Heterogeneity Mapping of Protein Expression in Tumors using Quantitative Immunofluorescence
In This Article
Summary
Here we describe a method to quantify molecular heterogeneity in histological sections of tumor material using quantitative immunofluorescence, image analysis, and a statistical measure of heterogeneity. The method is intended for use in clinical biomarker development and analysis.
Abstract
Morphologic heterogeneity within an individual tumor is well-recognized by histopathologists in surgical practice. While this often takes the form of areas of distinct differentiation into recognized histological subtypes, or different pathological grade, often there are more subtle differences in phenotype which defy accurate classification (Figure 1). Ultimately, since morphology is dictated by the underlying molecular phenotype, areas with visible differences are likely to be accompanied by differences in the expression of proteins which orchestrate cellular function and behavior, and therefore, appearance. The significance of visible and invisible (molecular) heterogeneity for prognosis is unknown, but recent evidence suggests that, at least at the genetic level, heterogeneity exists in the primary tumor1,2, and some of these sub-clones give rise to metastatic (and therefore lethal) disease.
Moreover, some proteins are measured as biomarkers because they are the targets of therapy (for instance ER and HER2 for tamoxifen and trastuzumab (Herceptin), respectively). If these proteins show variable expression within a tumor then therapeutic responses may also be variable. The widely used histopathologic scoring schemes for immunohistochemistry either ignore, or numerically homogenize the quantification of protein expression. Similarly, in destructive techniques, where the tumor samples are homogenized (such as gene expression profiling), quantitative information can be elucidated, but spatial information is lost. Genetic heterogeneity mapping approaches in pancreatic cancer have relied either on generation of a single cell suspension3, or on macrodissection4. A recent study has used quantum dots in order to map morphologic and molecular heterogeneity in prostate cancer tissue5, providing proof of principle that morphology and molecular mapping is feasible, but falling short of quantifying the heterogeneity. Since immunohistochemistry is, at best, only semi-quantitative and subject to intra- and inter-observer bias, more sensitive and quantitative methodologies are required in order to accurately map and quantify tissue heterogeneity in situ.
We have developed and applied an experimental and statistical methodology in order to systematically quantify the heterogeneity of protein expression in whole tissue sections of tumors, based on the Automated QUantitative Analysis (AQUA) system6. Tissue sections are labeled with specific antibodies directed against cytokeratins and targets of interest, coupled to fluorophore-labeled secondary antibodies. Slides are imaged using a whole-slide fluorescence scanner. Images are subdivided into hundreds to thousands of tiles, and each tile is then assigned an AQUA score which is a measure of protein concentration within the epithelial (tumor) component of the tissue. Heatmaps are generated to represent tissue expression of the proteins and a heterogeneity score assigned, using a statistical measure of heterogeneity originally used in ecology, based on the Simpson's biodiversity index7.
To date there have been no attempts to systematically map and quantify this variability in tandem with protein expression, in histological preparations. Here, we illustrate the first use of the method applied to ER and HER2 biomarker expression in ovarian cancer. Using this method paves the way for analyzing heterogeneity as an independent variable in studies of biomarker expression in translational studies, in order to establish the significance of heterogeneity in prognosis and prediction of responses to therapy.
Protocol
1. Tissue preparation
- Microtomy
- Section formalin fixed paraffin embedded tumor blocks at 4 μm thickness using a rotary microtome.
- Place the sections onto positively charged microscopic slides.
- Incubate slides in a 37 °C oven overnight.
- Storage
- Store sections in a -18 °C to -25 °C freezer, in order to prevent loss of antigenicity.
2. Immunofluorescence of tissue sections
- Dewaxing and Rehydration
- Dewax slides in xylene twice for 5 min.
- Rehydrate slides in 99%, 99%, 80%, 50% of ethanol and running tap water for 2 min of each in order.
- Antigen Retrieval
- Perform the heat induced antigen retrieval, using 0.15 mM Sodium Citrate, pH 6.0 buffer in a pressure cooker for 5 min in the microwave.
- Cool down slides for 20 min.
- Rinse slides in 0.05% PBST for 5 min on the rocker.
- Blocking procedure
- Treat sections in 3% hydrogen peroxide for 10 min.
- Rinse slides in 0.05% PBST for 5 min on the rocker.
- Add the sections to Sequenza Immunostaining rack.
- Treat sections in Serum free protein block for 10 min.
- Primary antibody incubation
- Incubate Primary antibody diluted to optimal dilution in Dako antibody diluent for 1 hour at room temperature.
- Rinse in 0.05% PBST three times for 5 min each.
- Epithelial mask (Cytokeratin) incubation
- Incubate mouse anti-pan cytokeratin, diluted 1:50 in Dako antibody diluent overnight at 4 °C.
- Epithelial mask visualization
- Prepare a 1:25 dilution of the goat anti-mouse Alexa555 secondary antibody in the pre-diluted Envision goat-rabbit HRP antibody solution. Incubate slides in the dark for 1.5 hrs at room temperature.
- Rinse in 0.05% PBST three times for 5min each.
- Target visualization
- Transfer the slides from the Sequenza to the humidity chamber.
- Combine Target signal amplification diluent (HistoRx tub E) and the Cy5 Tyramide (HistoRx tube F) at 1:50 concentration. Vortex to mix thoroughly. Incubate slides in the dark for 10 min at room temperature.
- Rinse in 0.05% PBST three times for 5 min each.
- Dehydrate slides in 80% ethanol for 1 min.
- Air dry slides in the dark.
- Counterstaining and coverslipping
- Apply DAPI mounting medium on the coverslips and place the coverslips over the tissue sections.
- Dry slides overnight in the dark.
- After the slides have dried, secure with nail polish to ensure long term preservation and keep in a 4 °C refrigerator.
3. Image capture using whole section scanning
- Load up to five slides in the slide cassette of the Aperio ScanScope FL slide scanner.
- For each slide, select the general region to image using the interface in the ScanScope Console. Select the region to encompass all the tissue on the slide, regardless of staining pattern or other observable aspects of the sample.
- Using the ScanScope Console, for each channel (DAPI, Cy3 and Cy5) determine an optimal exposure as follows:
- Select a region of tissue to interrogate - this area should, ideally, be in the tumor region of tissue to provide the best representation.
- Using the ScanScope Console autoexposure feature, evaluate the exposure time suggested for each channel, along with image focus.
- Repeat over up to 3-5 regions of each sample to confirm stability of the value.
- Select an additional region (as shown by a blue diamond on the ScanScope console image) away from the tissue, yet still within the coverslipped region of the slide, to define a background area where flat field correction images will be acquired for each filter.
- Review these images prior to image acquisition. If the images appear to have high background or other image artifacts, the image region should be moved and new images acquired.
- The acquired digital slide images are uploaded automatically into the Aperio Spectrum database.
4. AQUA automated image analysis
- View the digital whole slide images in the Spectrum database and annotate tumor areas of interest in the context of the entire tissue and staining pattern(s).
- Select the slide to analyze using AQUAnalysis from the digital slide list in the Spectrum database by double clicking the thumbnail image (alternatively, select the check box next to the image and then select 'View Images' from the menu at the top of the list). This automatically opens the ImageScope software and color merged image of the tissue sample.
- Use this software to navigate the image using the tools provided.
- Using the accompanying H&E (either the physical slide or an annotated image), annotate the region of interest on the fluorescent image. Multiple regions of interest, individual circled areas, can be selected on a single sample. There is also a 'negative' tool which is used to exclude areas within a selected region (i.e. damaged tissue, areas of poor histology, non-tumor areas, debris, etc.).
- Save the annotation layer(s) on the image.
- Return to the Spectrum main menu and select the check box next to the image that was just annotated. Select 'Analyze' from the menu strip across the top of the list.
- In the Analysis window which will come up, select "HistoRx AQUA Analysis" from the pull-down menu.
- If there are multiple layers of annotations, use the check boxes to select the appropriate layer(s) for analysis.
- Press the 'Analyze' button when ready to begin.
- At this point, a minimized console window will appear in the Windows taskbar. This will display the progress of the transfer of images from the Spectrum database to the local PC (the 'pull' operation).
- Once the data is transferred, AQUAnalysis will launch. The user can then use all of the internal functions of the software to view, navigate and analyze images.
- The annotated region of the image from the Spectrum database is broken into 512x512 pixel image tiles - each of which will be scored individually.
- For this study, an unsupervised AQUA scoring algorithm, based on data clustering, was used8. In this algorithm, AQUA scores are generated as follows:
- The pan-cytokeratin image signal is thresholded above background and those pixels are used to define a tumor 'mask' which is then used for subsequent calculations. The high -expression pan-cytokeratin pixels also define the cytoplasmic/non-nuclear regions of the tumor cells.
- Pixels with high signal in the DAPI channel that are within the tumor mask are identified as nuclear in nature.
- The clustering algorithm also examines pixels which have low intensity for either DAPI staining or cytokeratin expression and attempts to partially attribute them to either the nuclear or cytoplasmic compartments, or background.
- Once all pixels are assigned to one of the tumor compartments or background, the intensity of signal from the target protein (ER or HER2) is then calculated within each of the compartments and normalized to the size of the compartments, thus producing AQUA scores.
- Images which did not have sufficient tumor representation were not scored (as defined by those with less than 5% of pixels representing tumor). These regions also produce a 'Final Result' value called 'Fail' and can thus be removed in subsequent statistical analyses.
- Additionally, if upon review, an operator identified fields which were not suitable for scoring due to sample or imaging artifacts (e.g. folded tissue, debris), those fields were redacted from scoring and produced a 'Final Result' called 'Fail'.
- The results of AQUA scoring are tables of scores associated with each 512x512 pixel tile in the region of interest within a whole tissue section sample. These files, in .CSV format, can then be used for subsequent statistical analysis.
5. Statistical Analysis: All statistical analyses are performed in SPSS (IBM)
- Where possible, for large operations, generate SPSS syntax code files (.sps) to perform data manipulations and analysis steps. Example syntax code is provided as supplementary files.
- Read data files into SPSS. Remove unused variables in the raw data (e.g. instrument specific parameters, AQUA scores from non-utilized compartments).
- Where regions have a 'Final Result' of 'Fail' (see above), mark these values as SYSMIS, which removes them from the numerical analyses.
- Aggregate all data for all slides for each marker (ER or HER2) into a single master data set in SPSS format. Use this master file for all subsequent calculations.
- Using an SPSS syntax file (one for each marker: ER or HER2 (see supplementary syntax files) data was further validated and Simpson's Indices of diversity calculated as follows:
- Generate a master analytical file, in order to prevent modification of the master data set previously generated.
- Validate the master data set against the raw data and images generated.
- Produce summary statistics for AQUA scores for each sample (count of fields, mean score, median score, minimum score, maximum score, and standard deviation of scores for sample).
- Check a random sample of results against the original raw data output files and against the raw image data scored.
- To generate the heat map images (shown in Figures 2 and 3):
- 'Bin' the AQUA score data (using the 'Visual Binning' function in SPSS) into eight individual levels. For ER, nuclear scores were used, for HER2, cytoplasmic scores were used.
- Derive bins such that each bin represents an equal percentage of available cases from the entire data set.
- Use the SPSS function 'Split File' so that all subsequent output would be on a 'per slide/sample' level.
- Plot each region (corresponding to a 512x512 tile) by its x,y coordinates and color with a value corresponding to the bin into which it was allocated (see figures 2 and 3).
- To generate heterogeneity scores, code was written to generate a Simpson's index of diversity using AQUA scores.
- The Simpson's index of diversity, as used here, is defined as:
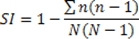
Where N is the total number of scores/fields used for a given sample and n is the number of scores/fields in a given group of standardized scores (produced as described below).
- The Simpson's index of diversity, as used here, is defined as:
- Use the master analytical file described above for data source. In the examples shown, all ER calculations used the nuclear AQUA score and HER2 calculations used the cytoplasmic AQUA score. Perform the calculations below for each sample/slide.
- Since fluorescence data is subject to variance instability, such that error magnitude propagates with intensity, apply logarithmic (base 2) normalization to all AQUA scores.
- Further transform the normalized (log transformed) data into z-(standard) scores.
- The Z-score transformation is calculated as follows: Z = (AQUA score - average of all scores for a sample)/standard deviation of scores for the sample. This translates the distribution of scores into a distribution of deviations around the central value of zero deviation.
- Bin these Z-score values into six groups (<-2, -2 - -1, -1 - 0, 0 - 1, 1 - 2, >2).
- Calculate the number of values allocated into each bin and sum for each bin. Use this value, along with the total number of fields, to calculate a value for each of the six bins used for the index.
- Sum these values and subtract from one to produce the index of diversity.
- The diversity index provides an indicator of the spread of scores around the mean for a particular sample. Thus, higher indices indicate a broader spread of scores, or increased heterogeneity of expression. Conversely, lower indices, indicate less variation in expression measurement and homogeneous expression in the sample. This is illustrated in Figures 2 and 3.
6. Representative Results:
Targeted therapy with relatively low toxicity agents, such as tamoxifen and trastuzumab, are not standard of care for ovarian cancer patients. There is good evidence that platinum-taxane first-line chemotherapy is superior to other chemotherapy regimens for ovarian cancer9-13, and while 70-80% of patients initially respond, most will relapse and die of their disease. Measurement of biomarkers which might predict response to alternative therapies would be useful in selecting individualized therapy for each patient, and since chemotherapy exerts selection pressure on a tumor, surviving tumor cells may possess different characteristics and represent a subpopulation of the tumor before therapy; quantifying this change might therefore be useful. We applied our heterogeneity mapping approach to ER and HER2 expression in ovarian cancer specimens before and after chemotherapy, in order to measure quantitative changes in expression, and assess the relationship between biomarker expression before and after therapy. Both ER and HER2 demonstrate marked heterogeneity within individual tumors, and in some tumors, the Simpson's index changes after therapy from relatively high heterogeneity to low heterogeneity, represented by decreasing Simpson's index scores (see Figures 2 and 3). This supports the notion that clonal selection occurs in response to cytotoxic chemotherapy; more importantly, this may be exploited using targeted therapy against the residual population, in order to better personalize therapy for cancer patients.
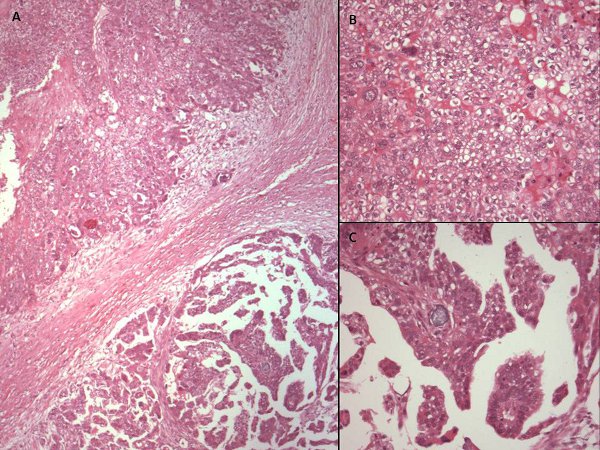
Figure 1. Hematoxylin and eosin stained photomicrographs of different areas of the same ovarian cancer showing variable morphology. (A) x40 photomicrograph shows two adjacent areas of tumor with varying morphology. High power views (x200) reveal cells with cytoplasmic clearing and a solid pattern of growth in the upper part of the tumor (B), while the lower part of the tissue (C) has more homogeneous eosinophilic cytoplasm and a papillary growth pattern. This tumor would, however, be classified as serous papillary histological subtype for the purposes of further management.
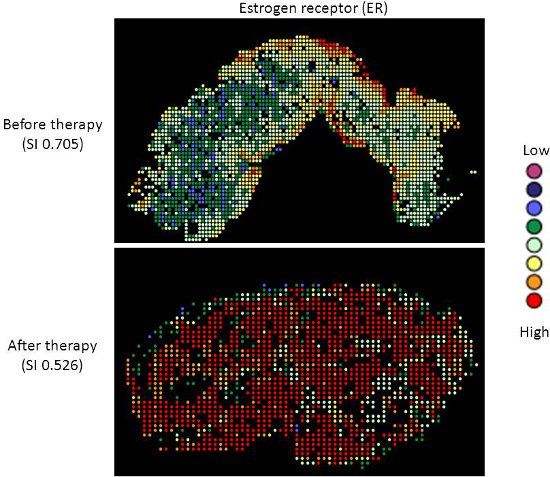
Figure 2. Heterogeneity heatmaps of ER protein expression in cytokeratin-positive ovarian cancer tissue, before (upper panel) and after (lower panel) therapy.
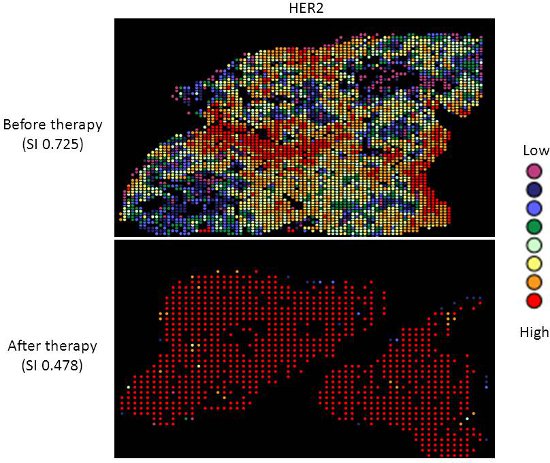
Figure 3. Heterogeneity heatmaps of HER2 protein expression in cytokeratin-positive ovarian cancer tissue, before (upper panel) and after (lower panel) therapy.
Discussion
The method described herein permits quantification of molecular heterogeneity in standard formalin-fixed, paraffin-embedded histologic sections of tumor material. The method permits a value to be assigned to the degree of heterogeneity in a tissue section, so that this may then be taken into account as a variable in biomarker development and analysis. While in general it is preferable that tissue biomarkers are homogeneous with respect to expression, so that the assay is less susceptible to sampling error or bias, it might under some circumstances be useful to quantify heterogeneity as a parameter. For instance, as shown in the illustrative example for ER and HER2, common biomarkers show considerable heterogeneity of expression and it is currently unknown whether this represents an independent prognostic or predictive factor with respect to clinical outcome or response to therapy, respectively. Likewise, as illustrated, therapy itself might dynamically alter the constituent populations of cells, and therefore measurement of target enrichment might be useful in guiding treatment decisions.
However, the technique is limited by the same factors which limit traditional histopathological or biomarker (such as immunohistochemical) analyses. The result is dependent on the type, size, and quality of the tissue being analyzed, and therefore a single tissue section may not be representative of the entire tumor. Indeed, the Simpson's index may be unduly biased by size of the tissue (number of frames captured and AQUA scores measured). The immunogenicity of the epitopes being measured may be subject to uncontrollable pre-analytical factors which artifactually alter their expression across the tissue section (such as cold ischemia time, or the length of fixation, and edge artifact). This is particularly a problem for phospho-epitopes, which are notoriously labile14, 15. Therefore the technique might be better suited to resection specimens rather than small biopsies, and performed on multiple sections rather than just one. Ultimately, 3D imaging techniques may offer an opportunity to better quantify this parameter.
The technique as presented may also be limited by biological factors, such as variability in expression of the cytokeratin masking epitope. This might be of particular concern in those cancers, including ovarian and breast cancer, which undergo epithelial-to-mesenchymal transition (EMT) or are enriched for non-epithelial components or stem cells16, as has recently been shown to occur in chemotherapy-treated ovarian cancer in vitro17, and in breast cancer patients in response to therapy18. This limitation may be overcome using alternative masking antibodies (or cocktails of antibodies), such as cytokeratin plus vimentin, which is upregulated in EMT16. In addition, since other components of the tissue (the microenvironment), such as fibroblasts or vascular endothelial cells are also increasingly being targeted by biological therapies, the technique may also be expanded to to score these components, using specific masks for compartments of interest, such as PECAM/ CD31 to stain for vascular endothelial cells.
Although the adaptation of the Simpson's index has been used as a measure of heterogeneity in the current protocol due to its simplicity, other measures of heterogeneity could be used, such as simple measurements of central tendency (mean, variance, median, etc). Additionally, the Simpson's index calculations could be modified to better suit a particular test population. The use of Z-score transformations allows the scoring to be more applicable for multiple markers since the heterogeneity is based on deviation around the mean for a data set. However, the overall range of scoring can be limited in this type of transformation. Some designs may be better suited to simply bin the actual AQUA scores for all samples of a particular marker set. The selection of the number of bins and cutoffs is also a limitation in the range of values which can result and may be optimized for alternative experimental designs.
We have used ER and HER2 in the current study as candidate biomarkers, since they are already used in the clinic, help answer relevant clinical questions with respect to the efficacy of targeted therapies, and are known to exhibit considerable heterogeneity and change in primary and distant disease19. However, the optimal marker of heterogeneity remains to be determined. Other possibilities might include cytogenetic markers of clonality, such as those previously used in interphase FISH studies20. The approach requires further validation in large, well annotated clinical cohorts or clinical trials in order to further establish the relevance of this parameter in cancer biology.
Disclosures
JC, MG, CJ, and CS are employees of HistoRx, Inc.
Acknowledgements
This work was supported in part by the Scottish Funding Council (Grant Number HR07005; http://www.sfc.ac.uk/), Medical Research Scotland (http://www.medicalresearchscotland.org.uk/), the Cancer Research UK Experimental Cancer Medicine Centre (http://www.cancerresearchuk.org/), and Breakthrough Breast Cancer (http://breakthrough.org.uk/).
Materials
| Name | Company | Catalog Number | Comments |
| Name of the reagent | Company | Catalogue number | Comments |
| ScanScope FL | Aperio | ScanScope FL | Used as equipped from vendor |
| Spectrum | Aperio | Spectrum | Configuration and catalog numbers will vary on how local institutions set up their database and IT structure. |
| AQUAnalysis, in conjunction with AQUAjustment | HistoRx | V2.3 | The AQUAjustment package allows AQUAnalysis to use images stored in Spectrum. |
| HER2 | Dako | A0485 | 1 in 400 dilution |
| mouse anti-pan cytokeratin | Dako | M3515 | 1 in 50 dilution |
| Antibody diluent | Dako | S0809 | |
| Envision goat-rabbit HRP | Dako | K4003 | |
| Protein block | Dako | X0909 | |
| Goat anti-mouse Alexa555 | Invitrogen | A21422 | |
| DAPI mounting medium | Invitrogen | P36931 | |
| Cy5 Tyramide | HistoRx | AQ-EMR1-00001 | |
| Sequenza Immunostaining Center | Thermo Scientific Shandon | 73300001 | Used here for bench-top immunofluorescence staining |
References
- Gutierrez, M. L., Sayagues, J. M., Del Mar, A. M. Cytogenetic heterogeneity of pancreatic ductal adenocarcinomas: identification of intratumoral pathways of clonal evolution. Histopathology. 58, 486-497 (2011).
- Yachida, S., Jones, S., Bozic, I. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 467, 1114-1117 (2010).
- Gutierrez, M. L., Sayagues, J. M., Del Mar, A. M. Cytogenetic heterogeneity of pancreatic ductal adenocarcinomas: identification of intratumoral pathways of clonal evolution. Histopathology. 58, 486-497 (2011).
- Yachida, S., Jones, S., Bozic, I. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 467, 1114-1117 (2010).
- Liu, J., Lau, S. K., Varma, V. A. Molecular mapping of tumor heterogeneity on clinical tissue specimens with multiplexed quantum dots. ACS Nano. 4, 2755-2765 (2010).
- Camp, R. L., Chung, G. G., Rimm, D. L. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat. Med. 8, 1323-1327 (2002).
- Simpson, E. H. Measurement of biodiversity. Nature. 163, 688-68 (1949).
- Gustavson, M. D., Bourke-Martin, B., Reilly, D. M. Development of an unsupervised pixel-based clustering algorithm for compartmentalization of immunohistochemical expression using Automated Quantitative Analysis. Appl. Immunohistochem. Mol. Morphol. 17, 329-337 (2009).
- . Paclitaxel plus carboplatin versus standard chemotherapy with either single-agent carboplatin or cyclophosphamide, doxorubicin, and cisplatin in women with ovarian cancer: the ICON3 randomised trial. Lancet. 360, 505-515 (2002).
- McGuire, W. P., Hoskins, W. J., Brady, M. F. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N. Engl. J. Med. 334, 1-6 (1996).
- Muggia, F. M., Braly, P. S., Brady, M. F. Phase III randomized study of cisplatin versus paclitaxel versus cisplatin and paclitaxel in patients with suboptimal stage III or IV ovarian cancer: a gynecologic oncology group study. J. Clin. Oncol. 18, 106-115 (2000).
- Piccart, M. J., Bertelsen, K., James, K. Randomized intergroup trial of cisplatin-paclitaxel versus cisplatin-cyclophosphamide in women with advanced epithelial ovarian cancer: three-year results. J. Natl. Cancer. Inst. 92, 699-708 (2000).
- Cannistra, S. A. Cancer of the ovary. N. Engl. J. Med. 351, 2519-2529 (2004).
- Baker, A. F., Dragovich, T., Ihle, N. T. Stability of phosphoprotein as a biological marker of tumor signaling. Clin. Cancer. Res. 11, 4338-4340 (2005).
- Espina, V., Edmiston, K. H., Heiby, M. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol. Cell. Proteomics. 7, 1998-2018 (2008).
- Klymkowsky, M. W., Savagner, P. Epithelial-mesenchymal transition: a cancer researcher's conceptual friend and foe. Am. J. Pathol. 174, 1588-1593 (2009).
- Kurrey, N. K., Jalgaonkar, S. P., Joglekar, A. V. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem. Cells. 27, 2059-2068 (2009).
- Creighton, C. J., Li, X., Landis, M. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. U. S. A. 106, 13820-13825 (2009).
- Aitken, S. J., Thomas, J. S., Langdon, S. P., Harrison, D. J., Faratian, D. Quantitative analysis of changes in ER, PR and HER2 expression in primary breast cancer and paired nodal metastases. Ann. Oncol. 21, 1254-1261 (2010).
- Sayagues, J. M., Abad, M. M., Melchor, H. B. Intratumoural cytogenetic heterogeneity of sporadic colorectal carcinomas suggests several pathways to liver metastasis. J. Pathol. 221, 308-319 (2010).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2025 MyJoVE Corporation. All rights reserved