BEST: Barcode Enabled Sequencing of Tetrads
In This Article
Summary
Barcode Enabled Sequencing of Tetrads (BEST) replaces the manual processes of isolating, disrupting and spacing tetrads. BEST isolates tetrads by fluorescence-activated cell sorting onto agar plates, separates the spores by agitation with glass beads, and determines which randomly arrayed colonies were derived from the same original tetrad using molecular barcodes.
Abstract
Tetrad analysis is a valuable tool for yeast genetics, but the laborious manual nature of the process has hindered its application on large scales. Barcode Enabled Sequencing of Tetrads (BEST)1 replaces the manual processes of isolating, disrupting and spacing tetrads. BEST isolates tetrads by virtue of a sporulation-specific GFP fusion protein that permits fluorescence-activated cell sorting of tetrads directly onto agar plates, where the ascus is enzymatically digested and the spores are disrupted and randomly arrayed by glass bead plating. The haploid colonies are then assigned sister spore relationships, i.e. information about which spores originated from the same tetrad, using molecular barcodes read during genotyping. By removing the bottleneck of manual dissection, hundreds or even thousands of tetrads can be isolated in minutes. Here we present a detailed description of the experimental procedures required to perform BEST in the yeast Saccharomyces cerevisiae, starting with a heterozygous diploid strain through the isolation of colonies derived from the haploid meiotic progeny.
Introduction
Meiotic gene mapping, commonly known as tetrad analysis, is central to yeast genetics. Briefly, two haploid strains, each containing a single copy of each chromosome, are mated to form a heterozygous diploid strain with two copies of each chromosome, one from each parent. Starving diploid cells for nitrogen induces meiosis (sporulation) in which chromosomes duplicate, undergo rearrangement, and divide into four haploid cells (spores or segregants) with new combinations of alleles from each parent. The four spores resulting from a single meiosis can be isolated by a manual process called tetrad dissection.
Conventional tetrad dissection2,3 which has changed relatively little since the original publication in 19374, has three goals. First, cells that have undergone meiosis (tetrads) must be isolated away from a background of vegetative cells. In conventional analysis, this is accomplished by a researcher who, using a microscope, visually identifies tetrads on an agar plate and employs a micromanipulator apparatus visually identifying tetrads on an agar plate and using a micromanipulator to move the tetrad onto a cell-free area of the plate. Next, the four spores in the tetrad must be physically separated to prevent interspore mating. Spores within a tetrad are held together by both an ascus, the remnant of the cell wall of the original diploid cell, and a set of interspore bridges5. In conventional tetrad analysis the ascus is removed by enzymatic digestion, and a researcher uses the micromanipulator to break the interspore bridges and tease apart the spores. Finally, the individual spores are arrayed in a gridded pattern that maintains the relationship between the spores, i.e. all progeny in a row of four are sister spores of the same original tetrad. An experienced yeast researcher can complete the process of dissecting 10 tetrads (a generally accepted minimum number per cross) in approximately 15 min.
We have developed a new high-throughput tetrad genotyping method, which we call Barcode Enabled Sequencing of Tetrads (BEST)1. BEST expands upon current methods in high-throughput genetics by enabling the generation and genotyping of large numbers of progeny that are isolated, genotyped, and maintained as individuals in a manner that allows the sister spore relationships of all four meiotic products to be recovered. This is accomplished using three main strategies (Figure 1). First is the introduction of a reporter construct that labels cells that have undergone meiosis with GFP and allows them to be isolated by fluorescence-activated cell sorting (FACS). Second is the incorporation of a highly complex DNA barcode (a string of 15 random nucleotides) in a form that is transmitted to all four spores of a tetrad and can be read by DNA sequencing of the recombinant progeny and thus identifies sister spores from the same tetrad. Third is the genotyping step, and BEST is compatible with numerous genotyping platforms that capture both a set of genomic markers as well as the tetrad-specific barcode. We employ a restriction site-associated DNA tag sequencing (RAD-seq)6 method that directs genome sequencing to specific restriction sites1, thereby capturing a consistent 2-3% of the genome of the progeny strains as well as the plasmid-borne barcode. The recovery of tetrad relationships along with the empirically-derived genotyping data from the cross allows the accurate inference of missing information, including the status of markers with low sequence coverage and the complete genotype of inviable (and therefore unrecoverable) individuals. Here, we describe the application of the method in the most commonly used microorganism for meiotic mapping, the yeast S. cerevisiae. However, with minor substitutions of organism-specific reagents, e.g. different sporulation-specific proteins fused to GFP, the method should be readily transferrable to other microorganisms, including organisms in which meiotic mapping is significantly more labor intensive or currently intractable.
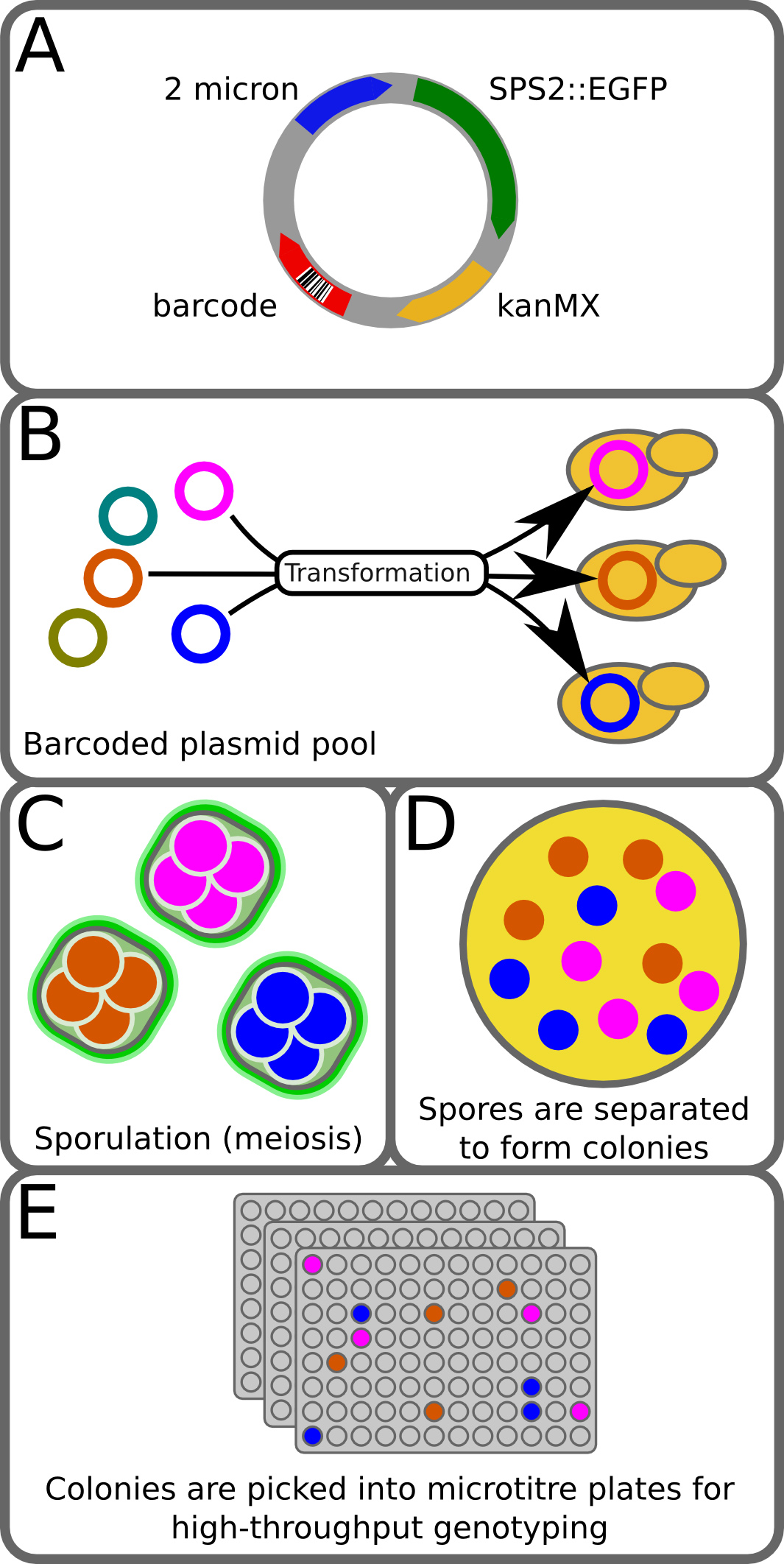
Figure 1. BEST method. (A) The pCL2_BC plasmid is a 2-micron plasmid with a sporulation-specific GFP fusion, resistance to G418, and a 15-mer random barcode. Construction of the plasmid library is described in Ludlow et al1. (B) A pool of barcoded plasmids is transformed into the diploid strain from a cross. (C) Transformants are then grown on selection and ~10,000 transformed colonies are pooled and sporulated. During meiosis each spore of a tetrad inherits a copy of the barcoded plasmid. (D) Tetrads are separated from unsporulated cells by FACS and collected on agar plates, where they are digested and disrupted to allow each spore to form a colony. (E) Colonies are then picked into 96-well plates, phenotyped, and genotyped. During genotyping the plasmid barcode is read and used to identify the four members of each tetrad. This figure has been modified from Ludlow et al1. Please click here to view a larger version of this figure.
Protocol
1. Strain Construction and Characterization
- Construct the heterozygous diploid strain to be used in the cross by mating haploid strains and selecting diploids2 or using a naturally occurring heterozygous strain. The strains should not harbor GFP.
- Confirm that the strain is unable to grow on YPD2 + 200 µg/ml G418.
- Confirm that the strain is able to sporulate. BEST is compatible with low sporulation efficiency crosses, but low sporulation frequency increases FACS sorting time required.
- Set up an O/N culture of the strain in 5 ml YPD and grow with agitation at 30 °C.
- Wash 0.5 ml of the O/N culture three times with PBS (pH 7.4).
- Pellet the washed cells, remove the supernatant, and resuspend in 2 ml sporulation medium7.
- Agitate cells at RT and check daily for tetrad formation using a phase contrast light microscope with a 40X objective.
- Estimate the spore viability of the cross by conventional dissection2,3 of a small number (~20) of tetrads. Spore viability in crosses between different strain backgrounds varies widely and unpredictably. However, a priori expectations of spore viability can be used to evaluate the efficiency of a BEST experiment prior to the labor- and cost-intensive steps of saving and genotyping the strains. Therefore, this step is highly recommended.
2. Transformation of Diploids with Library of Barcoded Plasmids
- Insert the pCL2_BC barcoded plasmid into the heterozygous diploid strain using the transformation protocol described in Gietz and Woods8 with the following modifications.
- After cells have been incubated with the transformation mixture at 30 °C for 20 min, add DMSO to 8% of the volume of the transformation mix. Addition of DMSO has been found to improve transformation efficiency in some strain backgrounds9.
- After the heat shock step, pellet the cells with a brief centrifugation, decant the transformation mixture, and wash cells gently in YPD. Then, resuspend the cells in 1 ml YPD and allow them to recover at RT for 3 hr without agitation.
- Select transformants by plating onto YPD + G418 (200 µg/ml) agar plates. To ensure a complex pool of plasmid barcodes, harvest a large number of colonies, e.g. 5 plates with ~2,500 colonies per plate.
- Once colonies are visible (2-3 days of growth at 30 °C), pool the transformants by carefully scraping them off the plate into liquid YPD + G418 (200 µg/ml) + 15% glycerol. Vortex this cell suspension, transfer to a cryopreservation vial, and freeze at -80° C. Make a separate frozen stock for each plate. These frozen stocks serve as the starting strains for Step 3.1 and can also be used to generate more tetrads at a later date, if desired.
3. Sporulating the Library of Transformed Cells to Obtain Barcoded Tetrads
- Revive cells from the frozen stocks created in 2.3 by O/N growth in 5 ml YPD + G418 (200 µg/ml) at 30 °C with agitation.
- Wash the cells from the O/N cultures 3x in PBS (pH 7.4) and then resuspend the cells in sporulation medium7 plus 200 µg/ml G418.
- Agitate sporulation cultures at RT and monitor sporulation progress daily until new tetrads have stopped forming.
- Once the culture contains well-formed tetrads and relatively few dyads, remove cultures from agitation and leave at RT for at least 7 days. While some crosses may be amenable to on plate digestion and disruption as soon as tetrads have formed (assessed as described in 7.3), the additional incubation (7-10 days in our experiments) is essential for other crosses. Therefore, this step is highly recommended.
4. Establish the FACS Gates for Tetrad Sorting
The meiotically expressed SPS2-GFP fusion on the pCL2_BC plasmid allows tetrads in the sporulation culture to be detected and isolated from the culture by fluorescence. The following protocol has been developed for a FACSAria II.
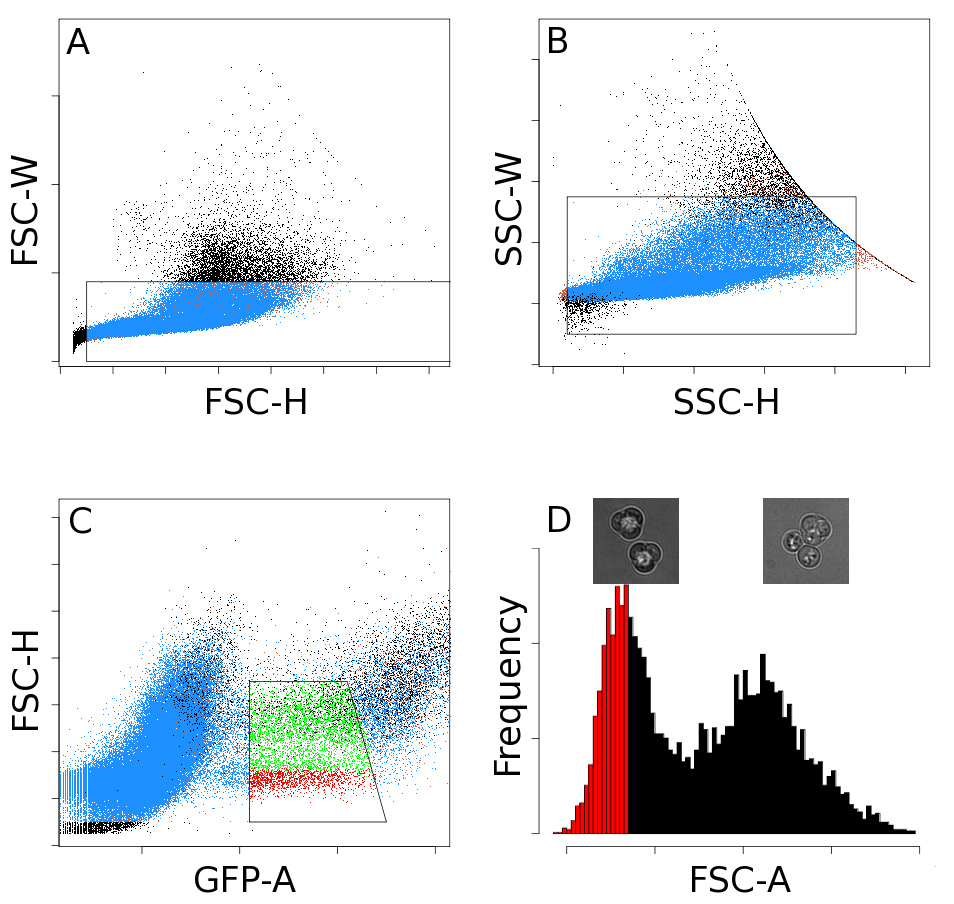
Figure 2. FACS gates. A series of four sequential gates is used to isolate tetrads. The gates shown in (A) and (B) help reduce the number of clumps. Fluorescent events are selected with the gate in (C). The histogram in (D) has two peaks. Events in the left peak are primarily tetrads (left inset image), and events in the right peak are generally tetrads with an attached bud (right inset image). A final gate is applied to this histogram to select events which are mostly tetrads (red bars). This figure has been modified from Ludlow, et al1.
- Load the sporulated culture onto a FACS sorter and set up two forward scatter and side scatter gates to exclude debris, clumped cells, and multiple cells per droplet (Figures 2A and 2B).
- There should be a population of fluorescent cells, which when using the SPS2-GFP reporter construct include dyads and tetrads10. Gate the GFP vs. FSC to include these fluorescent events (Figure 2C).
- Depending on the strain background it may be necessary to include an additional gate to remove clumps that include tetrads and other cells. If this is necessary, inspect an FSC histogram of the fluorescent events and specify a gate that includes events with lower FSC (Figure 2D, gated events are in red).
- Check the efficiency of the gating regime by sorting ~1,500 tetrads onto a microscope slide and counting the proportion of tetrads to non-tetrads using a phase contrast microscope at 400X magnification. The percent of tetrads can be accurately estimated by counting only ~200 sorted events, but sorting ~1,500 reduces the amount of time spent searching for cells on the microscope slide. Alternatively, the ratio of tetrads to non-tetrads can be assessed during Step 4.5.
- Periodically check that the sorter is accurately reporting the number of events by sorting 25 tetrads onto a microscope slide and examining the cells contained in the droplet using a phase contrast microscope at 400X magnification. There should be 25 tetrads in the droplet. Repeat 3-4x to assess the variance of the FACS instrument.
5. Sorting Tetrads Onto an Agar Plate
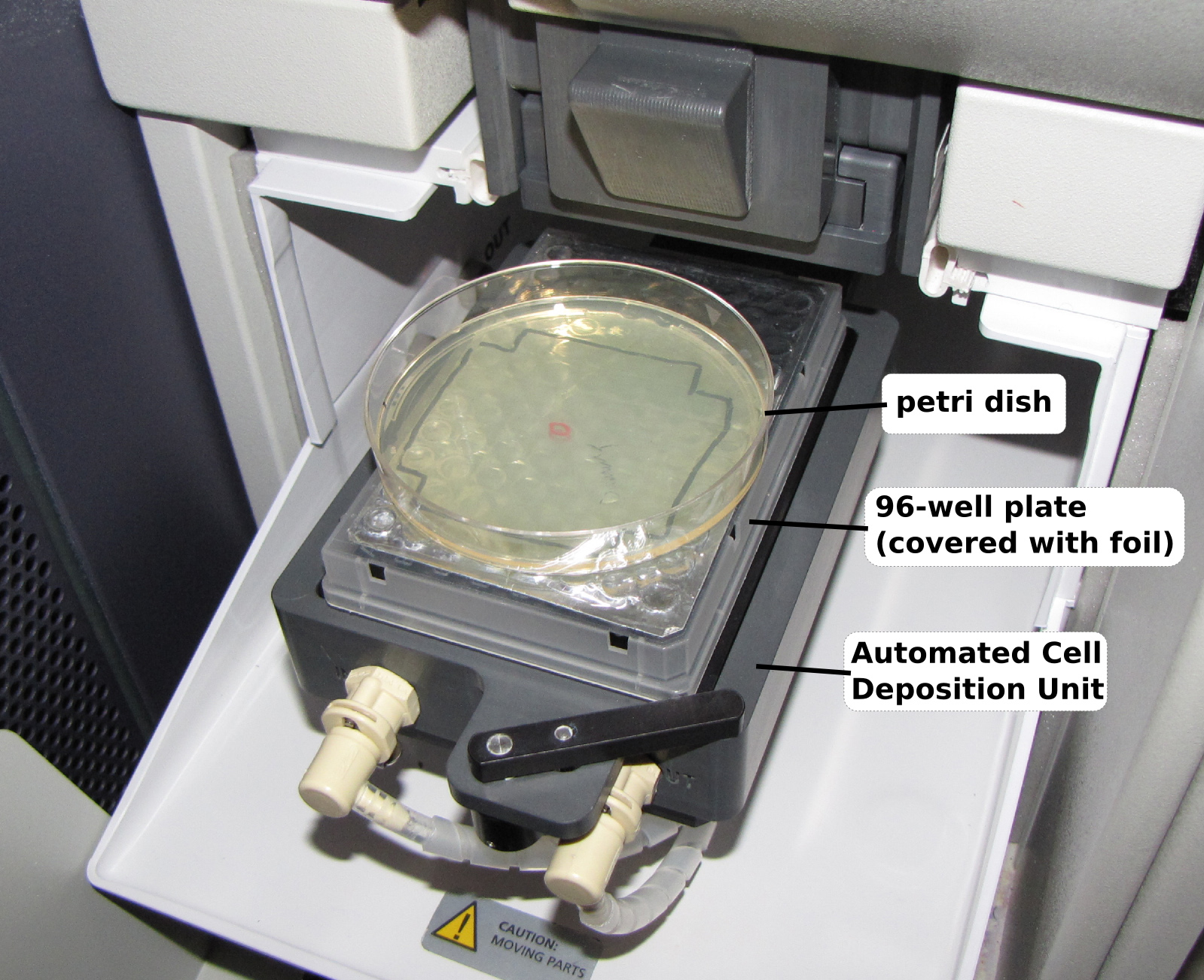
Figure 3. Placement of the agar plate on the FACS sorter. To sort tetrads into a small drop of the zymolyase solution placed in the middle of a standard (circular) agar plate, a standard (100 mm rectangular) 96-well plate is first placed on the stage of the cell sorter and the sorter is programmed to deposit tetrads into a specific well (in this case D5). Then, the circular agar plate is placed on top of the 96-well plate and aligned so that the drop of zymolyase is centered over the specified well.
- Warm the desired number of YPD + G418 (200 µg/ml) plates for at least 1.5 hr in a 37 °C incubator positioned near the FACS sorter.
- A key feature of the sorting protocol is the ability to accurately target FACS sorted tetrads to a specific location on an agar plate that contains a pool of zymolyase solution. To do this, place a “landmark” 96-well plate on the automated cell deposition unit (ACDU) of the sorter. Prepare a sort layout that will sort 25 tetrads into one well of a 96-well format. Choose a well near the middle of the plate, e.g. well D5. Figure 3 shows a Petri dish and 96-well plate loaded onto a cell sorter. Sorting 25 tetrads per plate yields well-separated colonies.
- Bring a stack of 10 agar plates from the 37 °C incubator to the sorter.
- Take one agar plate and pipette 25 μl of zymolyase solution (1 mg/ml in 0.7 M sorbitol) onto the agar near the center of the plate. Then, place the plate on the landmark 96-well plate, aligning the zymolyase droplet over the target well from Step 5.2, e.g. well D5.
- Start the sort and deposit 25 tetrads into the middle of the zymolyase droplet on the agar plate. Then, remove and cover the plate.
- Repeat Steps 5.4 and 5.5 until 25 tetrads have been sorted into zymolyase droplets on each plate in the stack.
- Return the stack of tetrad-containing plates to the 37 °C incubator.
- Repeat Steps 5.3 through 5.7 until all plates have tetrads.
6. Separating Spores
- Incubate each stack of tetrad-containing agar plates at 37 °C for 1.5 hr.
- Remove plates from the incubator, add 15-25 sterile 3 mm glass beads per plate, and shake the plates as a stack (or as two stacks of 5 plates) for 3-5 min. The best spore separation is achieved by moving the plates rigidly from side to side or front to back. Do not move the plates such that the beads "swirl" around the outside edge of the plate. Leave the beads on the plates when finished.
- Place the stack of plates with beads face up in a 30 °C incubator. Retrieve the next stack plates from the 37 °C incubator, and repeat Step 6.2.
- Incubate plates O/N at 30 °C.
7. Harvesting Colonies
- After an O/N incubation at 30 °C, small colonies should have formed on the plates. Carefully remove the plates from the incubator without disturbing the glass beads.
- Remove the glass beads by carefully and quickly inverting the plates over a deep container. The container should be deep enough to catch the beads without allowing them to bounce up and hit the agar plate. Tapping the bottom of the plate while it is inverted can help dislodge beads that are stuck to the plate. These beads can be reused by thorough washing with soap, rinsing well with deionized water and sterilizing by autoclaving.
- Count the number of colonies on each plate and use the number of colonies to estimate the efficiency of spore separation and recovery. For example, 25 tetrads per plate for a cross with near 100% viability could yield a total of 100 colonies per plate.
- From plates with sufficient numbers of colonies (>65 for a high-viability crosses), transfer cells from each colony into a separate well of a 96-well plate containing YPD liquid + G418. Also, make note of which wells contain colonies from the same agar plate; this information will be used to aid the tetrad assignment software.
- Use the 96-well liquid plates to seed cultures for genotyping and for saving strains as frozen glycerol stocks. Details on RAD-tag sequencing and sequence analysis can be found in Ludlow et al1.
Results
BEST can greatly improve the speed with which spores from tetrads can be isolated. As an example of the throughput this method can deliver, one researcher performed the FACS sorting and spore separation (Steps 5.1 through 6.3) with 149 agar plates in 3 hr1. An equivalent yield (approximately 3,725 tetrads) would require more than 90 hr of manual dissection. However, as with many high throughput methods, the current iteration of the method has some loss of efficiency compared to manual dissection. This loss manifests as a reduced number of colonies recovered compared to the expectation calculated based on the spore viability of the cross, determined by manual dissection (Step 1.4). Assuming that the causes of spore loss (Discussion) affect spores at random, estimations can be made of how many recovered colonies will be members of tetrads in which 1, 2, 3, or 4 spores have been recovered (Figure 4).
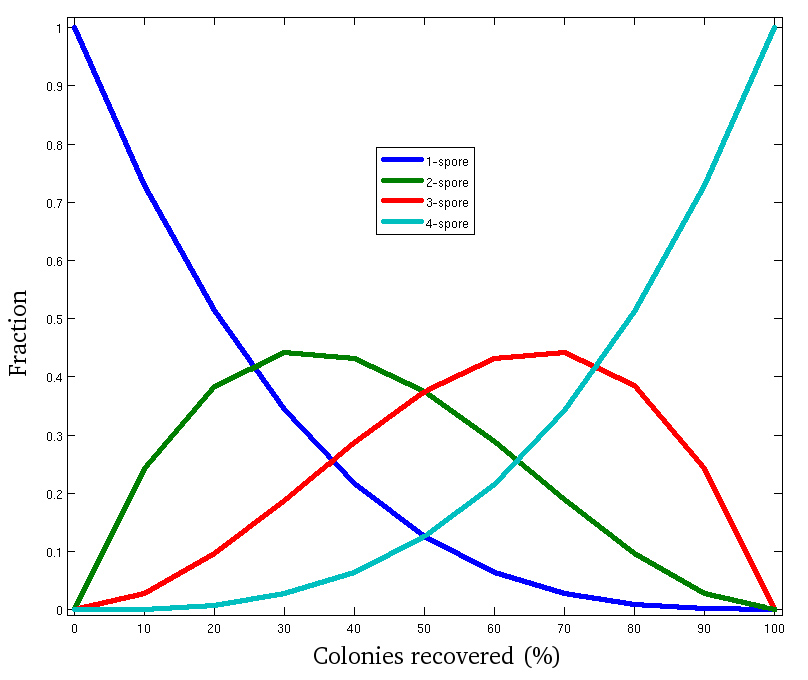
Figure 4. Simulation of method efficiency at the tetrad level. Failure to successfully recover spores (as colonies) may be due to spore inviability, spore adhesion to the glass beads, failure to separate spores from one another, etc. The Binomial distribution was used to calculate the expected fraction of tetrads (with at least one successfully recovered spore) producing 4, 3, 2 or 1 colonies based on the proportion of all spores successfully recovered. This graph can serve as a rough guide to the number of strains that will ultimately be assembled into 3- or 4- spore tetrads. As the number of colonies per plate increases, so does the number of complete tetrads recovered.
In the experiment described above, 25 tetrads from a cross with spore viability close to 100% (as determined by manual dissection)1 were plated on 149 agar plates. In this experiment, the maximum number of colonies observed per plate was 80, indicating that 20 spores failed to form discrete colonies. The mean number of colonies per plate was 62. Colonies from the 61 plates containing 65 colonies or more were harvested and sequenced. Of the 3,700 strains that passed sequencing quality control, 72% could be computationally assigned into 3- or 4- spore tetrads.
Discussion
Many areas of genetics research, ranging from complex trait mapping11 to the study of mechanisms underlying DNA recombination12, require extremely large numbers of progeny and high volumes of DNA sequencing. Techniques that marry the power of conventional approaches with the capacity of high throughput DNA sequencing have the potential to enable studies that were previously intractable. Although several high-throughput yeast genetics methods have been applied effectively to specific problems, each has limitations that fall short of the broad applicability of conventional tetrad analysis. BEST addresses these challenges by employing a high-throughput approach combining tetrad dissection and genotyping the progeny of yeast crosses. In conjunction with multiplexed RAD-tag sequencing, BEST offers an inexpensive way to glean genotype information for each progeny strain while preserving tetrad relationships by means of unique tetrad barcodes. Of all currently used high throughput methods, BEST most closely recapitulates the information provided by a manually dissected yeast cross.
BEST has two key features. The first is the use of FACS to rapidly isolate tetrads away from unsporulated cells in the culture (Protocol Steps 5-6). The ability to isolate thousands of fluorescent events (tetrads) gives BEST its greatest gain in throughput. This automation provides an even greater advantage over manual dissection for crosses with low sporulation efficiency, due to the increased time needed to visually identify rare events. The second key feature is the use of a highly complex molecular barcode (Protocol Step 2) that is transmitted to each sister spore of a tetrad. Because these barcodes can be used to computationally reconstruct the tetrad relationships between spores that have been randomly distributed across an agar plate, the need to manually array cells in an ordered grid is alleviated. Finally, because the molecular barcode and the sporulation-specific GFP reporter gene are physically linked (on the same plasmid), only tetrads that have retained the molecular barcode are selected by FACS sorting.
There are two considerations that govern the length of time that cultures should be incubated in the sporulation medium. First, because the sporulation specific reporter (SPS2-GFP) is expressed early in meiosis, GFP alone does not distinguish complete 4-spore tetrads from cells that have not completed meiosis (i.e. dyads). While additional FACS gating may decrease the number of fluorescent, incomplete tetrads, the easiest way to decrease these unwanted events is simply monitoring the sporulation culture (Step 3.3) and proceeding once new tetrads have stopped forming. In our experiments, the frequency of dyads sorted is less than 1%. Second, for some crosses additional time spent in sporulation medium at RT without agitation (Step 3.4) significantly improves the separation of tetrads by the glass beads during plating ((Protocol Step 6). In fact, this step is absolutely necessary for the disruption of some crosses.
Like all high throughput methods, BEST is “noisier” than the corresponding conventional approach. The modest reduction in efficiency of BEST compared to conventional tetrad dissection could result from combinations of several factors. One factor is the increased loss of otherwise viable spores, which could result from adhesion to the glass beads during spreading or increased spore death due to the mechanical stresses of the process. A second factor could be simple plasmid loss during meiosis. A third factor could be an effective decrease in usable colonies resulting from colonies with mixed genotypes, an estimated ~5% of colonies in our pilot crosses1. It is likely that these colonies represent failures of sister spore separation. Because the rapid fluorescence sorting and spore separation permit the collection of enormous numbers of tetrads, BEST is well equipped to overcome decreases in efficiency on this scale. While future iterations of BEST could increase efficiencies approaching that of manual dissection, it is perhaps more likely that the current exponential trajectory of DNA sequencing capacity will rapidly compensate for this level of usable strain loss. Regardless, the ability to perform tetrad analysis on the scale to which BEST can be applied will enable studies that are currently unfeasible by manual methods.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by an NIH/NHGRI Genome Scholar/ Faculty Transition Award (K22 HG002908) to A.M.D. and a strategic partnership between the Institute for Systems Biology and the University of Luxembourg. Requests for strains or plasmids should be directed to Aimée Dudley (aimee.dudley@gmail.com).
Materials
| Name | Company | Catalog Number | Comments |
| FACSAria II (Special order unit) | BD Biosciences | - | Other cell sorters should work for this procedure. The key factors are: (1) a laser/detection system for the fluorescent reporter being used (488nm laser to excite EGFP in our case), (2) the ability to sort single events, (3) the ability to sort onto microscope slides for assessing the sort quality and (4) the ability to sort onto petri dishes (which may need |
| Automated Cell Deposition Unit (for FACSAria II) | BD Biosciences | 643155 (if purchased separately from the FACSAria instrument) | |
| Zymolyase 100T | Seikagaku Biobusiness | ||
| G418 | A.G. Scientific | http://www.agscientific.com/molecular-biology/gene-expression/g-418-sulfate-solid.html | |
References
- Ludlow, C. L., et al. High-throughput tetrad analysis. Nat Methods. 10, 671-675 (2013).
- Rose, M., Winston, F., Hieter, P. . Methods in Yeast Genetics A Laboratory Course Manual. , (1990).
- Morin, A., Moores, A. W., Sacher, M. Dissection of Saccharomyces cerevisiae asci. J Vis Exp. 27, (2009).
- Winge, O., Laustsen, O. On two types of spore germination, and on genetic segregations in Saccharomyces demonstrated through single spore cultures. C.R. Trav. Lab. Carlsberg Ser. Physiol. 24, 263-315 (1937).
- Coluccio, A., Neiman, A. M. Interspore bridges: a new feature of the Saccharomyces cerevisiae spore wall. Microbiology. 150, 3189-3196 (2004).
- Baird, N. A., et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 3, (2008).
- Tong, A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294, 2364-2368 (2001).
- Gietz, R. D., Woods, R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87-96 (2002).
- Hill, J., Donald, K. A., Griffiths, D. E. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 19, 5791-57 (1991).
- Gerke, J. P., Chen, C. T., Cohen, B. A. Natural isolates of Saccharomyces cerevisiae display complex genetic variation in sporulation efficiency. Genetics. 174, 985-997 (2006).
- Bloom, J. S., Ehrenreich, I. M., Loo, W. T., Lite, T. L., Kruglyak, L. Finding the sources of missing heritability in a yeast cross. Nature. 494, 234-237 (2013).
- Fogel, S., Mortimer, R., Lusnak, K., Tavares, F. Meiotic gene conversion: a signal of the basic recombination event in yeast. Cold Spring Harb Symp Quant Biol. 43 Pt 2, 1325-1341 (1979).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved