauveria bassiana (Hypocreales: Clavicipitaceae) in tobacco soils and potential as biocontrol agents of Spodoptera litura (Lepidoptera: Noctuidae). Environmental entomology. 48 (1), 147-155 (2019).
- Muñiz-Reyes, E., Guzmán-Franco, A. W., Sánchez-Escudero, J., Nieto-Angel, R. Occurrence of entomopathogenic fungi in tejocote (C rataegus mexicana) orchard soils and their pathogenicity against R hagoletis pomonella. Journal of Applied Microbiology. 117 (5), 1450-1462 (2014).
- Lacey, L. A., et al. Goettel Insect pathogens as biological control agents: Back to the future. Journal of Invertebrate Pathology. 132, 1-41 (2015).
- Humber, R. A. . Manual of techniques in insect pathology. , 153-185 (1997).
- Rehner, S. A., Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 97 (1), 84-98 (2005).
- Quandt, C. A., et al. Phylogenetic-based nomenclatural proposals for Ophiocordycipitaceae (Hypocreales) with new combinations in Tolypocladium. IMA fungus. 5 (1), 121-134 (2014).
- Shah, F. A., Wang, C. S., Butt, T. M. Nutrition influences growth and virulence of the insect-pathogenic fungus Metarhizium anisopliae. FEMS Microbiology Letters. 251 (2), 259-266 (2005).
- Ignoffo, C. Environmental factors affecting persistence of entomopathogens. Florida Entomologist. , 516-525 (1992).
- Rodrigues, I. W., Forim, M., Da Silva, M., Fernandes, J., Batista Filho, A. Effect of ultraviolet radiation on fungi Beauveria bassiana and Metarhizium anisopliae, pure and encapsulated, and bio-insecticide action on Diatraea saccharalis. Advances in Entomology. 4 (3), 151-162 (2016).
- Paula, A. R., Ribeiro, A., Lemos, F. J. A., Silva, C. P., Samuels, R. I. Neem oil increases the persistence of the entomopathogenic fungus Metarhizium anisopliae for the control of Aedes aegypti (Diptera: Culicidae) larvae. Parasites and Vectors. 12 (1), 1-9 (2019).
- Morley-Davies, J., Moore, D., Prior, C. Screening of Metarhizium and Beauveria spp. conidia with exposure to simulated sunlight and a range of temperatures. Mycological Research. 100 (1), 31-38 (1996).
- Rangel, D. E., Braga, G. U., Flint, S. D., Anderson, A. J., Roberts, D. W. Variations in UV-B tolerance and germination speed of Metarhizium anisopliae conidia produced on insects and artificial substrates. Journal of Invertebrate Pathology. 87 (2-3), 77-83 (2004).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone