A Visual Assay to Monitor T6SS-mediated Bacterial Competition
In This Article
Summary
We describe a qualitative assay to monitor bacterial competition mediated by the Pseudomonas aeruginosa type VI secretion system (T6SS). The assay relies on the survival/killing of Escherichia coli target cells carrying a lacZ-reporter. This technique is adjustable to assess the bactericidal/bacteriostasis activity of T6SS-proficient microorganisms.
Abstract
Type VI secretion systems (T6SSs) are molecular nanomachines allowing Gram-negative bacteria to transport and inject proteins into a wide variety of target cells1,2. The T6SS is composed of 13 core components and displays structural similarities with the tail-tube of bacteriophages3. The phage uses a tube and a puncturing device to penetrate the cell envelope of target bacteria and inject DNA. It is proposed that the T6SS is an inverted bacteriophage device creating a specific path in the bacterial cell envelope to drive effectors and toxins to the surface. The process could be taken further and the T6SS device could perforate other cells with which the bacterium is in contact, thus injecting the effectors into these targets. The tail tube and puncturing device parts of the T6SS are made with Hcp and VgrG proteins, respectively4,5.
The versatility of the T6SS has been demonstrated through studies using various bacterial pathogens. The Vibrio cholerae T6SS can remodel the cytoskeleton of eukaryotic host cells by injecting an "evolved" VgrG carrying a C-terminal actin cross-linking domain6,7. Another striking example was recently documented using Pseudomonas aeruginosa which is able to target and kill bacteria in a T6SS-dependent manner, therefore promoting the establishment of bacteria in specific microbial niches and competitive environment8,9,10.
In the latter case, three T6SS-secreted proteins, namely Tse1, Tse2 and Tse3 have been identified as the toxins injected in the target bacteria (Figure 1). The donor cell is protected from the deleterious effect of these effectors via an anti-toxin mechanism, mediated by the Tsi1, Tsi2 and Tsi3 immunity proteins8,9,10. This antimicrobial activity can be monitored when T6SS-proficient bacteria are co-cultivated on solid surfaces in competition with other bacterial species or with T6SS-inactive bacteria of the same species8,11,12,13.
The data available emphasized a numerical approach to the bacterial competition assay, including time-consuming CFU counting that depends greatly on antibiotic makers. In the case of antibiotic resistant strains like P. aeruginosa, these methods can be inappropriate. Moreover, with the identification of about 200 different T6SS loci in more than 100 bacterial genomes14, a convenient screening tool is highly desirable. We developed an assay that is easy to use and requires standard laboratory material and reagents. The method offers a rapid and qualitative technique to monitor the T6SS-dependent bactericidal/bacteriostasis activity by using a reporter strain as a prey (in this case Escherichia coli DH5α) allowing a-complementation of the lacZ gene. Overall, this method is graphic and allows rapid identification of T6SS-related phenotypes on agar plates. This experimental protocol may be adapted to other strains or bacterial species taking into account specific conditions such as growth media, temperature or time of contact.
Protocol
1. Bacterial Strains and Cultures
- Engineer an Escherichia coli recipient cell (Prey, P) by transforming (using standard CaCl2 treatment or electroporation)15 E. coli DH5α cells with a plasmid allowing the a-complementation of the lacZ gene (Table 1). Plate the transformed cells on Luria-Bertani agar plates (LBA, 1.5% agar) containing 5-bromo-4-chloro-indolyl-β-D-galactopyranoside (X-gal) at 40 mg/ml final concentration and appropriate antibiotic. Incubate at 37 °C overnight and the following morning select for blue transformants (see §1.3).
- Grow the Pseudomonas aeruginosa donor cells that are T6SS-active (D+) or T6SS-inactive (D-) (Table 1) on LBA overnight at 37 °C. In the example presented here we used a P. aeruginosa retS strain possessing a constitutively active H1-T6SS16 and an isogenic mutant with a deletion of the H1-T6SS gene cluster (Table 1).
- The next day, prepare an overnight liquid culture in aerated flask by inoculating a bright blue clone among the E. coli transformants (P) from the plate described in §1.1, in 5 ml of tryptic soy broth (TSB) supplemented with the appropriate antibiotic. Grow under agitation at 37 °C. Proceed equally with a single colony of (D+) and of (D-) from the plate described in §1.2.
2. Competition Assay
- Prepare LBA plates for the next day experiment. Make sure these plates are properly dried (either near the Bunsen burner or in laminar flow cabinet). Prepare an "Assay-input" (A-Input) plate for the strains (D+), (D-), and (P). Prepare an "Assay-output" (A-output) plate for the strains (D-+P) and (D++P). Divide and label the plates accordingly.
- After overnight growth (see §1.3), measure the optical density (OD600nm) of the input bacterial culture (D-, D+ and P) and calculate the volume required to obtain a cell density equivalent to 1 unit OD600nm of each strain. Each "input" cell culture (D-, D+ and P) is initially collected in a sterile 1.5 ml Eppendorf tubes.
- Centrifuge the bacterial samples at 13,000 rpm during 1 min at room temperature and discard the supernatant.
- Resuspend the pellets of the D-, D+, and P cultures, in 100 μl of fresh TSB by gentle pipetting and inoculate 10 ml of each corresponding strain as a single spot on the "A-input" plate (prepared in §2.1).
- Inoculate the "A-output" plate (prepared in §2.1). More precisely, mix gently 30 μl of (D+) with 30 μl (P) and 30 μl of (D-) with 30 ml (P) in two separate Eppendorf tubes (Note that the cultures used are those described in §2.4). Inoculate 20 μl of the mixes (D+/P) and (D-/P) as individual spots on the "A-output" plate. Allow the spots to dry nearby a Bunsen burner and place the plate in an incubator at 37 °C for an appropriate period of time during which the bacterial killing is taking place. In the case of P. aeruginosa, an efficient bacterial killing is observed after 5 hr incubation when comparing a T6SS active with a T6SS defective strain.
3. Qualitative Observation of the Bacterial Killing
- Prepare LBA plates containing 40 μg/ml X-gal for the readout of the assay. The plates will be called "Readout input" or "R-input" to spot isolated bacteria or "Readout output" or "R-output" to spot mixed bacterial cultures and thus read the killing performance. These plates need also to be properly dried. Divide the plate in four equal parts on its back and annotate these parts 0, 10-1, 10-2 and 10-3, designating the dilutions of bacterial culture to spot on the agar plates (see §3.3). The dilution will allow a semi-quantitative evaluation of the blue/white bacteria ratio within the same spot.
- Collect with a sterile loop the individual bacterial spots from the "A-input" plate (see §2.4) and "A-output" plate (see §2.5) and resuspend each spot in distinct 1.5 ml Eppendorf tubes containing 1 ml of TSB. Place in an Eppendorf shaker block during 30 min to resuspend efficiently the bacteria.
- Prepare a series of 5 times 3 Eppendorf tubes containing 900 μl of TSB and proceed for each resuspended spot to ten fold serial dilutions up to 10-3. Make sure to change pipette tips and to vortex 5 seconds between each dilution step.
- Proceed to the spotting of your bacterial dilutions prepared in §3.3 on well-dried LBA plates containing 40 μg/ml X-gal by starting with the most diluted to the undiluted suspension, making these the "R-input" plates for the "A-input" spots and the "R-output" plates for the "A-output" spots (Figure 2). Vortex briefly each tube before proceeding to the spotting to keep a homogenous bacterial suspension. For reproducibility of the results, spot 20 μl in triplicate within a quadrant (Figure 2). The relative dryness of your plate is important at this stage since the spots need to remain individually separated on the plate and not glide towards each other.
A quantification of the competition assay is also possible at this stage of the experiment. Following the step 3.3, spread 100 μl of the dilution 10-3 on LBA plates containing 40 μg/ml X-gal. For reproducibility of the results, proceed to a plating in triplicate for each spot from the "A-output" plate. Place the dilution plates in a 37 °C incubator overnight (corresponding to 16 hr incubation). Proceed to the counting of the blue colonies (the P cells that remained alive). Typical results obtained after 5 hr of competition are shown in Figure 3. - Leave the plate open (no lid) within a sterile zone to allow the absorption of the liquid in excess within each spot.
- Place the plate in a 37 °C incubator overnight. During this incubation time, the killing is still occurring in the mix (D+/P) since both strains are still in contact.
- Take a picture or scan your plates for the output analysis (Figure 2). Where spots remain largely blue it indicates that E. coli has not been killed by P. aeruginosa. That is the case when E. coli is mixed with a T6SS-defective P. aeruginosa strain (D-) (Figure 2, plate at the bottom right).
Representative Results
Typical results are shown in Figure 1 with the strains and reagents described in Table 1. The plates shown in this figure were scanned after an overnight incubation. The "Readout-Input" plates show a serial dilution pattern for the strains used in this assay. As expected, the E. coli prey spots (P) overexpressing the lacZ gene appear blue on media supplemented with X-gal, while the donor P. aeruginosa strains (D+, T6SS active) and (D-, T6SS inactive) remain white. The "Readout-output" plates on which the mix between the prey and a T6SS active strain (D+/P) has been spotted show the disappearance of the blue prey thus indicating it has been killed. This demonstrates the ability of the donor to outcompete the prey. The persistence of the blue color on the (D-/P) plate demonstrates the inability of an inactive T6SS donor to kill the blue prey.
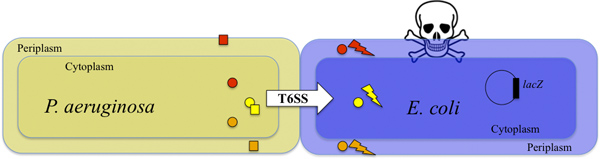
Figure 1. Killing of E. coli by T6SS-proficient P. aeruginosa. P. aeruginosa injects toxins into the E. coli target cell in a T6SS-dependent manner (shown by the white arrow). Two toxins Tse1 and Tse3 (orange and red circles) are injected into the E. coli periplasm and degrade the peptidoglycan9. The Tse2 toxin (yellow circle) is injected into the E. coli cytoplasm and has a bacteriostatic activity8,10. The combined action of the toxins kills the target cells (flash of lightning and skull). The survival of target cells can be detected by monitoring the activity of the produced β-galactosidase (see also Figure 2). P. aeruginosa is protected against the activity of the toxins by the immunity proteins Tsi1, Tsi2 and Tsi3 (orange, yellow and red squares, respectively)8,9,10.
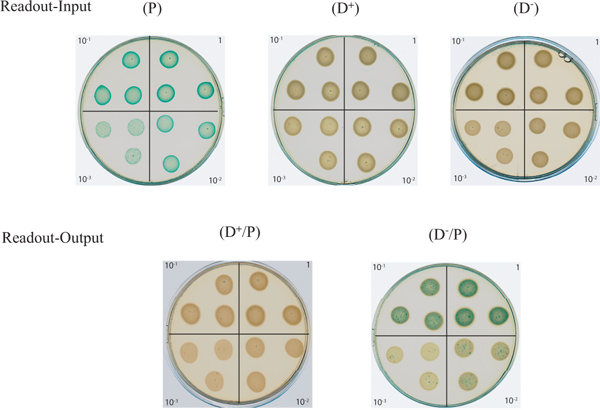
Figure 2. Agar plate assay to monitor T6SS-dependent bacterial killing. In this figure are shown on the upper part the "Readout-Input" plates consisting of the serial dilutions of the D-, D+, and P input cells. The P input cells are blue due to the α-complementation of the lacZ gene and thus produced β-galactosidase that cleaves X-gal. In the lower part is shown the "Readout-Output" plates consisting of serial dilutions of the bacterial mix between an active (D+/P),or an inactive (D-/P), T6SS donor P. aeruginosa strain with the E. coli prey. Click here to view larger figure.
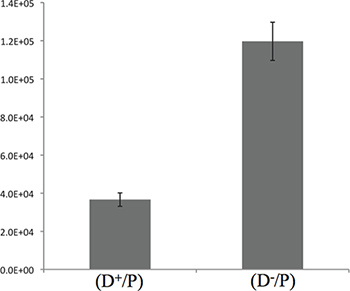
Figure 3. Quantification of the killing of E. coli by P. aeruginosa after 5 hr incubation. The graph presents the CFU counting of E. coli described in the 3.4 step. The results presented here show a 3-fold difference between the T6SS+ and the T6SS- strains, suggesting that most of the killing is taking place during the 5 initial hours of contact.
Discussion
The method presented in this article allows a visual observation of T6SS-mediated bactericidal/bacteriostasis activity. The assay is performed on the surface of an agar plate. It has been previously shown that T6SS-dependent killing assay performed with mixed bacterial liquid culture is not efficient, likely because of the lack of steady contact between the two bacteria8. The T6SS is believed to operate with a mechanism akin to the one used by bacteriophages to inject DNA into target cells17. In liquid culture, the tube-like structure of the T6SS may break more easily, inter-bacterial contact can be lost, and the toxins are not efficiently delivered.
In terms of incubation times, the 5 initial hours of contact that we describe between the donor strain and the prey are sufficient to observe bacterial killing between P. aeruginosa and E. coli, as illustrated in Figure 3. Nevertheless, it is advisable to adjust the incubation time by performing a kinetic in order to optimize the experimental conditions.
Since this method is a color based technique, the output results can be compromised by the pigmentation of the donor strain. For instance, in the case of P. aeruginosa, some strains produce high levels of colored pigments such as pyocyanin and pyoverdine, which can interfere with the assay readout, making the distinction from the prey relatively difficult. Other chromogenic β-galactosidase substrates, such as the magenta-gal or the red-gal, can be used instead of the X-gal (Table 1).
The competition assay can make use of other reporter genes for the readout. For instance, similar assay has also been performed by using green fluorescent protein-labeled preys12.
Our assay, while not quantitative, gives a good indication of the T6SS activity since it is based on the survival or the killing of a reporter prey. This technique presents the advantage of being easy and convenient to evaluate the bactericidal/bacteriostasis activity of T6SSs from any bacterial species. So far, the activity of the T6SS has been shown against Gram-negative bacteria and no clear example of T6SS-sensitive Gram-positive bacteria has been reported yet12. It is also obvious that incompatibility in the culture of the different bacterial species to test (e.g. growth temperature, oxygenation, specific media) is to be considered.
Our assay can also be used to evaluate which of the T6SS components are absolutely essential since even traces of a secreted toxin might be sufficient to kill the prey. Even weak activity of the T6SS could then clearly be detected by our assay as compared to standard procedure testing T6SS-dependent secretion using culture supernatant and western blot analysis. However, a proper colony-forming unit (CFU) counting is still required for accurate quantification of this T6SS activity.
Acknowledgements
This work was funded by the Wellcome trust grant WT091939MA. Alain Filloux is supported by the Royal Society.
Materials
| Name | Company | Catalog Number | Comments |
| Name of Reagent/Material | Company | Catalogue Number | Comments |
| P. aeruginosa PAKΔretS (D+) (active T6SS) | Lab strain | Described in Reference 16 | |
| P. aeruginosa PAKΔretSΔH1-T6SS (D-)(inactive T6SS) | This study | The H1-T6SS cluster (encompassing the genes PA0070 to PA0095) has been deleted by allelic exchange following the procedure described in Reference 18. The mutator fragment was generated with the following set of primers: The Up fragment primers: 5'-ATGGTCAACGACATGGAGCTGGAG-3', and 5'CGAGGCCGATCAGGCCTTCAGAACTGA-3'. The Down fragment primers : 5'-TCAGGCCTTCAGAACTGAAGCGGCGCA-3', 5'-GGTGGCGTTCAACAGTTCCATGTC-3' | |
| E.coli DH5α | Invitrogen | 18258-012 | F- φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rk-, mk+) phoA supE44 λ- thi-1 gyrA96 relA1 |
| pBluescript II SK(+) | Agilent | 212205 | This vector expresses the α peptide of β-galactosidase used for α-complementation. |
| X-gal | Invitrogen | 15520-018 | Use at 40 μg/ml |
| Luria Bertani agar | Merck-chemicals | 1.10283.0500 | |
| TSB (casein soya bean broth) | Oxoid | CM109 | |
| Vortex shaker Genius 3 | IKA | 3340000 | |
| Scanner | Epson | V700 | |
| Spectrophotometer | WPA Biowave | CO8000 Cell Density Meter | |
| Magenta-gal | Bioworld | 30350001-1 (715241) | |
| Red-gal | Research organics | 1364c | |
Table 1. Strain, plasmid, material and reagent used. |
References
- Filloux, A., et al. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 154 (6), 1570 (2008).
- Cascales, E., Cambillau, C. Structural biology of type VI secretion systems. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 367 (1592), 1102 (2012).
- Leiman, P. G., et al. Morphogenesis of the T4 tail and tail fibers. Virol. J. 7, 355 (2010).
- Ballister, E. R., et al. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl. Acad. Sci. U.S.A. 105 (10), 3733 (2008).
- Leiman, P. G., et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106 (11), 4154 (2009).
- Pukatzki, S., et al. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104 (39), 15508 (2007).
- Ma, A. T., et al. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe. 5 (3), 234 (2009).
- Hood, R. D., et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 7 (1), 25 (2010).
- Russell, A. B., et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 475 (7356), 343 (2011).
- Li, M., et al. Structural basis for type VI secretion effector recognition by a cognate immunity protein. PLoS Pathog. 8 (4), e1002613 (2012).
- Zheng, J., et al. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One. 6 (8), (2011).
- Schwarz, S., et al. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6 (8), e23876 (2010).
- Murdoch, S. L., et al. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J. Bacteriol. 193 (21), 6057 (2011).
- Boyer, F., et al. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources?. BMC Genomics. 10, 104 (2009).
- Maniatis, T., et al. . Molecular cloning: A Laboratory Manual. , (1982).
- Mougous, J. D., et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 312 (5779), 1526 (2006).
- Records, A. R., et al. The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol. Plant Microbe Interact. 24 (7), 751 (2011).
- Hachani, A., et al. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J. Biol. Chem. 286 (14), 12317 (2011).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved