An Anaerobic Biosensor Assay for the Detection of Mercury and Cadmium
In This Article
Summary
Here, we present a protocol to use an anaerobic whole-cell microbial biosensor to evaluate how different environmental variables affect the bioavailability of Hg and Cd to bacteria in anoxic environments.
Abstract
Mercury (Hg) bioavailability to microbes is a key step to toxic Hg biomagnification in food webs. Cadmium (Cd) transformations and bioavailability to bacteria control the amount that will accumulate in staple food crops. The bioavailability of these metals is dependent on their speciation in solution, but more particularly under anoxic conditions where Hg is methylated to toxic monomethylmercury (MeHg) and Cd persists in the rhizosphere. Whole-cell microbial biosensors give a quantifiable signal when a metal enters the cytosol and therefore are useful tools to assess metal bioavailability. Unfortunately, most biosensing efforts have so far been constrained to oxic environments due to the limited ability of existing reporter proteins to function in the absence of oxygen. In this study, we present our effort to develop and optimize a whole-cell biosensor assay capable of functioning anaerobically that can detect metals under anoxic condition in quasi-real time and within hours. We describe how the biosensor can help assess how chemical variables relevant to the environmental cycling of metals affect their bioavailability. The following protocol includes methods to (1) prepare Hg and Cd standards under anoxic conditions, (2) prepare the biosensor in the absence of oxygen, (3) design and execute an experiment to determine how a series of variable affects Hg or Cd bioavailability, and (4) to quantify and interpret biosensor data. We show the linear ranges of the biosensors and provide examples showing the method's ability to distinguish between metal bioavailability and toxicity by utilizing both metal-inducible and constitutive strains.
Introduction
Mercury (Hg) is a global pollutant and its bioavailability to Hg methylating microbes is the first step towards its biomagnification through food webs and its possible neurotoxic effects in human and wildlife1. It is currently thought that microbial Hg methylation is an intracellular process that requires: i) the species of Hg to be bioavailable2,3,4,5,6,7 and ii) for the cell to be physiologically capable of methylating Hg8,9,10. Cadmium (Cd) bioaccumulates in organisms, but does not biomagnifies in foodwebs and is widely used in industrial and commercial processes that commonly cause acute exposures in people and the environment11. Although microbes play several key roles in the fate of Hg in the environment, most studies on Cd geochemistry and ecotoxicology focus on microbial eukaryotes12. Consumption of agricultural crops (e.g., rice) is the main source of direct exposure to cadmium; in this case, the bioavailability of Cd to microbes of the rhizosphere directly influences the amount plants can accumulate13.
Hg transport pathways are complex and possibly involve an active transport step14. When a transporter is involved, recent work suggested that HgII uses a ZnII or MnII transporter7,15,16,17. Whereas CdII is hypothesized to be accidentally transported into the cytosol through divalent metal transport pathways (particularly MnII or ZnII), mechanisms of CdII transport inside the cells remain speculative, and no Cd-specific transport pathway has been identified13,18. Regardless of the nature of the transporters involved, three mechanistic factors ultimately determine the ability of metals to enter a cell: i) the metal speciation in solution2,6,15,16,17,19,20,21,22,23, ii) the biophysiochemical properties of the cell membrane17,24,25,26,27,28,29,30,31, and iii) the ability for the metal to access a transport site7,32. Cd and Hg are unlikely to exists as free ions under microbial physiologically relevant conditions due to their high affinity for Dissolved Organic Matter (DOM), chelating contaminants (e.g., EDTA), or reduced sulfur moieties33,34,35 (CdII can exist as a free ion or form ion-pairs in the absence of these ligands). There is a lack of efficient methods in determining how these metal species are bioavailable under conditions relevant to their fate in the environment. For instance, Hg is methylated under anaerobic conditions14, and both cadmium and Hg are soft metals (or class B cations), requiring that their speciation be investigated under conditions that maintain the integrity of reduced sulfur groups.
Microbial biosensors are bacterial cells that emit a quantifiable signal in response to the intracellular concentrations of a metal, in this case Hg or Cd. As such, they are ideal tools to understand how metals enter a cell36, provided that exposure conditions are carefully controlled for. Hg biosensors typically contain gene fusions between the regulatory circuitry of the mer-operon (including genes encoding for the transcription regulator MerRas well as the operator and promoter regions of the operon), and reporting genes (e.g., lux, gfp, lac genes). When mercury is present in the cytoplasm, it will bind to MerR, resulting in transcription of the reporting genes and subsequent signal production19,37. Specific Cd biosensors are usually designed using the cadC, cadAC, zntA or zntR encoded transcription regulators38, but it is worth noting that MerR has a lower, yet quantifiable affinity to Cd5. Due to aerobic restriction of most commonly used luminescent or fluorescent reporter proteins, until recently microbial biosensors remained unable to offer insights into the biotransformation of metals under anoxic conditions. This makes anaerobic detection of metals bioavailability very difficult over a range of conditions relevant to their environmental fate, specifically in the presence of redox sensitive ligands (e.g., sulfide and thiols)4,5,39.
To alleviate the methodological hurdle of live imaging in the absence of oxygen, Drepper et al. (2007) have developed a flavin-based fluorescent protein (FbFp), based on light oxygen voltage domain of SB2 protein from P. putida. This protein family is able to fluoresce in the absence of oxygen40. Building on the work of Drepper et al., our lab designed an anaerobic biosensor allowing for the study of Hg bioavailability under oxic and anoxic conditions and over a wide range of salinity 17. In the current paper, we describe how to prepare and use the biosensor to test environmental variables' influence on Hg or Cd bioavailability. Although we developed the biosensor for HgII, we chose to perform experiments with CdII as a means to draw the reader's attention to the fact that biosensors may also respond to multiple stressors that are likely to co-occur in environmental matrices; in this case CdII was investigated because it is known to bind to the transcriptional regulator MerR5. Here, we show representative calibration and functional linear ranges with respect to either metal. We also give an example when the biosensor's results are conclusive (MgII and MnII on Hg bioavailability) and inconclusive (ZnII on Hg bioavailability).
Protocol
1. Growth Media and Exposure Media Preparation
- To make 250 mL of growth medium:
Note: If trace elements #1 solution and trace elements #2 solution are already prepared, skip to step 1.1.5.- Prepare trace elements #1 solution in a clean volumetric flask (200 mL) to contain the final molarities of 1.5 x 10-3 M Na2MoO4, 6.5 x 10-4 M Na2SeO4, 5 x 10-3 M H3BO3, and 0.1 M NaOH.
CAUTION: Strong bases (NaOH) are corrosive. Make sure when weighing the reagents to ensure that the final molarity represents the key trace elements; Mo, Se, and B. - Under a sterile field, filter sterilize using a 0.22 µm polyethersulfone syringe filter in a clean/sterile plastic/Polytetrafluoroethylene (PTFE) bottle.
- Prepare trace elements #2 solution in a clean volumetric flask (200 mL) to contain the final molarities of 0.01 M MnSO4, 5 x 10-4 M ZnSO4, 3.25 x 10-3 M CoCl2, 6.25 x 10-3 M NiCl2, and 0.1 M H2SO4.
CAUTION: Strong acids (H2SO4) are corrosive. Make sure when weighing the reagents to ensure that the final molarity represents the key trace elements; Mn, Zn, Co, and Ni. - Under a sterile field, filter sterilize using a 0.22 µm polyethersulfone syringe filter in a clean glass bottle.
- Under a sterile field, in a clean/sterile glass bottle (250 mL minimum); add 200 mL ultrapure water, 42.5 mL of M9 Minimal Salts (5x; see Table of Materials), 2.5 mL of 2 M Glucose, 125 µL of 2 M MgSO4, 1200 µL of 0.6 M Thiamine HCl from a frozen (-20 ˚C) aliquot, 1.25 mL of 4 M NaNO3, 770 µL of 0.075 M L-leucine/ L-isoleucine/ valine solution, 250 µL trace elements #1, 250 µL of trace element #2, and 250 µL of 0.01 M EDTA sodium salt.
Note: All reagents in step 1.1.5 should be prepared prior and filter sterilized using a 0.22 µm polyethersulfone syringe filter. Ultrapure water may be sterilized using an autoclave. - Take the glass bottle now containing solution from step 1.1.5., loosely cap it and cycle it through the anaerobic chamber air lock.
- In the anaerobic chamber, add 15 µL of 0.225 M FeSO4 in 0.2 M H2SO4, cap bottle tightly, and shake until all white precipitates disappear.
Note: All reagents in step 1.1.7 should be prepared prior and filter sterilized using a 0.22 µm polyethersulfone syringe filter. Store the 0.225 M FeSO4 in 0.2 M H2SO4 in the anaerobic chamber. - Remove cap from bottle, wait 1 minute for air to exchange, replace the cap from bottle, and then shake vigorously. Repeat this step once.
- Fasten cap tightly onto the bottle, remove from anaerobic chamber, and store bottle in fridge (4 ˚C) until use.
- Growth medium should be remade once a week by repeating steps 1.1.5 to 1.1.9.
- Prepare trace elements #1 solution in a clean volumetric flask (200 mL) to contain the final molarities of 1.5 x 10-3 M Na2MoO4, 6.5 x 10-4 M Na2SeO4, 5 x 10-3 M H3BO3, and 0.1 M NaOH.
- To make 100 mL of exposure medium in 2 x 50 mL conical sterile polypropylene centrifuge tubes:
Note: We recommend that exposure medium be prepared on the day of the exposure assay to minimize the risk of contamination, however it can be made in advance and stored in the fridge.- In the anaerobic chamber, to each 50 mL centrifuge tube add 42 mL of anaerobic ultrapure water, 350 µL of 1 M Sodium Beta-Gylcerophosphate from a frozen (-20 ˚C) aliquot, 2 mL of 1 M MOPS free acid, 50 μL of 1 M (NH4)2SO4, 125 μL of 2 M Glucose, 300 μL of 2.5 M KOH, and 200 μL of 2.5 M NaOH.
Note: All reagents in step 1.2.1 should be prepared prior and filter sterilized using a 0.22 µm polyethersulfone syringe filter. Ultrapure water may be heat sterilized using an autoclave. 2.5 M NaOH and 2.5 M NaOH must be stored in plastic/PTFE bottles. All reagents listed must be stored in the anaerobic chamber except Sodium Beta-Gylcerophosphate, which is to be stored in a (-20 ˚C) freezer. In general, it is good practice to leave all plastic or glass parts and containers for several days in the anaerobic chamber to ensure that no traces of oxygen remain. - Cap the 50 mL centrifuge tubes, shake well, and remove a 10 mL aliquot to measure the pH. The measured pH of this exposure media must always measure 7.00 ± 0.02 at 25 ˚C. If the pH does not measure within this range, readjust the volume of added 2.5 M KOH to correct for this.
Note: Never titrate the solution to correct for the pH with a pH probe. pH probes represent a source of contamination for the exposure medium. The pH must always be a product of the added reagents in step 1.2.1.
- In the anaerobic chamber, to each 50 mL centrifuge tube add 42 mL of anaerobic ultrapure water, 350 µL of 1 M Sodium Beta-Gylcerophosphate from a frozen (-20 ˚C) aliquot, 2 mL of 1 M MOPS free acid, 50 μL of 1 M (NH4)2SO4, 125 μL of 2 M Glucose, 300 μL of 2.5 M KOH, and 200 μL of 2.5 M NaOH.
- Prepare a 5-10 mM NaNO3 stock solution and leave in anaerobic chamber to be used during the time of the exposure assay.
Note: It is easier to dilute a more concentrated primary NaNO3 standard as opposed to making a 5-10 mM NaNO3 primary standard.
2. Preparation of Mercury and Cadmium Standards.
- Preparing a 4-8 µM Mercuric (HgII) solution in 0.2 M H2SO4.
- Prepare a Hg2+ standard by making a millimolar (1-10 mM) HgCl2, HgNO3 or HgSO4 solution in 0.2 M H2SO4.
CAUTION: Mercury is highly toxic and H2SO4 is corrosive. Operate in fume hoods with all required personal protective equipment. - In a PTFE bottle, dilute standard from 2.1.1. into 0.2 M H2SO4 to obtain a concentration within the range of 4-8 µM Hg2+.
Note: The acid used must be analytical grade H2SO4. - The Hg concentration from 2.1.2 should be validated using a mercury analyzer (see Table of Materials).
Note: Other methods for validating the Hg concentration may be used.
- Prepare a Hg2+ standard by making a millimolar (1-10 mM) HgCl2, HgNO3 or HgSO4 solution in 0.2 M H2SO4.
- Preparing a 10 µM CdII solution in 10 mM H2SO4.
- Prepare a primary standard of CdCl2 (10-50 mM range for accurate weighing of powder) in 0.1 M H2SO4.
- In a series of serial dilutions in clean acid rinsed 20 mL borosilicate glass vials with black phenolic screw caps with PTFE faced rubber liners, dilute the standard from 2.2.1 to 10 µM Cd2+. Ensure that the concentration of H2SO4 in every subsequent serial dilution remains 10 mM.
3. Preparation of the Biosensor for Anaerobic Exposure Assay
- Plate the mercury inducible biosensor (E. coli NEB5α harboring PUC57merR-PpFbFp) and the constitutively expressed biosensor (E. coli NEB5α harboring PUC19Balch-PpFbFp) from -80 ˚C cryostock onto Lysogeny Broth plates containing 120 ug/mL ampicillin. See our previously published work for details on the production of these biosensors17.
- At 4:30-5 PM, inoculate a culture in 10 mL of LB (+ amp) and grow overnight.
Note: Starting from a plate it takes 2 days to prepare the anaerobic culture for the exposure assay (i.e., a culture started on Monday afternoon (4:30-5 PM) will be ready for the exposure assay at around noon on Wednesday). The following steps are the required microbiological techniques necessary to prepare the cultures on the day of the Exposure assay.- Take one colony from the plate culture and add to 10 mL Lysogeny Broth (LB) with 21 µL of a 100 mg/mL stock solution of ampicillin sodium salt (final concentration is 210 ug/mL amp) in a sterile culture tube.
- Place the culture into an incubator/shaker and grow overnight at +37 ˚C with shaking at 220 rpm.
- The next morning at 9-10 AM, resuspend the culture and grow anaerobically throughout the day (20% inoculum).
- Bring the culture from the incubator (step 3.2.1) and the growth medium (step 1.1.9) into the anaerobic chamber.
- Add 8 mL of fresh growth medium and ampicillin (210 μg/mL) into a sterile Balch tube.
- Collect 2 mL of the overnight grown culture and transfer into a 2 mL Microcentrifuge tube. Centrifuge at 10,000 RCF step for 90 seconds, dump supernatant and resuspend in 2 mL of fresh growth medium. Add the resuspended culture to the Balch tube containing 8 mL of fresh growth medium and ampicillin.
- Using sterile technique, carefully place a rubber stopper on the Balch tube. Remove from the anaerobic chamber and place in an incubator/shaker and grow anaerobically until 3-5 PM at +37 ˚C with shaking at 220 rpm.
- Between 3 and 5 PM, perform a 1% anaerobic inoculum into fresh growth medium and grow overnight.
- Bring the Balch tube (step 3.3.4) into the anaerobic chamber along with growth medium. Add 100 µL of the culture to 10 mL of fresh growth medium (1% inoculum) with amp (210 ug/mL amp) in a sterile Balch tube.
- Using sterile technique, carefully place a rubber stopper on the Balch tube. Remove from the anaerobic chamber, place in an incubator/shaker, and grow overnight at +37 ˚C with shaking at 220 rpm.
- Between 9 and 10 AM, resuspend the culture to grow anaerobically throughout the day (20% inoculum).
- Bring the culture from the incubator (step 3.4.1) and the growth medium (step 1.1.9) into the anaerobic chamber.
- Add 8 mL of growth medium and ampicillin (210 ug/mL amp) into a sterile Balch tube.
- Collect 2 m of the overnight grown culture and transfer into a 2 mL Microcentrifuge tube. Centrifuge at 10,000 x g for 90 seconds, dump supernatant, and resuspend in 2 mL of fresh growth medium. Add the resuspended culture to the Balch tube containing 8 mL of fresh growth medium and ampicillin.
- Using sterile technique, carefully place a rubber stopper on the Balch tube. Remove from chamber and place in an incubator/shaker and grow anaerobically at +37 ˚C with shaking at 220 rpm.
- Monitor the growth of the culture using a spectrophotometer (step 3.5.4) until an OD600 of 0.6 is reached. Be sure to vortex culture prior to each OD reading.
Note: This step takes 3-4 hours, and the exposure medium (step 1.2) should be prepared during this time as well as the exposure assay (step 4.1). - Stop the growth when the culture reaches an OD600 of 0.6 (±0.1) (3-4 hours expected growth).
- Bring tube in the anaerobic chamber (step 3.6) and transfer culture into 2 x 2 mL Microcentrifuge tubes. Centrifuge at 10,000 x g for 90 seconds, dump supernatant, and resuspend in 2 x 2 mL of fresh exposure medium.
- Repeat washing step 3.7.1. once to remove any trace of the growth medium.
- Combine both microcentrifuge tubes of cell culture (step 3.7) into a 7 mL PTFE standard vial to obtain Biosensor Stock to be used in the Exposure Assay (step 4).
Note: Be sure to mix the Biosensor Stock by thoroughly yet gently pipetting back and forth prior to use. The method may be paused here for up to an hour.
4. Exposure Assay
- Designing a plate layout.
Note: Be sure to have all pipetting values calculated and stocks prepared prior to starting the exposure assay. How to properly design an experiment and what controls to include is detailed in the text. In addition, the experiment should not be started if there is oxygen in the anaerobic chamber indicated on the anaerobic monitor.- Design the plate layout according to a 96 well template. To run experiments in technical replicates of 3, this will allow for 32 different treatments, which is best represented by a 4 x 8 grid to set up vials (see Figure 1).
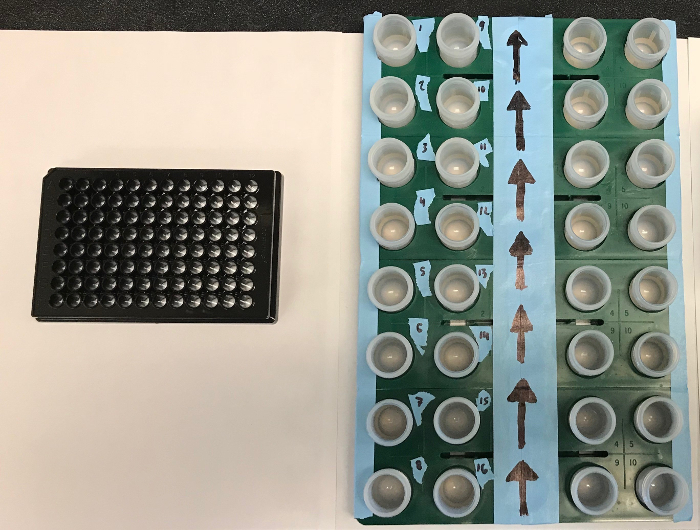
Figure 1: A 96 well plate (left) and a corresponding 4 x 8 grid containing PTFE vials (right) to be transferred to the plate. Please click here to view a larger version of this figure.
Note: When testing for the role of a variable on Hg uptake with the mercury inducible biosensor; two treatments are required for each variable: the treatment (biosensor + Hg + variable + nitrate) and its treatment blank (biosensor + variable + nitrate). When testing for the role of a variable on the physiology of the cell using the constitutive biosensor, two treatments are required for each variable: the treatment (biosensor + Hg + variable + nitrate) and treatment blank (biosensor + variable + Hg). Mercury may be replaced with Cadmium. Hg or Cd will become the variable when performing a calibration curve. The constitutive and inducible biosensors do not need to be run at the same time (in the same plate layout). A template example for the plate layout and corresponding 4 x 8 grid when testing a concentration range of magnesium (variable) is provided in Table 1).
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| a | Hg induced biosensor + Hg + Nitrate + 0 mM Mg | Hg induced biosensor + Nitrate + 0 mM Mg | Constitutive biosensor + Hg + Nitrate + 0 mM Mg | Constitutive biosensor + Hg + 0 mM Mg | ||||||||
| b | Hg induced biosensor+ Hg + Nitrate + 0.1 mM Mg | Hg induced biosensor + Nitrate + 0.1 mM Mg | Constitutive biosensor+ Hg + Nitrate + 0.1 mM Mg | Constitutive biosensor + Hg + 0.1 mM Mg | ||||||||
| c | Hg induced biosensor + Hg + Nitrate + 1 mM Mg | Hg induced biosensor + Nitrate + 1 mM Mg | Constitutive biosensor + Hg + Nitrate + 1 mM Mg | Constitutive biosensor + Hg + 1 mM Mg | ||||||||
| d | Hg induced biosensor + Hg + Nitrate + 10 mM Mg | Hg induced biosensor + Nitrate + 10 mM Mg | Constitutive biosensor + Hg + Nitrate + 10 mM Mg | Constitutive biosensor + Hg + 10 mM Mg | ||||||||
| e | ||||||||||||
Table 1: An example plate layout for using the biosensor to test Hg bioavailability (5 nM) over a gradient of Magnesium (0-10 mM)
- Set up the 4 x 8 grid according to the assay plate layout.
- Place 7 mL PTFE standard vials in the tray.
Note: PTFE vials should be acid washed/heat sterilized prior to use. Vials should only be handled by manipulating the outside of the vial. - To each vial, add the exposure medium volume corresponding to each treatment.
Note: In an exposure with a total volume of 2,000 µL (2 mL), the added exposure medium will be: exposure medium µL = 2,000 µL – treatment (blank) µL. (e.g., exposure medium µL = 2,000 – 100 µL (biosensor) – 40 µL (nitrate) – 100 µL (Hg) – 100 µL (variable (e.g., MgSO4)) = 1660 µL). Be sure that the volume added of the tested variable does not exceed 5% (100 µL) of the final volume. - To each vial, add the corresponding volume of the solution of the chemical variable to be tested according to the plate layout.
- From step 1.3, add nitrate to each vial so that the final concentration is 200 µM (40-80 µL). Exclude this step for constitutive biosensor treatment blanks.
- From step 2.1 or 2.2, add Hg (5 nM when testing for a variable) or Cd (300 nM when testing for a variable) to the vials according to the plate layout. Exclude this step for mercury inducible biosensor treatment blanks.
- When using Hg, take the 4-8 µM stock and shake well. Dilute the solution in exposure medium in a 7 mL PTFE vial to 100-250 nM to make a working Hg solution. From this working solution add Hg to the required vials. In this case a calibration curve of Hg ranging from 0 to 12.5 nM.
Note: When testing for a variable’s effect on Hg or Cd bioavailability, make sure that the [Hg] or [Cd] remains constant across all treatments. When adding Hg or Cd, be sure to use one pipette tip but never touch the exposure medium in the vials.
- When using Hg, take the 4-8 µM stock and shake well. Dilute the solution in exposure medium in a 7 mL PTFE vial to 100-250 nM to make a working Hg solution. From this working solution add Hg to the required vials. In this case a calibration curve of Hg ranging from 0 to 12.5 nM.
- Place 7 mL PTFE standard vials in the tray.
- Shake in an orbital motion manually.
Note: The experiment may be paused now depending on the time required for Hg or Cd to speciate in solution. If left for more than an hour, place PTFE caps on the PTFE vials to prevent evaporation/contamination. - Gently pipette Biosensor Stock back and forth to ensure homogeneity. Add 100 µL of Biosensor Stock to each vial. Shake orbitally manually.
- Prepare the plate reader to warm up to read with the following criteria: Temperature at 37˚C, kinetic run for 10 hours with reads every 2.5-5 minutes with orbital shaking in between reads, and fluorescence measurements with a fluorescence excitation of 440 nm and an emission of 500 nm.
- Pipette 200 µL from each PTFE vials in the 4 x 8 grid into the corresponding wells of the 96 well plate (Black, 96-Well Clear-Bottom Nonbinding Surface Microplates). Pipette back and forth 5 times before transferring each 200 µL.
Note: Instead of discarding the pipette tip, leave the pipette tip in the PTFE vial to keep track of pipetting progress. - Place the 96 well plate into the tray of the plate reader, then place the lid on the 96 well plate and begin the assay.
5. Quantifying the Data
- The fluorescence of each treatment at each time point must be corrected for each individual well noise and blanked to the treatment blank.
- The fluorescence for each time point (t) of each treatment (T) must be translated to account for the initial fluorescence (t0) of the first time point of each well, then averaged across the 3 treatments replicates (r1-r3). This treatment average must then be blanked to the average treatment blank (TB) translated in the same manner (Equation 1).
Fluorescence (t) = average(Tr1(t) – Tr1(t0), Tr2(t) – Tr2(t0), Tr3(t) – Tr3(t0)) – average(TBr1(t) – TBr1(t0), TBr2(t) – TBr2(t0), TBr3(t) – TBr3(t0)) (1)
Note: This should be made as a spreadsheet function. Proper propagation of error should also be calculated for each time point.
- Graph the corrected fluorescence of each treatment as a function of time
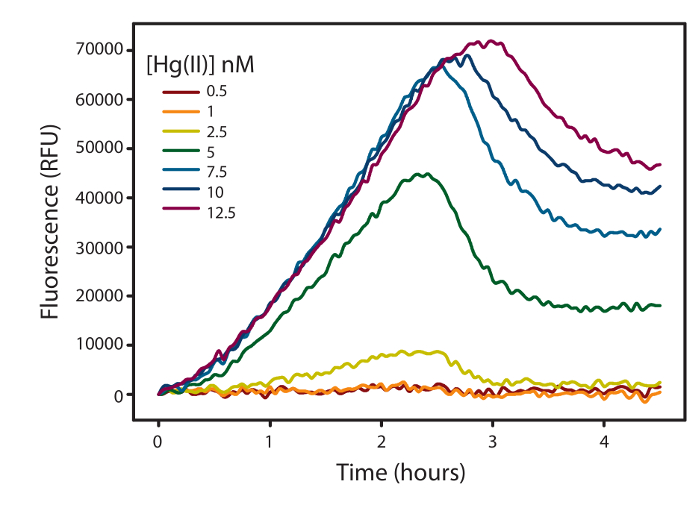
Figure 2: Corrected fluorescence data as a function of time. Fluorescence measured as relative fluorescence units (RFU) emitted by E. coli NEB5α harboring the pUC57merR-Pp (Inducible strain) over time with the addition of HgII (0-12.5 nM) under anaerobic conditions. Fluorescence was the average of 3 technical replicates at 37 ˚C. Please click here to view a larger version of this figure.
Note: There is no 0 nM Hg value on the graph, and all other Hg concentrations have been blanked to the 0 nM Hg as a treatment blank. Therefore, 0 nM Hg represents the x axis and any positive fluorescence represents fluorescence from Hg given any variable. It is optional to not blank the fluorescence in this manner, but the fluorescent curves will give misleading fluorescence curves if the variable tested has background fluorescence (i.e., dissolved organic matter itself will fluoresce and if there is no treatment blank containing just cells and the dissolved organic matter, increasing dissolved organic matter concentration will increase the fluorescent signal).
- Quantify the fluorescence peak. A quantifiable fluorescence peak will typically occur after 2.5-4 hours, representing the energy expended from the consumption of all 200 µM nitrate as a terminal electron acceptor. This peak should be quantified and represents the final fluorescence value of that specific treatment.
Note: In the event no fluorescence peak is observed (i.e., no Hg bioavailability), the fluorescence of treatment at the time point of the fluorescence peak of the control (i.e., no added variable or 5nM Hg) should be quantified. Alternatively, if there is fluorescence induction in the treatment, but fluorescence does not produce a well defined peak, there is likely contamination of a higher affinity electron acceptor such as O2 and measures should be taken to remove traces of oxygen from all solutions and the experiment must be performed again.
Representative Results
Once the fluorescence peaks have been quantified according to step 5.3, the result of the fluorescence peaks can be graphed according to the variable concentration illustrating how that variable affects the relative bioavailability of either Hg or Cd. For example, the calibration curve of fluorescence over [HgII] from Figure 2 will yield the inducible data presented in Figure 3A. For HgII calibration, the curve will always contain 3 components for the Hg-inducible strain; a threshold response of about 1-2 nM HgII before fluorescence signal production is linearly proportional to [HgII], the linear range where 5 nM HgII will reliably always be in the center of that range, and a plateau where increasing [HgII] will no longer increase fluorescence signal. No change in signal production on the constitutive strain shows that toxicity from [HgII] does not affect signal production. For CdII in Figure 3B, there are always 2 components for the inducible strain; a linear range where 200-300 nM CdII will reliably always be in the center of the linear range and a plateau. A decrease in fluorescence signal with increasing [CdII] shows that higher Cd concentrations are toxic to the cells and can explain a decrease in the inducible fluorescence production after the plateau at 1000 nM CdII. Therefore, when testing Hg or Cd bioavailability with respect to an environmental variable, we suggest using 5 nM for HgII and 300 nM for CdII.
In some instances, signal production can be properly attributed to Hg or Cd bioavailability, but in other cases, signal production can be affected by variation in the physiological state of the biosensor cell host (e.g., the metal of interest or environmental conditions tested are toxic). In Figure 4A, 5 nM Hg bioavailability was tested over a gradient of Zn (0-10 µM). In both Hg-inducible and constitutive strains, there is a similar decrease in signal with increasing Zn concentrations. Therefore, one cannot discriminate whether the signal results from lowered bioavailability or is a result of Zn toxicity. In Figure 4B and 4C, 5nM Hg bioavailability was tested over a gradient of MgII (0-10 mM) and MnII (0-10 µM). Increasing MgII and MnII concentrations decreased the fluorescence signal of the inducible strain. On the other hand, the constitutive strain did not show a decrease in fluorescence with increasing MgII and MnII concentrations (MgII and MnII are beneficial for the cells in the production of the FbFp, as demonstrated by an increase in the fluorescence signal). This demonstrates that the cells are viable and the fluorescence decrease of the Hg-inducible strain results from a decrease in Hg bioavailability. This data emphasizes how important it is for all biosensor assays to also provide constitutive measurements of overall cell fitness.
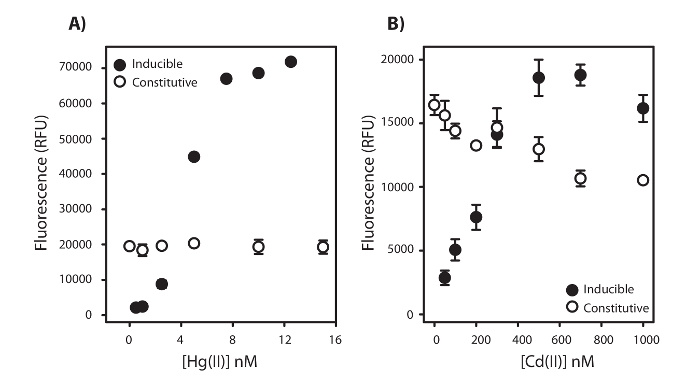
Figure 3: Linear ranges of the biosensor with Mercury and Cadmium. Maximum fluorescence measured as relative fluorescence units (RFU) ± 1 Standard Deviation emitted by E. coli NEB5α harboring the pUC57merR-Pp (Hg-Inducible) and pUC19Balch-Pp (Constitutive) with the addition of A) HgII (0-15 nM) and B) CdII (0-1,000 nM) under anaerobic conditions. Fluorescence was the average of 3 technical replicates at 37 ˚C. Please click here to view a larger version of this figure.
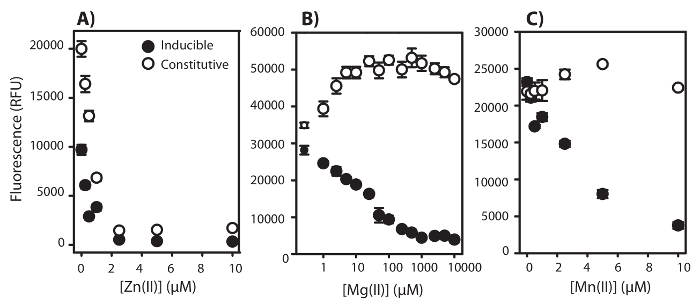
Figure 4: Example of an inconclusive result with Zinc and a conclusive result with Magnesium and Manganese. Maximum fluorescence measured as relative fluorescence units (RFU) ± 1 Standard Deviation emitted by E. coli NEB5α harboring the pUC57merR-Pp (Hg-Inducible) and pUC19Balch-Pp (Constitutive) with the addition of A) ZnII (0-10 µM), B) MgII (0-10 mM), and C) MnII (0-10 µM) under anaerobic conditions. [HgII] was set to 5 nM for all treatments and fluorescence was the average of 3 technical replicates at 37 ˚C. Please click here to view a larger version of this figure.
Discussion
Microbial biosensors are useful tools to identify mechanisms by which metals are interacting with microbes. Here, we describe a method that can anaerobically quantify HgII and CdII bioavailability to a Gram-negative host cell (E. coli) and give a quantifiable result within a few hours. One of the major strengths of this protocol is that it allows extensive control of metal speciation in the exposure medium by avoiding strong binding ligands or components that may lead to metal precipitation. Metal speciation has been modelled and tested in this exposure medium using PHREEQC17, however other metal speciation software may be deployed. In the case of no added ligands, Hg speciation is expected to be present as 97% Hg(OH)2 and 3% Hg(NH3)22+, while Cd speciation will be present as 59% Cd-β-Glycerophosphate, 25% Cd2+, and 16% CdSO4. Using a simple thermodynamic modelling software, the user can design exposure media and test for the bioavailability of the metal of interest. In addition, the biosensor host cell (E. coli NEB5α) is viable over a wide range of pH (5-8.5) and NaCl concentration (0-0.55 M)17.
Hg methylation is an anaerobic process, and the protocol outlined in this study does not have a requirement for oxygen, allowing for more accurate description of anaerobic metabolism on metal bioavailability. This is important because the presence of oxygen alters gene expression profiles48,49 and hence, potential transport pathways; therefore this method presents an advantage over currently exising aerobic alternatives. The biosensing construct presented here can potentially be used with other anaerobic hosts that may be more relevant for mercury methylation (e.g., Geobacter, Desulfovibrio), but maybe less tractable than E. coli. One current limitation of the approach presented here is that our limit of detection has not yet reached pM levels, contrary to existing aerobic systems4,19,37. It is however important to note that to achieve these low detection limits several steps need to be taken44: i) ligand addition is required to ensure that Hg remains in solution and does not adsorb onto the microbial cell wall (Hg will be irreversible bound to cell surface thiols preventing its bioavailability25,27; see the threshold response for Hg in Figure 3A), ii) modifications to cell density, or iii) modify the genetic construct to include transport proteins of the mer-operon (namely merT and merP), increasing Hg flux inside the cell50,51. These modifications would be beneficial in detecting low concentrations of Hg, but not necessarily ideal when assessing environmentally relevant situations. Whereas previous cadmium biosensors primarily exist as a "proof of principle", they were designed in complex media that do not allow the investigator to assess the role of speciation on bioavailability41,42,43,44.
The biosensor is an incredibly useful tool in determining mechanisms in which metal species are bioavailable. Because the host organism is not a Hg-methylator, it may only be used to develop a model for how Hg may enter Gram-negative bacteria and not a definitive rationale for how Hg-methylators acquire Hg. Other methods exist for determining Hg bioavailability, such as methylation, as an outcome of uptake or the use of a mass balance approach10,15,20,45,46. That being said, the method presented here offers the advantage of quasi real time bioavailability data in viable cells. We do not recommend that this method be used to quantify total Hg or Cd levels in an environmental matrix. Despite the proposed use of biosensors to determine metal concentrations in the environment36, many more readily available standard methods are available such as ICP-MS, FAAS (for Cd analysis) or Cold vapor atomic absorption spectroscopy (for Hg analysis). The biosensor can however be used to determine if a given environmental matrix has the potential to enhance or hamper bioavailability; this is achieved by performing standard additions.
The pH of the exposure media may be altered to anywhere within the range of 5 and 8.5, provided the MOPS Free acid (buffer) is exchanged with an alternate free acid of a buffer with the appropriate pKa (please see list of appropriate buffers (Ferreira et al. (2015)47) and adjusted with KOH to the appropriate pH when making the exposure media. In addition, the method is not limited to Hg and Cd, but could be extended to other metals using other transcription regulators.
The exposure assay may be modified to explore the influence of other electron acceptors such as O2 and fumarate on Hg or Cd bioavailability. Minor modifications to the method to utilize both O2 and fumarate as electron acceptors are available upon request.
In summary we would like to emphasize the following points: I) It is imperative that the concentrations of Cd or Hg stocks are known in step 2, as these will be used to calibrate the biosensor. II) On the exposure day, the growth of cells must be stopped at an OD600 of 0.6 (± 0.1) and that care is taken when resuspending the cell cultures, as the biosensor is calibrated to this cell density. III) Lastly, it is important that the exposure medium is made meticulously on the exposure day. To ensure the success of the protocol, multiple cultures should be grown simultaneously (to circumvent the possibility of growth failure) and the growth medium should be remade weekly (to circumvent the metastability of the media and possible contamination). It should also be noted that biological replicates (multiple cell cultures) express variability when it comes to signal production. Although the fluorescent responses may vary from culture to culture, the fluorescent trends in response to a given variable should remain the same throughout numerous biological replicates.
Acknowledgements
We would like to thank comments from two anonymous reviewers as well members of the Poulain Lab for insightful discussion on the development of the anaerobic biosensor. An Early Researcher Award from the Province of Ontario and a Discovery Grant and an accelerator supplement from the Natural Sciences and Engineering Research Council of Canada to A.J.P. funded this study.
Materials
| Name | Company | Catalog Number | Comments |
| 7 ml standard vial, rounded interior | Delta Scientific | 200-007-20 | 34 recommended |
| Vial tray, 21 mm openings | Delta Scientific | 730-2001 | 4 for (4 x 8 grid) |
| 24 mm Closure | Delta Scientific | 600-024-01 | 32 recommended |
| LID CORNER NOTCH BLK STR CS/50 | Corning | Corning 3931 | |
| Corning 96- well clear-bottom nonbinding surface microplate | Corning | Corning 3651 | |
| Anaerobic Chamber (glovebox) | The air in the anaerobic chamber should be (97 % N2 ± 2 % and 3 % H2 ± 2 %) | ||
| Palladium catalyst | Converts O2 to H2O in the anaerobic chamber. Not required but recommended. | ||
| Microplate reader | |||
| 450 (±10) nM filter for the microplate reader | |||
| 500 (±10) nM filter for the microplate reader | |||
| Anaerobic culture vial (balch tubes) + rubber stoppers | |||
| Spectrophotometer | Modified for anaerobic culture tubes | ||
| MA-3000 (mercury analyzer) | |||
| pH probe | Any pH probe will work | ||
| 50 mL conical sterile polypropylene centrifuge tubes. | |||
| 0.22 µm polyethersulfone syringe filter + syringe | |||
| Sterile/clean glass bottles | For growth media and standards | ||
| Sterile/clean plastic or PTFE bottles | For alkali solutions (NaOH/KOH) | ||
| Reagents powders: Na2MoO4, Na2SeO4, H3BO3, NaOH, KOH, MnSO4, ZnSO4, CoCl2, NiCl2, ampicillin sodium salt, Difco M9 Minimal Salts, Glucose, MgSO4, Thiamine HCl, NaNO3, l-leucine, l-isoleucine valine, EDTA sodium salt, FeSO4, Sodium Beta-Gylcerophosphate, Mops free acid, (NH4)2SO4, Hg(NO3)2, CdCl2 | |||
| Sterile Milli-Q water | Autoclaved or filter sterilized is fine. | ||
| Lysogeny Broth | |||
| Analytical grade H2SO4 |
References
- Futsaeter, G., Wilson, S. The UNEP Global Mercury Assessment: Sources, Emissions and Transport. E3S Web of Conferences. 1, 36001 (2013).
- Barkay, T., Gillman, M., Turner, R. R. Effects of dissolved organic carbon and salinity on bioavailability of mercury. Applied and Environmental Microbiology. 63 (11), 4267-4271 (1997).
- Ndu, U., Mason, R. P., Zhang, H., Lin, S., Visscher, P. T. Effect of inorganic and organic ligands on the bioavailability of methylmercury as determined by using a mer-lux bioreporter. Applied and Environmental Microbiology. 78 (20), 7276-7282 (2012).
- Golding, G. R., et al. Evidence for facilitated uptake of Hg(II) by Vibrio anguillarum and Escherichia coli under anaerobic and aerobic conditions. Limnology and Oceanography. 47 (4), 967-975 (2002).
- Golding, G. R., Sparling, R., Kelly, C. A. Effect of pH on intracellular accumulation of trace concentrations of Hg(II) in Escherichia coli under anaerobic conditions, as measured using a mer-lux bioreporter. Applied and Environmental Microbiology. 74 (3), 667-675 (2008).
- Chiasson-Gould, S. A., Blais, J. M., Poulain, A. J. Dissolved Organic Matter Kinetically Controls Mercury Bioavailability to Bacteria. Environmental Science & Technology. 48 (6), 3153-3161 (2014).
- Schaefer, J. K., Szczuka, A., Morel, F. M. M. Effect of Divalent Metals on Hg(II) Uptake and Methylation by Bacteria. Environmental Science & Technology. 48 (5), 3007-3013 (2014).
- Compeau, G., Bartha, R. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Applied and Environmental Microbiology. 50 (2), 498-502 (1985).
- Compeau, G. C., Bartha, R. Effect of salinity on mercury-methylating activity of sulfate-reducing bacteria in estuarine sediments. Applied and Environmental Microbiology. 53 (2), 261-265 (1987).
- Gilmour, C. C., Henry, E. A., Mitchell, R. Sulfate stimulation of mercury methylation in freshwater sediments. Environmental Science & Technology. 26 (11), 2281-2287 (1992).
- Rani, A., Kumar, A., Lal, A., Pant, M. Cellular mechanisms of cadmium-induced toxicity: a review. International Journal of Environmental Health Research. 24 (4), 378-399 (2014).
- Liu, F., Fortin, C., Campbell, P. G. C. Can freshwater phytoplankton access cadmium bound to low-molecular-weight thiols?. Limnology and Oceanography. 62 (6), 2604-2615 (2017).
- Shahid, M., Dumat, C., Khalid, S., Niazi, N. K., Antunes, P. M. C., de Voogt, P. . Reviews of Environmental Contamination and Toxicology. 241, 73-137 (2017).
- Hsu-Kim, H., Kucharzyk, K. H., Zhang, T., Deshusses, M. A. Mechanisms regulating mercury bioavailability for methylating microorganisms in the aquatic environment: a critical review. Environmental Science and Technology. 47 (6), 2441-2456 (2013).
- Szczuka, A., Morel, F. M. M., Schaefer, J. K. Effect of Thiols, Zinc, and Redox Conditions on Hg Uptake in Shewanella oneidensis. Environmental Science and Technology. 49 (12), 7432-7438 (2015).
- Schaefer, J. K., et al. Active transport, substrate specificity, and methylation of Hg(II) in anaerobic bacteria. Proceedings of the National Academy of Sciences of the United States of America. 108 (21), 8714-8719 (2011).
- Stenzler, B. R., Hinz, A., Ruuskanen, M. O., Poulain, A. J. Ionic strength differentially affects the bioavailability of neutral and negatively charged inorganic Hg complexes. Environmental Science and Technology. , (2017).
- Sigel, A. . Cadmium: From Toxicity to Essentiality. , (2013).
- Barkay, T., Turner, R. R., Rasmussen, L. D., Kelly, C. A., Rudd, J. W. Luminescence facilitated detection of bioavailable mercury in natural waters. Methods in Molecular Biology. 102, 231-246 (1998).
- Zhang, T., et al. Methylation of mercury by bacteria exposed to dissolved, nanoparticulate, and microparticulate mercuric sulfides. Environmental Science and Technology. 46 (13), 6950-6958 (2012).
- Ndu, U., Mason, R. P., Zhang, H., Lin, S., Visscher, P. T. Effect of inorganic and organic ligands on the bioavailability of methylmercury as determined by using a mer-lux bioreporter. Applied and Environmental Microbiology. 78 (20), 7276-7282 (2012).
- Moreau, J. W., et al. The Effect of Natural Organic Matter on Mercury Methylation by Desulfobulbus propionicus 1pr3. Frontiers in Microbiology. 6 (1389), (2015).
- Schaefer, J. K., Morel, F. M. M. High methylation rates of mercury bound to cysteine by Geobacter sulfurreducens. Nature Geoscience. 2 (2), 123-126 (2009).
- Dunham-Cheatham, S., Farrell, B., Mishra, B., Myneni, S., Fein, J. B. The effect of chloride on the adsorption of Hg onto three bacterial species. Chemical Geology. 373, 106-114 (2014).
- Dunham-Cheatham, S., Mishra, B., Myneni, S., Fein, J. B. The effect of natural organic matter on the adsorption of mercury to bacterial cells. Geochimica et Cosmochimica Acta. 150, 1-10 (2015).
- Mishra, B., Shoenfelt, E., Yu, Q., et al. Stoichiometry of mercury-thiol complexes on bacterial cell envelopes. Chemical Geology. 464, 137-146 (2017).
- Sara Anne, T., Qing, M., Jean-François, G. Probing changes in Hg(II) coordination during its bacterial uptake. Journal of Physics: Conference. 712 (1), 012078 (2016).
- Macek, T. . Microbial Biosorption of Metals. , (2011).
- Ledin, M., Pedersen, K., Allard, B. Effects of pH and Ionic Strength on the Adsorption of Cs, Sr, Eu, Zn, Cd and Hg by Pseudomonas Putida. Water, Air, and Soil Pollution. 93 (1-4), 367-381 (1997).
- Daughney, C. J., Fein, J. B. The effect of ionic strength on the adsorption of H+, Cd2+, Pb2+ and Cu2+ by Bacillus subtilis and Bacillus licheniformis: A surface complexation model. Journal of Colloid and Interface Science. 198 (1), 53-77 (1998).
- Borrok, D. M., Fein, J. B. The impact of ionic strength on the adsorption of protons, Pb, Cd, and Sr onto the surfaces of Gram negative bacteria: testing non-electrostatic, diffuse, and triple-layer models. Journal of Colloid and Interface Science. 286 (1), 110-126 (2005).
- Daguené, V., et al. Divalent Base Cations Hamper HgII Uptake. Environmental Science & Technology. 46 (12), 6645-6653 (2012).
- Turner, D. R. Speciation and cycling of arsenic, cadmium, lead and mercury in natural waters. Lead, Mercury, Cadmium and Arsenic in the Environment. , 175-186 (1987).
- Liu, G., Cai, Y., O'Driscoll, N. . Environmental Chemistry and Toxicology of Mercury. , (2011).
- North, A. E., Sarpong-Kumankomah, S., Bellavie, A. R., White, W. M., Gailer, J. Environmentally relevant concentrations of aminopolycarboxylate chelating agents mobilize Cd from humic acid. Journal of Environmental Sciences. 57, 249-257 (2017).
- Van Der Meer, J. R., Belkin, S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nature Reviews Microbiology. 8 (7), 511-522 (2010).
- Rasmussen, L. D., Turner, R. R., Barkay, T. Cell-density-dependent sensitivity of a mer-lux bioassay. Applied and Environmental Microbiology. 63 (8), 3291-3293 (1997).
- Magrisso, S., Erel, Y., Belkin, S. Microbial reporters of metal bioavailability. Microbial Biotechnology. 1 (4), 320-330 (2008).
- Golding, G. R., Kelly, C. A., Sparling, R., Loewen, P. C., Barkay, T. Evaluation of mercury toxicity as a predictor of mercury bioavailability. Environmental Science and Technology. 41 (16), 5685-5692 (2007).
- Drepper, T., et al. Reporter proteins for in vivo fluorescence without oxygen. Nature biotechnology. 25 (4), 443-445 (2007).
- Kumar, S., Verma, N., Singh, A. K. Development of cadmium specific recombinant biosensor and its application in milk samples. Sensors and Actuators B: Chemical. 240, 248-254 (2017).
- Yoon, Y., et al. Simultaneous detection of bioavailable arsenic and cadmium in contaminated soils using dual-sensing bioreporters. Applied and Environmental Microbiology. 100 (8), 3713-3722 (2016).
- Kang, Y., Lee, W., Jang, G., Kim, B. G., Yoon, Y. Modulating the sensing properties of Escherichia coli-based bioreporters for cadmium and mercury. Applied and Environmental Microbiology. 102 (11), 4863-4872 (2018).
- Hynninen, A., Tõnismann, K., Virta, M. Improving the sensitivity of bacterial bioreporters for heavy metals. Bioengineered Bugs. 1 (2), 132-138 (2010).
- Graham, A. M., Bullock, A. L., Maizel, A. C., Elias, D. A., Gilmour, C. C. Detailed assessment of the kinetics of Hg-cell association, Hg methylation, and methylmercury degradation in several Desulfovibrio species. Applied and Environmental Microbiology. 78 (20), 7337-7346 (2012).
- Graham, A. M., Aiken, G. R., Gilmour, C. C. Dissolved Organic Matter Enhances Microbial Mercury Methylation Under Sulfidic Conditions. Environmental Science & Technology. 46 (5), 2715-2723 (2012).
- Ferreira, C. M. H., Pinto, I. S. S., Soares, E. V., Soares, H. M. V. M. (Un)suitability of the use of pH buffers in biological, biochemical and environmental studies and their interaction with metal ions-a review. RSC Advances. 5 (39), 30989-31003 (2015).
- Salmon, K., Hung, S. P., Mekjian, K., Baldi, P., Hatfield, G. W., Gunsalus, R. P. Global gene expression profiling in Escherichia coli K12: the effects of oxygen availability and FNR. Journal of Biological Chemistry. 278 (32), 29837-29855 (2003).
- Salmon, K. A., Hung, S. P., Steffen, N. R., Krupp, R., Baldi, P., Hatfield, G. W., Gunsalus, R. P. Global Gene Expression Profiling in Escherichia coli K12 effects of oxygen availability and ArcA. Journal of Biological Chemistry. 280 (15), 15084-15096 (2005).
- Larose, C., Dommergue, A., Marusczak, N., Coves, J., Ferrari, C. P., Schneider, D. Bioavailable mercury cycling in polar snowpacks. Environmental Science & Technology. 45 (6), 2150-2156 (2011).
- Omura, T., Kiyono, M., Pan-Hou, H. Development of a specific and sensitive bacteria sensor for detection of mercury at picomolar levels in environment. Journal of Health Science. 50 (4), 379-383 (2004).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved