Immunohistochemical Detection of 5-Methylcytosine and 5-Hydroxymethylcytosine in Developing and Postmitotic Mouse Retina
In This Article
Summary
The objective of this report is to describe the protocol for robust immunohistochemical detection of epigenetic markers, 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) in developing and postmitotic mouse retina.
Abstract
The epigenetics of retinal development is a well-studied research field, which promises to bring a new level of understanding about the mechanisms of a variety of human retinal degenerative diseases and pinpoint new treatment approaches. The nuclear architecture of mouse retina is organized in two different patterns: conventional and inverted. Conventional pattern is universal where heterochromatin is localized to the periphery of the nucleus, while active euchromatin resides in the nuclear interior. In contrast, inverted nuclear pattern is unique to the adult rod photoreceptor cell nuclei where heterochromatin localizes to the nuclear center, and euchromatin resides in the nuclear periphery. DNA methylation is predominantly observed in chromocenters. DNA methylation is a dynamic covalent modification on the cytosine residues (5-methylcytosine, 5mC) of CpG dinucleotides that are enriched in the promoter regions of many genes. Three DNA methyltransferases (DNMT1, DNMT3A and DNMT3B) participate in methylation of DNA during development. Detecting 5mC with immunohistochemical techniques is very challenging, contributing to variability in results, as all DNA bases including 5mC modified bases are hidden within the double-stranded DNA helix. However, detailed delineation of 5mC distribution during development is very informative. Here, we describe a reproducible technique for robust immunohistochemical detection of 5mC and another epigenetic DNA marker 5-hydroxymethylcytosine (5hmC), which colocalizes with the "open", transcriptionally active chromatin in developing and postmitotic mouse retina.
Introduction
Epigenetic regulation of development and postmitotic homeostasis of mouse retina is an exciting area of research, which promises to bring a novel understanding of biological mechanisms controlling retinogenesis and cell fate determination, cell type-specific metabolic function as well as cell pathologies, cell death and regeneration1,2,3,4,5,6,7,8,9,10,11,12,13. Chromatin undergoes dynamic changes in development, as well as in response to variable external signals whether it is stress, metabolic or cell death stimuli6,14,15,16,17,18. In mouse nuclei, chromatin is divided into active euchromatin and inactive heterochromatin6,19,20. In interphase nucleus, euchromatin resides in the inner region of the nucleus whereas heterochromatin lines the nuclear periphery and nucleolus. This pattern is called conventional and is highly conserved in eukaryotes. In contrast, rod nuclei in nocturnal animals such as mouse and rat have inverted pattern where the nucleus is occupied by the heterochromatin in the center surrounded by euchromatin forming the outermost shell6,18,20. At birth, rod nuclei have conventional nuclear architecture. Inversion of rod nuclei occurs during development by remodeling of conventional nuclear architecture. During this process, peripheral heterochromatin separates from the nuclear periphery and chromocenters fuse together to form a single chromocenter in the center of the nucleus6,18.
Genomic DNA methylation participates in controlling local chromatin conformation21. DNA methylation occurs at the 5' position of cytosine residues of CpG dinucleotides that are enriched in the promoter region of many genes in mouse genomes22,23,24,25,26 as well as in intergenic and intron regions, which carry regulatory elements27. Methylation of CpG islands in proximal promoters21,24 and CpG sequences in gene enhancers27,28 is important in regulating the dynamic state of bivalent chromatin, containing many developmental genes/transcription factors playing important role in development, cancer and aging29,30,31,32,33,34. Three DNA methyltransferases (DNMTs) participate in DNA methylation during mouse development35. All three DNMTs are expressed during mouse retinal development36 and retina-specific changes in Dnmt genes impact retinal development2,7. Hydroxymethylcytosine (5hmC) is the oxidative product of 5-methylcytosine (5mC) demethylation. The three Ten-Eleven Translocation (TET) enzymes participate in 5mC demethylation37,38,39. Hydroxymethyl-C modification is enriched in promoters, gene bodies and intergenic genomic sequences within gene rich and actively transcribed regions27,40.
There is a variety of described approaches for determining changing DNA methylation patterns in cells41,42,43,44,45,46, some of them enabling quantifying global changes of DNA methylation while others focusing on precise changes in CpG-rich regions within promoters and enhancers. Methods for detecting and quantification of 5hmC were also reported47, including by immunohistochemistry13. Immunohistochemical techniques, which enable robust monitoring of 5mC and 5hmC changes in nuclei of developing and postmitotic cells, are very informative for delineating chromatin dynamics in situ in response to external or internal changes impacting the cells4,13,48,49. However, this approach is prone to underestimating DNA methylation and hydroxymethylation changes due to technical difficulties, associated with detecting cytosine modifications in histological preparations. This is because the nonpolar DNA bases, including 5mC and 5hmC-modified cytosine, are hidden within the center of the double-stranded DNA helix50, and require unmasking. This is especially challenging in frozen histological preparations, which may rapidly lose informative organization of the original tissue when harsh treatment is applied.
The mouse retina is an excellent model to dissect the contribution of DNA methylation to neural development and postmitotic homeostasis. There are only 6 types of neurons (rod and cone photoreceptors, amacrine, bipolar, horizontal and ganglion cells), one glial cell type (Muller glia) and one neuroepithelial cell type (retinal pigment epithelium)51. Retinal cell types were reported to have distinct patterns of DNA methylation18,19, which recently has been studied at single-base resolution1,5,9,12. We recently reported immunohistochemistry application to delineating 5mC pattern of distribution within nuclei of retinal cells and changes in the nuclei of adult mouse retina, where all three DNA methyltransferases have been removed by conditional targeting7. Compared to a number of reports, where the presence of DNA methylation in retinal cells could be viewed only as a positive or negative signal in hypermethylated cells undergoing apoptosis, our approach enabled detecting specific chromatin arrangement within the retinal cell nuclei, which we and several groups reported and discussed earlier5,6,7,18,19,36,52. Here, we describe the immunohistochemical (IHC) technique of detecting 5mC and 5hmC in frozen paraformaldehyde-fixed histological sections in details as well as demonstrate the data, which we were able to reproduce from our original report7.
Protocol
All procedures with mice were performed in accordance with protocols approved by the Children's Hospital Oakland Research Institute animal care and use committee and adhered to the Association for Research in Vision and Ophthalmology (ARVO) statement for the use of animals in ophthalmic and vision research.
1. Tissue Preparation for Immunostaining
- Euthanize C57 BL/6J mice at embryonic day (E) 16, postnatal day (P) 0, P15 and P90 by carbon dioxide (CO2) followed by decapitation (E16 and P0 mice).
- Immediately enucleate mouse eyes punctured with 29G1/2 needle on the dorsal side of the eye. Incubate for 5 minutes (min) in 1 mL of 4% paraformaldehyde (PFA) in phosphate-buffered saline (1x PBS) pH 8.0 at room temperature.
- To make eye cups from P0, P15 and P90, dissect out cornea and lens with fine ophthalmic scissors while eyes are in the 4% PFA.
NOTE: Skip this step for E16 eyes as the eyes are too small. - Fix the eye cups (P0, P15 and P90) and the whole eyes (E16) in 4% PFA for 20 min at room temperature. Then wash the eye cups and whole eyes twice in 1 mL of 1x PBS for 10 min at room temperature.
- Cryoprotect the eye cups and whole eyes in 1xPBS, pH 8.0 containing 10% sucrose for 1 hour. Keep in 20% sucrose in 1x PBS for 1 hour and then saturate in 30% sucrose in 1x PBS overnight at 4 °C.
- The next day, embed the eye cups and the whole eyes in optimal cutting temperature compound (OCT), (500 µL) in cryomolds, snap-freeze in a dry ice/ethanol bath, and store at -80 °C.
- Remove the molds with embedded eye cups and whole eyes from -80 °C and allow to equilibrate to -20 °C for 1 hour before cryosectioning.
- Cut retinal cross sections (12 µm thick) parallel to the temporonasal axis through the optic nerve head using a cryostat at -20 °C.
- Mount retinal sections on microscope slides53 and store at -80 °C.
2. Immunostaining with 5-mC and 5-hmC Antibodies
- Encircle the retinal sections mounted on slides with a hydrophobic barrier pen. The hydrophobic barrier pen reduces the volume of antibody required to stain the tissue.
- Wash the retinal sections once with 200 µL of 1x PBS for 10 min.
- Permeabilize the retinal sections with 200 µL of 0.1% Triton X-100 in 1x PBS (PBS-T) for 10 min at room temperature.
- Denature retinal sections for 30 min with 200 µL of freshly made 2 N hydrochloric (HCl) acid in 1x PBS in a 37 °C incubator.
NOTE: Careful titration of HCl treatment is needed to optimize the 5mC signal. - Always include biological replicates (3-4) while optimizing 5mC signal54.
NOTE: We did antigen retrieval at 4 time points (15 min, 30 min, 1 h and 2 h) and found 30 min incubation is optimal for 5mC signal. Longer incubation with HCl degrades the structure of nuclei leading to inability to localize 5mC signal to the nucleus. - After denaturation, neutralize retinal sections by adding 100 µL of 0.1 M Tris-HCl (pH 8.3) on retinal sections for 10 min.
- Incubate the sections in 500 µL of blocking solution (5% preimmune normal goat serum in 0.1% Triton X-100 in 1x PBS) for 1 h at room temperature in a humidity chamber.
- After blocking, add anti-5mC; anti-5hmC antibodies diluted 1:500 in blocking solution to the retinal sections.
- Incubate retinal sections overnight at 4 °C in humidity chamber.
- The next day wash, retinal sections 3 times (10-15 min each time) with 0.1% PBS-T and then incubate in blocking solution with the corresponding secondary antibodies (Goat Anti-Rabbit and Goat Anti-Mouse IgG, diluted; 1:1000), containing DAPI ( 4', 6-diamidino-2-phenylindole) solution (1 µg/mL) at room temperature for 1 h in blocking solution.
NOTE: Avoid drying of sections during all stages of immunodetection. - Wash the slides three time with 1 mL of 0.1% PBS-T.
- After the last wash, mount the sections with aqueous mounting medium under a coverslip and seal with colorless nail polish.
3. Confocal Immunohistochemistry
- Orient the sections to have the retinal pigment epithelium side of the optic cup always facing one way (e.g., up), for consistency.
- Perform image capture of immunostained retinal sections at 63X (magnification of an objective lens) with 2X or 3X zoom.
- Generate multiple optical sections of each selected image (z-stacks) in all 3 channels (Red, Green and Blue, RGB) using 0.35 µm step.
NOTE: The final magnification of a specimen visualized at 10-20X (magnification of an objective lens) will be 63X, respectively, when combined with a (typically used) 10X ocular lens.
Visualize compressed optical stack to locate the 5mC and 5hmC signal
Representative Results
To determine the distribution of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) during retinal development and in postmitotic retina, we immunostained C57BL/6J mouse retinal section from E16, P0, P15 and P90 with antibodies specific to 5mC and 5hmC.
At E16 the 5mC staining was strong in chromocenters of the cell nuclei while weak staining was observed in the whole cell nuclei (Figure 1a). The 5hmC staining was confined to the cell nuclei and was absent from the chromocenters (Figure 1b).
In P0 retina, the 5mC signal was localized to the chromocenters of the cell nuclei and nuclear periphery (Figure 2a, arrow and arrowhead). We also observed weak staining of 5mC between the chromocenters of the cell nuclei. The 5hmC signal was strongly present in the cell nuclei except for the chromocenters (Figure 2b). We found more nuclei are stained with 5mC and 5hmC in inner neuroblast layer (INBL) (Figure 2c, dotted line)
In P15 retina, we found strong staining of 5mC and 5hmC in outer nuclear layer (ONL), inner nuclear layer (INL) and ganglion cell layer (GCL) (Figure 3). In ONL (rod photoreceptors), the 5mC signal was present in the chromocenters of the cell nuclei and nuclear periphery (Figures 3a-3c, 3g). The 5hmC signal was found in the whole cell nuclei except for the chromocenters (Figures 3d-3f). The transmission electron microscopy done on P15 retina shows distribution of heterochromatic domains in rod cell nuclei similar to that found with 5mC antibody signal (Figures 3h and 3i).
The 5mC staining in INL and GCL was strong in the chromocenters of the cell nuclei and weak staining was also observed in whole cell nuclei (Figures 3j-3l and 3r-3t). In contrast to 5mC, the 5hmC signal was localized to the whole cell nuclei except for the chromocenters in INL and GCL (Figures 3m-3o and 3u-3w). We observed strong 5hmC signal on the periphery of GCL cell nuclei.
In adult rod photoreceptors, the 5mC signal was restricted to the chromocenter and nuclear periphery of the cell nuclei (Figures 4a-4c) while 5hmc signal was confined to the nuclear periphery (Figures 4d-4f).
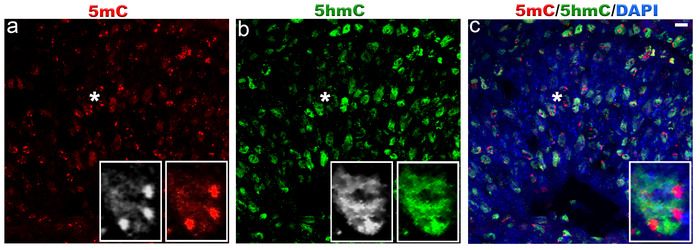
Figure 1. Localization of 5-methylcytosine and 5-hydroxymethylcytosine in mouse retina at embryonic day 16.
Immunostaining of C57BL/6J retina with antibodies to 5mC (red, a) and 5hmC (green, b). At E16, 5mC signal is very strong in chromocenters of the cell nuclei and also present in the whole cell nuclei at much lower level. 5hmC staining was exclusively confined to the cell nuclei. Panel c represents merged 5mC and 5hmC image. Nuclei are counterstained with DAPI (blue, c). Insets represent magnification of area shown with asterisk in the images. Scale bar: 5 µm. Please click here to view a larger version of this figure.
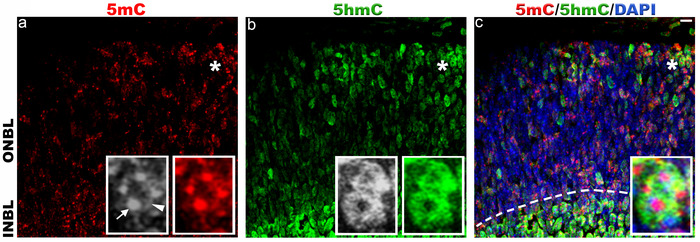
Figure 2. Immunohistochemical staining of 5-methylcytosine and 5-hydroxymethylcytosine in mouse retina at postnatal day 0.
Immunolabeling of C57BL/6J retina with antibodies to 5mC (red, a) and 5hmC (green, b). In P0 retina, 5mC and 5hmC are present in both outer neuroblast layer (ONBL) and inner neuroblast layer (INBL). a) 5mC signal is strongly present in the chromocenters (arrow) and nuclear periphery of the cell nuclei (arrowhead). Weak staining of 5mC is also observed in between the chromocenters of cell nuclei. b) 5hmC staining is confined to the cell nucleus and strong signal is observed in INBL (dotted line in panel c). Panel c represents merged 5mC and 5hmC image. Nuclei are counterstained with DAPI (blue, c). Insets represent magnification of area shown with asterisk in the images. Scale bar: 5 µm. Please click here to view a larger version of this figure.
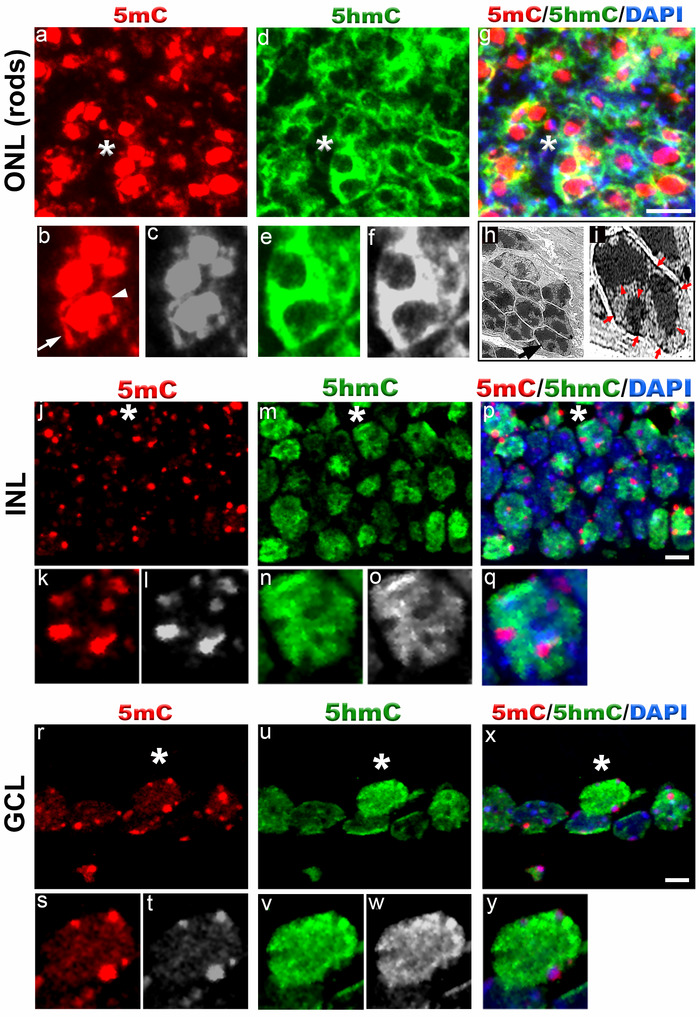
Figure 3. 5-methylcytosine and 5-hydroxymethylcytosine distribution in mouse retina at postnatal day 15.
Immunostaining of C57BL/6J retina with anti-5mC and 5hmC antibodies (a-g, j-y). In outer nuclear layer (ONL) (rod photoreceptors), 5mC signal (red, a) usually coincides with the chromocenters of the cell nuclei (arrowhead, b) and also localizes to the nuclear periphery (arrow, b). Panel b (red) and c (grey) represent magnification of area shown with asterisk in the image a. The 5hmC signal (green, d) is confined to the whole cell nuclei except for the chromocenters. Panel e (green) and f (grey) represent magnification of area shown with asterisk in the image d. Panel g represents merged 5mC and 5hmC image. Nuclei are counterstained with DAPI. Panel h and i are transmission electron microscope image of photoreceptor nuclei at postnatal day 15 showing distribution of heterochromatic domains. Black arrowhead in panel h points to single rod photoreceptor nucleus enlarged in panel i. Red arrowheads in panel i point to heterochromatic domains within rod photoreceptor nucleus while red arrows point to heterochromatin in the nuclear periphery. In INL (j-q) and GCL cell nuclei (r-y), the 5mC signal (red) is localized to the chromocenters present in the periphery and weak staining is also detected in the rest of the cell nuclei. Panel k (red) and i (grey) represent magnification of area shown with asterisk in the image j. The 5hmC staining (green) in INL and GCL cell nuclei is confined to the whole cell nucleus. Note strong 5hmC staining in the nuclear periphery of GCL cells. Panel p and x represent merged 5mC and 5hmC image while panel q and y represent magnification of area shown with asterisk in the image p and x respectively. Nuclei are counterstained with DAPI. Scale bar: 5 µm. Please click here to view a larger version of this figure.
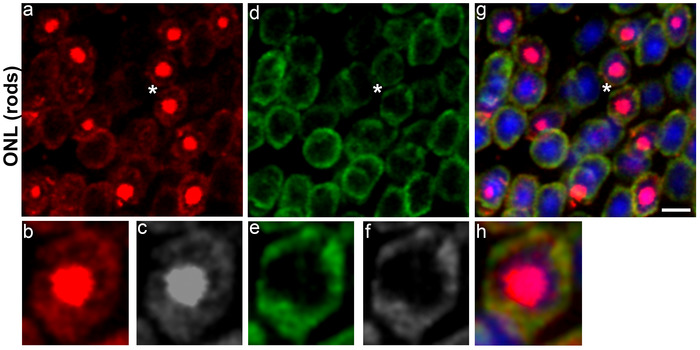
Figure 4. 5-methylcytosine and 5-hydroxymethylcytosine distribution in rod photoreceptor nuclei of C57BL/6J mouse retina at postnatal day 90.
The 5mC staining (red, a) is strongly present in the chromocenter of the cell nuclei and also weakly present in the nuclear periphery. Panel b (red) and c (grey) represent magnification of area shown with asterisk in the image a. The 5hmC staining (green, d) is exclusively confined to the nuclear periphery Panel e (green) and f (grey) represent magnification of area shown with asterisk in the image d. Panel g represents merged 5mC and 5hmC image while panel h is magnification of area shown with asterisk in the image g. Nuclei are counterstained with DAPI (blue). Scale bar: 5 µm. Please click here to view a larger version of this figure.
Discussion
DNA methylation and hydroxymethylation are dynamic and reversible DNA modifications, which modulatethe diverse range of biological mechanisms in a developing as well as in postmitotic cells. Here, we describe a reproducible technique for detecting 5mC and 5hmC DNA modifications in situ by immunohistochemical technique (using anti-5mC and anti-5hmC antibodies) in paraformaldehyde-fixed frozen sections of mouse retina, and provide guidelines for improving and standardizing the results, especially when comparing several different specimens. We earlier used this method to delineate 5mC changes in mouse retina with retina-specific targeting of Dnmt1, Dnmt3a and Dnmt3b DNA methyltransferase genes, and reported the (expected) depletion of 5mC marks in their retinas7.
The critical step in 5mC immunohistochemical detection is optimization of treatment of histological sections with HCl: less than 15 min pretreatment is not expected to produce reproducible results, whereas excessive treatment with HCl destroys the nuclear architecture. We found best result after 30 min incubation with HCl. We recommend to always use freshly diluted HCl and freshly made 4% PFA solution for DNA denaturation and retinal tissue fixation.
To troubleshoot the results, we recommend to include three to four biological replicates while optimizing 5mC signal. While each individual set of immunohistochemical staining may generate slightly different results (more or less bright heterochromatic regions), which is expected in immunohistochemical method, the 5mC and 5hmC nuclear distribution within the individual set of sections is expected to be very reproducible, for as long as the protocol is followed.
This technique also has limitation as we consistently observed variability in 5mC distribution in the photoreceptor cell nuclei within the same section. We found some nuclei showed strong 5mC signal while adjacent cell nuclei showed no 5mC signal. This is due to limitation in unmasking 5mC antigen by HCl treatment in frozen sections.
The significance of this technique enables 5mC localization without DNase and proteinase K treatment leading to better preservation of chromocenters. This technique is expected to work robustly on any sections from all vertebrate animal tissues, carrying 5mC, 5hmC marks, and processed with a similar approach, not only mouse retina, for as long as the protocol is followed.
Acknowledgements
This work was supported by a SBIR grant (1R44EY027654-01A1).
Materials
| Name | Company | Catalog Number | Comments |
| Supplies | |||
| 29G1/2 ULTRA-FINE | BD Biosciences | 329461 | |
| Ophthalmic scissor | Fine Science tools, Inc. | 15000-03 | |
| PAP pen | RPICORP.com | 195505 | |
| Microslide Superfrost plus | VWR | 48311-703 | |
| Reagent | |||
| Paraformaldehyde | Electron Microscope Science | 15710 | |
| 1x PBS | Corning | 21-031-CV | |
| Sucrose | Sigma | S9378 | |
| Tissue-Tek Optical cutting compound (OCT) | VWR | 4583 | |
| Triton X 100 | Sigma | T9284 | |
| 6 N HCl | VWR analytical | BDH7204-1 | |
| Tris-HCl pH 8.3 | Teknova | T1083 | |
| Goat serum | Jackson Immunoresearch | 005-000-121 | |
| 5mC | Genway Biotech | GWB-BD5190 | |
| 5hmC | Active Motif | 39791 | |
| LaminB1 | Abcam | Ab16048 | |
| Cy2 AffiniPure Goat Ant-Rabbit IgG | Jackson Immunoresearch | 111-225-1444 | |
| Cy3 AffiniPure Goat Ant-Mouse IgG | Jackson Immunoresearch | 115-585-166 | |
| DAPI | Thermo Fisher Scientific | D1306 | |
| Prolong Gold Antifade medium | Thermo Fisher Scientific | P36930 | |
| Equipment | |||
| MICRM HM 550 | Thermo Fisher Scientific | 388114 | |
| Confocal microscope | Zeiss |
References
- Merbs, S. L., et al. Cell-specific DNA methylation patterns of retina-specific genes. PLoS One. 7 (3), e32602 (2012).
- Nasonkin, I. O., et al. Conditional knockdown of DNA methyltransferase 1 reveals a key role of retinal pigment epithelium integrity in photoreceptor outer segment morphogenesis. Development. 140 (6), 1330-1341 (2013).
- Rhee, K. D., Yu, J., Zhao, C. Y., Fan, G., Yang, X. J. Dnmt1-dependent DNA methylation is essential for photoreceptor terminal differentiation and retinal neuron survival. Cell Death & Disease. 3, e427 (2012).
- Wahlin, K. J., et al. Epigenetics and cell death: DNA hypermethylation in programmed retinal cell death. PLoS One. 8 (11), e79140 (2013).
- Aldiri, I., et al. The Dynamic Epigenetic Landscape of the Retina During Development, Reprogramming, and Tumorigenesis. Neuron. 94 (3), 550-568 (2017).
- Eberhart, A., et al. Epigenetics of eu- and heterochromatin in inverted and conventional nuclei from mouse retina. Chromosome Research. 21 (5), 535-554 (2013).
- Singh, R. K., et al. Dnmt3a and Dnmt3b cooperate in photoreceptor and outer plexiform layer development in the mammalian retina. Experimental Eye Research. 159, 132-146 (2017).
- Farinelli, P., et al. DNA methylation and differential gene regulation in photoreceptor cell death. Cell Death & Disease. 5, e1558 (2014).
- Mo, A., et al. Epigenomic landscapes of retinal rods and cones. Elife. 5, e11613 (2016).
- Popova, E. Y., Barnstable, C. J., Zhang, S. S. Cell type-specific epigenetic signatures accompany late stages of mouse retina development. Advances in Experimental Medicine and Biology. 801, 3-8 (2014).
- Kim, J. W., et al. NRL-Regulated Transcriptome Dynamics of Developing Rod Photoreceptors. Cell Reports. 17 (9), 2460-2473 (2016).
- Wang, L., et al. Retinal Cell Type DNA Methylation and Histone Modifications Predict Reprogramming Efficiency and Retinogenesis in 3D Organoid Cultures. Cell Reports. 22 (10), 2601-2614 (2018).
- Perera, A., et al. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Reports. 11 (2), 283-294 (2015).
- Mimura, I., et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Molecular and Cellular Biology. 32 (15), 3018-3032 (2012).
- Nair, N., Shoaib, M., Sorensen, C. S. Chromatin Dynamics in Genome Stability: Roles in Suppressing Endogenous DNA Damage and Facilitating DNA Repair. Int J Mol Sci. 18 (7), (2017).
- Mekhail, K., Moazed, D. The nuclear envelope in genome organization, expression and stability. Nature Reviews Molecular Cell Biology. 11 (5), 317-328 (2010).
- Misteli, T., Soutoglou, E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nature Reviews Molecular Cell Biology. 10 (4), 243-254 (2009).
- Solovei, I., et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 137 (2), 356-368 (2009).
- Solovei, I., et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 152 (3), 584-598 (2013).
- Solovei, I., Thanisch, K., Feodorova, Y. How to rule the nucleus: divide et impera. Current Opinion in Cell Biology. 40, 47-59 (2016).
- Jaenisch, R., Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genetics. 33, 245-254 (2003).
- Cross, S. H., Charlton, J. A., Nan, X., Bird, A. P. Purification of CpG islands using a methylated DNA binding column. Nature Genetics. 6 (3), 236-244 (1994).
- Antequera, F., Bird, A. Number of CpG islands and genes in human and mouse. Proceedings of the National Academy of Sciences of the United States of America. 90 (24), 11995-11999 (1993).
- Antequera, F., Boyes, J., Bird, A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 62 (3), 503-514 (1990).
- Fouse, S. D., et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with. Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2 (2), 160-169 (2008).
- Elango, N., Yi, S. V. DNA methylation and structural and functional bimodality of vertebrate promoters. Molecular Biology and Evolution. 25 (8), 1602-1608 (2008).
- Lister, R., et al. Global epigenomic reconfiguration during mammalian brain development. Science. 341 (6146), 1237905 (2013).
- Colwell, M., et al. Evolutionary conservation of DNA methylation in CpG sites within ultraconserved noncoding elements. Epigenetics. 13 (1), 49-60 (2018).
- Lee, S. M., et al. Intragenic CpG islands play important roles in bivalent chromatin assembly of developmental genes. Proceedings of the National Academy of Sciences of the United States of America. 114 (10), E1885-E1894 (2017).
- Charlet, J., et al. Bivalent Regions of Cytosine Methylation and H3K27 Acetylation Suggest an Active Role for DNA Methylation at Enhancers. Molecular Cell. 62 (3), 422-431 (2016).
- Bernhart, S. H., et al. Changes of bivalent chromatin coincide with increased expression of developmental genes in cancer. Scientific Reports. 6, 37393 (2016).
- Curry, E., et al. Genes Predisposed to DNA Hypermethylation during Acquired Resistance to Chemotherapy Are Identified in Ovarian Tumors by Bivalent Chromatin Domains at Initial Diagnosis. Cancer Research. 78 (6), 1383-1391 (2018).
- Rakyan, V. K., et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Research. 20 (4), 434-439 (2010).
- Smith, Z. D., Meissner, A. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 14 (3), 204-220 (2013).
- Chen, T., Li, E. Structure and function of eukaryotic DNA methyltransferases. Current Topics in Developmental Biology. 60, 55-89 (2004).
- Nasonkin, I. O., et al. Distinct nuclear localization patterns of DNA methyltransferases in developing and mature mammalian retina. The Journal of Comparative Neurology. 519 (10), 1914-1930 (2011).
- Scourzic, L., Mouly, E., Bernard, O. A. TET proteins and the control of cytosine demethylation in cancer. Genome Medicine. 7 (1), 9 (2015).
- Gong, Z., Zhu, J. K. Active DNA demethylation by oxidation and repair. Cell Research. 21 (12), 1649-1651 (2011).
- Ito, S., et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 333 (6047), 1300-1303 (2011).
- Pastor, W. A., et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 473 (7347), 394-397 (2011).
- Suzuki, M. M., Bird, A. DNA methylation landscapes: provocative insights from epigenomics. Nature Reviews Genetics. 9 (6), 465-476 (2008).
- Kurdyukov, S., Bullock, M. DNA Methylation Analysis: Choosing the Right Method. Biology (Basel). 5 (1), (2016).
- Lisanti, S., et al. Comparison of methods for quantification of global DNA methylation in human cells and tissues). PLoS One. 8 (11), e79044 (2013).
- Armstrong, K. M., et al. Global DNA methylation measurement by HPLC using low amounts of DNA. Biotechnology Journal. 6 (1), 113-117 (2011).
- consortium, B. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nature Biotechnology. 34 (7), 726-737 (2016).
- Bock, C., et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nature Biotechnology. 28 (10), 1106-1114 (2010).
- Song, C. X., He, C. The hunt for 5-hydroxymethylcytosine: the sixth base. Epigenomics. 3 (5), 521-523 (2011).
- Abakir, A., Wheldon, L., Johnson, A. D., Laurent, P., Ruzov, A. Detection of Modified Forms of Cytosine Using Sensitive Immunohistochemistry. Journal of Visualized Experiments. (114), (2016).
- Santos, F., Dean, W. Using immunofluorescence to observe methylation changes in mammalian preimplantation embryos. Methods in Molecular Biology. 325, 129-137 (2006).
- Watson, J. D., Crick, F. H. The structure of DNA. Cold Spring Harbor Symposia on Quantitative Biology. 18, 123-131 (1953).
- Swaroop, A., Kim, D., Forrest, D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nature Reviews Neuroscience. 11 (8), 563-576 (2010).
- Kizilyaprak, C., Spehner, D., Devys, D., Schultz, P. In vivo chromatin organization of mouse rod photoreceptors correlates with histone modifications. PLoS One. 5 (6), e11039 (2010).
- Singh, R. K., Kolandaivelu, S., Ramamurthy, V. Early alteration of retinal neurons in Aipl1-/- animals. Investigative Ophthalmology & Visual Science. 55 (5), 3081-3092 (2014).
- Eberhart, A., Kimura, H., Leonhardt, H., Joffe, B., Solovei, I. Reliable detection of epigenetic histone marks and nuclear proteins in tissue cryosections. Chromosome Research. 20 (7), 849-858 (2012).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved