Immunization of Alpacas (Lama pacos) with Protein Antigens and Production of Antigen-specific Single Domain Antibodies
In This Article
Summary
A method for the production of single domain antibodies from alpacas, including immunization, blood collection, B-cell isolation, and selection is described.
Abstract
In this manuscript, a method for the immunization of alpaca and the use of molecular biology methods to produce antigen-specific single domain antibodies is described and demonstrated. Camelids, such as alpacas and llamas, have become a valuable resource for biomedical research since they produce a novel type of heavy chain-only antibody which can be used to produce single domain antibodies. Because the immune system is highly flexible, single domain antibodies can be made to many different protein antigens, and even different conformations of the antigen, with a very high degree of specificity. These features, among others, make single domain antibodies an invaluable tool for biomedical research. A method for the production of single domain antibodies from alpacas is reported. A protocol for immunization, blood collection, and B-cell isolation is described. The B-cells are used for the construction of an immunized library, which is used in the selection of specific single domain antibodies via panning. Putative specific single domain antibodies obtained via panning are confirmed by pull-down, ELISA, or gel-shift assays. The resulting single domain antibodies can then be used either directly or as a part of an engineered reagent. The uses of single domain antibody and single domain antibody-based regents include structural, biochemical, cellular, in vivo, and therapeutic applications. Single domain antibodies can be produced in large quantities as recombinant proteins in prokaryotic expression systems, purified, and used directly or can be engineered to contain specific markers or tags that can be used as reporters in cellular studies or in diagnostics.
Introduction
Alpacas, and other members of the camelid family, have become a popular source for generating antibodies for biomedical research1,2,3. Camelids have a unique immune system in that they produce both normal antibodies as well as heavy-chain only antibodies which allow the production of much smaller single domain antibodies, while retaining high specificity and high affinity. Thus, camelid antibodies provide a versatile reagent useful for a variety of purposes. A method to immunize alpaca and produce single domain antibodies to a variety of protein antigens is here described. These antibodies can be produced in camelids via a relatively straightforward process that only requires the immunization via small injections of the target protein (antigen) and a blood draw4. Subsequently, library construction, M13 phage display5 and panning versus the recombinant antigen6 is used to isolate and produce single domain antibodies with the desired characteristics. Because the process takes advantage of the power of phage display technology, five or more antigens can be simultaneously utilized per animal, thereby reducing the number of animals used and associated costs.
Typical mammalian antibodies or immunoglobulins are large molecules consisting of two types of chains (2 heavy chains and 2 light chains), which are linked together through disulfide bonds. These antibodies are relatively large, exhibiting molecular weights of ~140-190 kDa for the more abundant IgG species from humans, mice, goats, and rabbits. Due to their multi-subunit structure, high molecular weight, and disulfide bonds, immunoglobulins can be rather challenging to produce in large quantities, making their cost high. In the 1990's, it was discovered that camelids, which includes camels, llamas and alpacas make, in addition to the typical mammalian type immunoglobulin, a novel form of immunoglobulin that is simpler in structure, possessing only heavy chains7. These unique camelid antibodies possess the same ability to specifically bind foreign substances with high affinity but are only made up of one protein chain8. Furthermore, camelid antibodies can be experimentally truncated to an even smaller unit the VHH fragment, also called a single domain antibody9. Due to their small size, single domain antibodies are useful for a wide range of biomedical research applications and are available from a variety of academic and commercial sources1,10. An exception appears to be Western blotting since single domain antibodies are more often directed against conformation dependent epitopes, which are usually lost under the denaturing conditions used in Western blots.
Since they are made up of a single polypeptide chain, specific single domain antibodies can be selected and easily produced in large quantities as recombinant proteins in bacteria. Single domain antibodies can also be genetically engineered to contain specific markers or tags that can be used as "reporter groups" for diagnosing diseases11. This ease of manipulation coupled with their flexibility of use makes single domain antibodies an important resource for researchers at universities and elsewhere.
As part of a National Institutes of Health supported core, existing methods have been adapted to produce single domain antibodies from alpacas3,4. Alpacas have significant advantages in both ease of handling relative to larger camelid family members as well as accessibility. Alpacas are widely raised for fiber and meat and thus can be obtained regionally from local alpaca farmers, who can be identified through their websites or through state breeder associations. Two breeds of alpacas are available, Suri and Huacaya. Huacaya are more common and were used for this protocol, but the protocol is generally applicable to either breeds and also more widely applicable to other camelids.
Protocol
All procedures with the alpacas were performed in accordance with protocols (2017-2627 and 2018-2925) approved by the University of Kentucky's Institutional Animal Care and Use Committee (IACUC).
1. Generation of Antigen-specific Alpaca Antibodies
- Alpaca handling and general considerations
- Obtain appropriate institutional and/or regulatory approval.
- Obtain adult alpacas, less than 10 years of age with a body condition score of at least 3.0 (scale 1-5)12. Because they are herd animals, house a minimum of two but preferably three animals together.
NOTE: Males are recommended because, in general, they have a more even temperament and are easier to work with. - Check the health status of animals through a veterinary examination including the review of diet, housing, vaccination and parasite control programs13. Ensure that the animals are up to date with vaccinations appropriate to the local geographic region based on professional veterinary recommendations. Develop an ecto- and endoparasite control program in consultation with a veterinarian based on a review of the history of the herd of origin and specific local conditions13.
- Acclimatize for at least one week, including halter training and exposure to restraint.
- Collect a 4 mL blood sample to obtain serum for use as a pre-immunization control (see Step 1.4). Freeze the sera and store at -20 °C in 0.5 mL aliquots.
NOTE: At this time, additional blood can be taken for a complete blood cell (CBC) analysis to monitor the animals' initial health status.
- Antigen preparation
- Prepare purified protein antigens in phosphate-buffered saline (PBS) or HEPES-buffered saline (HBS) and concentrate to ≥1 mg/mL. Approximately 3 mg of protein is needed for the complete protocol, including the immunization, panning, and confirmation of clones.
NOTE: Camelid antibodies display high specificity and selectivity against protein antigens but are generally not well suited for unstructured proteins or peptide antigens which are generally unstructured. - Establish antigen purity, with purity >90% desired. Utilize an analytical method such as SDS-PAGE.
CAUTION: Significant contamination with other proteins can lead to problems during the selection where single domain antibodies can be isolated to contaminants rather than the target protein. - Prepare frozen aliquots of antigen that contains a total of 100 µg of antigen. 50 µL of a 2 mg/mL preparation is ideal. 100 µL of 1 mg/mL is the most dilute protein solution suitable for immunization. Alternatively, if the antigen cannot undergo freeze/thaw and is known to be stable in solution long term, it can be stored at 4 °C.
NOTE: To ensure the antigen can undergo a freeze/thaw cycle, test a 20 µL aliquot of the antigen should be dispensed into a thin walled PCR tube and snap frozen in liquid nitrogen. Thaw the tube, perform high speed centrifugation, and verify the quantitative recovery by comparing UV/VIS absorption before and after freezing or by running an SDS-PAGE gel. If feasible, the antigen should also be tested for the retention of biological activity. If the freeze thaw cycle is successful, the remaining antigen is snap frozen in 100 µL aliquots and stored at -80 °C. If there are issues with the freeze/thaw cycle, the protocol can be repeated with the addition of up to 20% glycerol.
- Prepare purified protein antigens in phosphate-buffered saline (PBS) or HEPES-buffered saline (HBS) and concentrate to ≥1 mg/mL. Approximately 3 mg of protein is needed for the complete protocol, including the immunization, panning, and confirmation of clones.
- Immunization
- Mix up to five antigens, 200 µg each, with adjuvant in a final volume of between 300 to 1,000 µL. The adjuvant should constitute ≥50% of the final volume using water as the diluent.
NOTE: If buffer is necessary, use HEPES, MOPS, or glycine. A pH between 5 and 6 has often proved to be more favorable than strictly neutral. Avoid polyvalent ions such as phosphate or citrate, since they can provoke coagulation. - Trim the alpaca's hair with electric clippers, in a crescent moon pattern along the base of its neck from shoulder to shoulder to expose the regions adjacent to the bow lymph nodes.
- Holding the alpaca by the neck, slowly inject the antigen subcutaneously along the base of the neck near the bow lymph nodes using a 22 G needle. Perform five injections of ~200 µL (or less) ~5 cm apart.
NOTE: The animals should be restrained by a second staff member during the injections. Injecting and bleeding alpacas calls for caution since they are prone to kicking, not just from behind but sideways. Having the animals close to each other, in a group, during the procedures is optimal. Given that they are herd animals, they are calmer in a group and less forceful restraint is needed during the procedures. - Monitor the animals for ~20 min post injection for signs of anaphylaxis (see Step 1.6). Monitor for localized inflammation at the injection sites weekly, which is not expected when using the recommended adjuvant. Ensure that the blood leak from the site of injection is not excessive.
- Perform five subsequent boosting injections weekly using 100 µg of each antigen in the mixture. Clipping hair at the injection sites may be required weekly depending on the season. In fall and winter, the animals' hair can grow rapidly.
- Mix up to five antigens, 200 µg each, with adjuvant in a final volume of between 300 to 1,000 µL. The adjuvant should constitute ≥50% of the final volume using water as the diluent.
- Drawing blood
- Clip and disinfect the collection site with alcohol swabs.
- Restrain the animal gently with the neck held upright and straight.
NOTE: Animals can be restrained manually as demonstrated in the video. If available, use of an alpaca chute to restrain the alpacas during bleeding and injections can ease handling. To use an alpaca chute, haltered alpacas are led into the chute and restrained with neck bars and straps with quick releases. Versions of the alpaca chute are available from commercial vendors. - Using the ventral projection of the transverse process of the vertebrae as a landmark, occlude the vein by applying pressure just medial to the ventral process.
NOTE: There is no distinct jugular furrow in adult alpaca. The jugular veins are protected by ventral projections of the transverse vertebral processes. This thick skin (e.g., 1 cm thick "fighting plate" over the middle 1/3 of neck in males) and neck musculature makes it difficult to visualize or palpate the jugular vein itself. Subsequently, the use of vertebral landmarks is the most helpful indicators for jugular location. - Insert the needle at a 45° angle slightly medial (~finger width) to the projection toward the center of the neck. Manipulate the needle until the blood flows.
NOTE: The jugular vein and carotid artery are close to each other. Venous blood can be quite red in color but is still darker than arterial blood. Following the collection, pressure should be applied to the site for 0.5-1 min (or longer if the carotid is accessed) until hemostasis is assured. - Draw 4-10 mL of blood for analytical purposes. Utilize a 1.5 inch, 20 G needle typically in conjunction with an appropriate sized syringe or blood collection tube. Utilize lavender top K2EDTA tubes for collecting the whole blood when purifying lymphocytes. Utilize gold top serum separation tubes (BD) when collecting the blood for serum production.
- Draw 50-100 mL of blood for production purposes. Use an additional 12-inch infusion extension set with a 19-20 G winged "butterfly" infusion set.
NOTE: An extension set configuration allows an assistant to fill multiple blood collection tubes, allowing the venipuncturist to focus on stabilizing the collection needle. The addition of the infusion set accommodates potential movement of the animal (3-5 min procedure) and gives a comfortable working distance for the operators.
- Immune monitoring
- Three to four days after the 3rd and 5th injections, perform a small test bleed from each animal to obtain sera for testing. Freeze and store the sera in 0.5 mL aliquots. Obtain additional blood samples at the same time to monitor the health of the animal, as discussed above.
- Confirm the presence of antigen-specific antibodies by ELISA using the sera obtained from test bleeds at pre-immune, three-week, and five-week time points. It is best to take a final bleed while the antibody titer is still increasing.
- Generate antigen affinity plates using surface treated 96-well plates. Dilute 0.1 µg of the antigen in 100 mM sodium carbonate buffer, pH 9.0. Add 100 µL of the solution to each well and incubate overnight at 4 °C. Ten dilutions of each antigen are tested in duplicate. A blank control well is prepared that is coated with carbonate buffer only.
- After the overnight incubation, invert the plate to remove the contents of the wells and wash three times with 0.1 mL of PBS, 0.1% Tween 20 (PBS-T).
- Block the wells by adding 0.1 mL of BSA (5 mg/mL) in PBS-T and incubating the plate for 1 h at room temperature.
- Wash the plate three times with PBS-T by pipetting the wash solution into the well and then removing the wash solution by the inversion of the plate.
- Prepare two-fold serial dilutions of 0.3 mL each of pre-immune and immune sera in PBS-T buffer using a starting dilution of 1/100, and up to 1/102,400.
- Add 100 µL of each dilution to the wells and allow to incubate for 2 h at room temperature.
- Remove the serum from each well and wash with 0.2 mL of PBS-T three times.
- Incubate each well with 0.1 mL of HRP-conjugated goat anti-llama IgG antibody at a dilution of 1:20,000 for 1 h.
NOTE: The immune response measured includes both conventional as well as heavy-chain only antibodies, which are present in approximately equal ratios in circulation3,14. - Remove the antibody solution and wash each well with 0.2 mL of PBS-T three times.
- Add 100 µL of chromogenic peroxidase substrate to the well and incubate at room temperature until the intensity of the two highest concentration points have become saturated, which usually takes 5-10 min.
- Add 100 µL of 0.1 M sulfuric acid to each well to stop the reaction.
- Measure the absorbance of each well at 450 nm using a plate reader.
- Plot the absorbance as a function concentration.
- Alpaca monitoring and potential complications
- Ensure appropriate alpaca monitoring throughout the procedure.
NOTE: The procedures are not expected to cause significant adverse effects. Animal well-being and health should be monitored daily by husbandry and/or research personnel during feeding and watering. Any animal with unalleviated experimentally-induced, or spontaneous natural morbidity should be referred for the evaluation by veterinary services with treatment(s) provided as appropriate, up to humane euthanasia, if necessary.
Potential adverse consequences associated with phlebotomy are bleeding, bruising, and hematoma. The animals should be monitored for 20 min post blood draw to look for the signs of bleeding. Additional pressure on the sites should be used. If bleeding does not stop, a veterinarian should be contacted.
Other potential adverse consequences associated with injections and bleeding include granuloma formation and inflammation. Granuloma formation is not expected and has not been observed at the University of Kentucky. Injection site inflammation (redness or swelling) could occur and should be brought to the attention of a veterinarian. The production bleed represents a potentially significant volume of blood, and the animals should be monitored to assure that they are stable and active after the blood collection.
Anaphylaxis and pyrogenic responses are the two most likely serious adverse consequences, but are very rarely observed. If it were to occur, Anaphylaxis would occur shortly after the antigen injections and be manifested by an increase in rectal temperature, weakness, vomiting, breathing problems, and/or diarrhea. Rectal temperature can be monitored after each injection to detect any increase in body temperature. If these are recognized, notify a veterinarian immediately for a consultation. Additionally, an emergency "Crash Kit" (e.g., epinephrine, corticosteroids, and antihistamine) prepared in consultation with a veterinarian should be available to treat animals suspected of having an adverse reaction.
- Ensure appropriate alpaca monitoring throughout the procedure.
2. Purifying Lymphocytes for Single Domain Antibody Library Construction
- Collect 50 mL of blood three to five days after the last injection (See Step 1.4).
NOTE: Rapid processing of blood and isolation of RNA is important to obtain a suitable library size15,3, which is very important for the success of panning. - Dilute 15 mL of the collected blood with 5 mL of PBS.
- Utilize a lymphocyte separation tube to isolate peripheral blood lymphocytes (PBLs) by density gradient centrifugation according to manufactures suggestions.
CAUTION: Manufacturer's instructions indicate that up to 25 mL of blood can be used, but this may saturate the system and prevent obtaining a clean lymphocyte preparation. - Add 5 mL of PBS prechilled to 4 °C to PBL.
- Spin for 20 min at 800 x g at 4 °C.
- Wash two times by adding 5 mL of PBS prechilled to 4 °C to PBL followed by the centrifugation for 20 min at 800 x g at 4 °C.
- Isolate total RNA using a spin-column based system, according to the manufacturer's instructions. Homogenize the cells by vertexing in appropriate buffer and applying to a biopolymer-shredding system. Use an appropriate number of mini or maxi-columns based on the cell input.
- Synthesize cDNA using reverse transcriptase by combining 10 µg of RNA with a mix of 0.1 µg of Oligo(dT)12-18 primers.
- Generate a bacteriophage library using established methods4. This involves cloning with restriction enzymes into the phage display vector pMES4 followed by the expression of the insert fused to gene III of the filamentous phage for the production of the phage solution.
3. Single Domain Antibody Panning
- Produce antigen affinity plates using surface treated 96-well plates. Dilute 0.25 µg of the antigen in 100 mM sodium carbonate buffer, pH 9.0. Add 100 µL of this solution to each well and incubate overnight at 4 °C. Coat two wells per antigen. Prepare a blank control well that is coated with carbonate buffer only.
NOTE: Plates can be produced and stored for up to one week at 4 °C if multiple rounds of selection will be performed. Also, this method utilizes direct absorption which does not require labeling while other methods use biotinylation or other types of labeling strategies. - Incubate the plate overnight. Invert to remove the contents of the wells and wash three times with 0.1 mL of PBS-T.
- Block the wells by adding 0.1 mL of BSA (20 mg/mL) in PBS and incubating the plate for 2 h at room temperature.
- Wash the plate three times with PBS-T followed by three times with PBS. Antigens can also be covalently labelled and used in capture systems such as, for example, streptavidin/biotin systems.
- Pre-treat the phage solution (100 µL) with 100 µL of 40 mg/mL BSA in PBS on a vibrating platform at room temperature for 30 min.
- Add the phage solution to the antigen coated wells and incubate with gentle agitation for 2 h at room temperature. Use multiple wells in order to ensure a total of 1011 phage for each antigen.
- Discard the unbound phage by inversion and then wash the wells five times with 200 µL of PBS-T followed by five times with PBS.
- Elute the bound phage by adding 0.1 mL of 0.25 mg/mL trypsin in PBS and incubating for 30 min at room temperature on the vibrating platform at 700 rpm.
- Transfer the solution to a vial containing 5 µL of 4 mg/mL 4-benzenesulfonyl fluoride hydrochloride(AEBSF) solution to quench trypsin activity.
- Grow TG1 phage display competent cells at 37 °C with shaking until OD600 = 0.5.
- Add 50 µL of eluted phage to 3 mL of the TG1 cells.
- Incubate at 37 °C without shaking for 30 min.
- Add 7 mL of LB media supplemented with 100 µg/mL ampicillin, 2% glucose.
- Shake overnight at 37 °C.
NOTE: To obtain the most diverse single domain antibodies, clones can be sequenced, tested for desired properties, and utilized after one round of panning. To obtain the highest affinity single domain antibodies, two rounds of panning should be utilized. - Plate the bacteria after overnight growth on LB agar plates supplemented with 100 µg/mL ampicillin, 2% glucose. Several serial dilutions will need to be tested, starting with two dilutions, plating 100 µL of 1/10,000 and 1/100,000 dilutions.
- Incubate the plate overnight at 37 °C.
- Pick the colonies and inoculate the wells of a sterile 96-well plate containing 100 µL of 2XYT with 100 µg/mL ampicillin, 2% glucose, 10% v/v glycerol per well.
- Incubate overnight at 37 °C without shaking.
- The next day, shake at 170 rpm at 37 °C for 60 min.
- Remove 2 µL of the media into PCR tubes. The remaining solution can be left in the plate and stored at 4 °C for 1-2 days or frozen and stored at -80 °C for later use.
- Perform PCR reaction using primers appropriate for the vector utilized. For pMES4, PCR screening of positive clones uses primers that flank the multiple cloning site are CCTATTGCCTACGGCAGCCGCTGGATTGTTATTAC and GGTGGTGTGAGGAGACGGTGACCTG. Utilize cycling conditions appropriate for the polymerase used, an annealing temperature of 57 °C, and 30 cycles.
- Analyze the PCR reaction by running a 1% (w/v) agarose gel. Colonies that are positive have single domain antibody gene inserts of ~500 bp. Ensure that positive clones are 20-50% of the sample.
NOTE: This method provides a means for clone selection and analysis that can be performed in most research laboratories. Alternatively, next-generation sequencing can be applied and provides large datasets which allow statistical and comparative analysis16. - Inoculate the positive clones from the plate into fresh tubes containing 7 mL of LB with 100 µg/mL ampicillin.
- Grow with shaking for overnight at 37 °C.
- Perform a miniprep on the culture to obtain plasmid DNA.
- Sequence positive clones using the standard sequencing primer pEX-Rev (CAGGCTTTACACTTTATGCTTCCGGC).
- Perform computational analysis on resulting single domain antibody sequences. Ensure that there are no stop codons or frame shifts.
- Classify the resulting sequences for similarity and diversity. The sequences that are repeatedly identified are most likely to be high affinity binding sequences, particularly after two rounds of selection.
- Express the single domain antibody using a BL21-derived expression strain of E. coil. Induce the expression through the addition of IPTG, growing overnight at 22 °C.
- Purify the single domain antibody using immobilized metal affinity chromatography (IMAC).
- Validate single domain antibody/antigen binding using a direct interaction assay such as pull-down, native gel electrophoresis, or ELISA.
- Verify application-specific single domain antibody performance, as desired for the specific experiment.
Representative Results
The protocol presented here was utilized to generate single domain antibodies against a range of protein antigens. Five antigens were utilized per alpaca. Immune monitoring indicated that the majority of antigens showed robust response beginning at three weeks (Figure 1). Production bleeds and library construction after approximately six weeks gives the best balance for the animals that have multiple antigens injected. Two rounds of panning were performed for each antigen, and isolated colonies screened (Figure 2). Sequencing of positive colonies identified diverse single domain antibody sequences for the different antigens. For examples, three unique clones were isolated to the reference antigen Maltose Binding Protein (MBP) (Figure 3A). As is frequently observed, the single domain antibodies contain significant sequence diversity and highly variable CDR3 length (Figure 3). These features are particularly important in single domain antibody/antigen interactions as seen in the reference single domain antibody/CX3CL1 complex17 (Figure 3B).
The majority of full-length single domain antibody sequences can be expressed and purified at 1-10 mg/L of culture (Figure 4A). Because of the diversity in CDR3 length, different single domain antibodies show slight variation in molecular weight. Confirmation of direct binding and application specific performance is critical. For example, after two rounds of selection against Antigen B, a panel of single domain antibodies were identified (Figure 2). Multiple identical sequences were identified, and the single domain antibody was produced and purified. It was then tested via direct pull down with antigen affinity resin and showed robust dose-dependent binding (Figure 4B). Importantly, the single domain antibody showed no binding to control resin. These data demonstrate a direct binding interaction, and one that is well suited for affinity capture in both pull-down and ELISA-based formats.
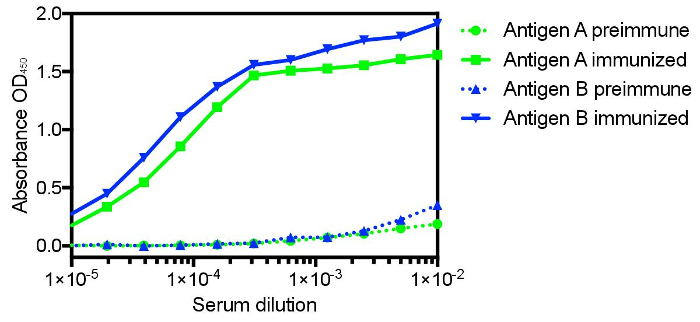
Figure 1: Representative immune monitoring of two distinct antigens showing the significant specific immune response to distinct antigens in the same animal. Many antigens show robust response as early as three weeks after commencing immunization, with the majority showing maximal response after six weeks. Six weeks also allows for affinity maturation and is the recommended time for the production bleed and B-cell isolation. Please click here to view a larger version of this figure.
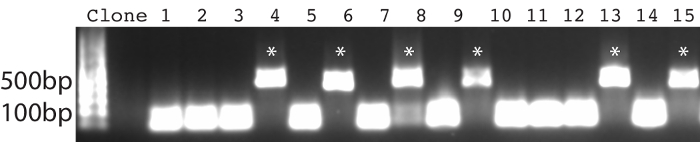
Figure 2: PCR-based confirmation of positive clones. Primers span the MCS of the pMES4 vector, and a positive nanobody clone produces an amplicon of ~500 bp (marked with *). Please click here to view a larger version of this figure.
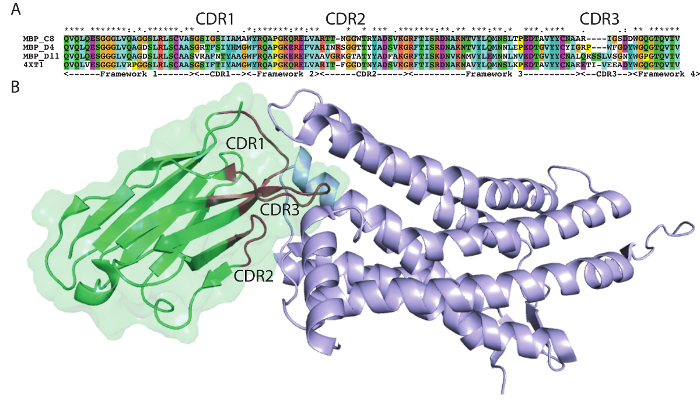
Figure 3: Antigen-specific sequences highlight alpaca single domain antibody diversity. (A) Alignment of select single domain antibody sequences isolated to MBP with a reference single domain antibody 4XT1, with framework and complementarity-determining regions (Kabat) highlighted below. (B) Structure of alpaca single domain antibody 4XT1 bound to CX3CL1 (PDB 4XT1), emphasizing the critical role of variable CDR loops in specific antigen binding. Please click here to view a larger version of this figure.
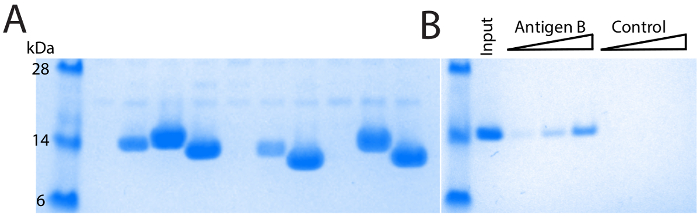
Figure 4: Purification and validation of single domain antibodies. (A) SDS-PAGE of IMAC purified single domain antibodies, comparing diverse single domain antibodies. Different molecular weights are primarily due to varying length of the CDR3 loop. Differing levels of expression are also observed, with a minority of single domain antibodies showing poor yields of purified protein. (B) Verification of direct binding using an antigen affinity pull-down. Robust dose-dependent single domain antibody binding is observed with antigen-coupled resin but not with control resin. Please click here to view a larger version of this figure.
Discussion
As noted, the high affinity and specificity of single domain antibodies, combined with their facile expression and stability, make them ideal reagents for the applications in biomedical research, as critical biochemical reagents, diagnostic tools, or therapeutic agents18. For commercial applications, there are some issues related to intellectual property that must be considered. Additionally, single domain antibodies can be engineered to contain specific markers or tags that can be used as reporters in cellular assays or in diagnostics. The key to these uses is the ability to produce specific single domain antibodies against the antigens of interest. A method is described here to produce single domain antibodies using alpacas, which produces a robust immune response against a wide variety of protein antigens. Considerations for animal handling and regulatory compliance are described. These are key for the success in this part of the process.
There are other strategies for producing single domain antibodies that do not require the immunization, but instead use semisynthetic libraries with different systems for the selection and iterative optimization of affinities19,20. However, the advantage of using libraries derived from immunized animals is the ability to reliably isolate highly enriched, diverse, and high-affinity single domain antibodies. Additionally, not only are significant sequence variations observed, but a significant majority of specific single domain antibodies isolated had unique insertions and deletions, particularly in CDR3.
Further, single domain antibodies can be produced via a straightforward process that only requires small injections of the antigen, and after this process, the animals can be returned to the herd. Of note, the animal can be freely used for fiber production, but must be excluded from use as meat (human food chain) due to the use of the test antigens and non-FDA approved adjuvant for single domain antibody production. When the procedure is completed, the animals should be monitored for one week, given a final veterinary exam, and then can be either returned to their home farm or rested for six months prior to another round of single domain antibody production.
Acknowledgements
This work was supported by the National Institutes of Health (P30GM103486).
Materials
| Name | Company | Catalog Number | Comments |
| GERBU FAMA adjuvant | Biotechnik, Heidelberg, Germany | 3003,6001 | |
| Serum collection tube | Becton Dickinson | 367983 | |
| Blood collection tube | Becton Dickinson | 366643 | |
| Vacutainer blood collection set | Becton Dickinson | 368652 | |
| Maxisorp Immuno plates | Nunc | 439454 | |
| BSA | Sigma-Aldrich | A7906 | |
| HRP-conjugated goat anti-llama IgG antibody | Bethyl Labs | A160-100P | |
| TMB reagent chromogenic peroxidase substrate | KPL | 50-76-03 | |
| Plate Reader | Spectramax | M5 | Any UV/VIS capable reader is acceptable |
| Uni-SepMAXI+ lyphocyte separation tube | Novamed | U-17 | |
| RNeasy Mini Kit | Qiagen | 74104 | |
| QIAshredder column | Qiagen | 79654 | |
| Superscript IV reverse transcriptase | Invitrogen | 18064014 | |
| AEBSF solution | Biosynth | A-5440 | |
| TG1 phage display competent cells | Lucigen | 60502 |
References
- Krah, S., et al. Single-domain antibodies for biomedical applications. Immunopharmacology and Immunotoxicology. 38 (1), 21-28 (2016).
- Rothbauer, U., et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nature Methods. 3 (11), 887-889 (2006).
- Maass, D. R., Sepulveda, J., Pernthaner, A., Shoemaker, C. B. Alpaca (Lama pacos) as a convenient source of recombinant camelid heavy chain antibodies (VHHs). Journal of Immunological Methods. 324 (1-2), 13-25 (2007).
- Pardon, E., et al. A general protocol for the generation of Nanobodies for structural biology. Nature Protocols. 9 (3), 674-693 (2014).
- Hertveldt, K., Belien, T., Volckaert, G. General M13 phage display: M13 phage display in identification and characterization of protein-protein interactions. Methods in Molecular Biology. 502, 321-339 (2009).
- Kushwaha, R., Schafermeyer, K. R., Downie, A. B. A protocol for phage display and affinity selection using recombinant protein baits. Journal of Visualized Experiments. (84), e50685 (2014).
- Hamers-Casterman, C., et al. Naturally occurring antibodies devoid of light chains. Nature. 363 (6428), 446-448 (1993).
- Muyldermans, S., et al. Camelid immunoglobulins and nanobody technology. Veterinary Immunology and Immunopathology. 128 (1-3), 178-183 (2009).
- Muyldermans, S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 82, 775-797 (2013).
- Muyldermans, S., Cambillau, C., Wyns, L. Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends in Biochemical Sciences. 26 (4), 230-235 (2001).
- Wang, Y., et al. Nanobody-derived nanobiotechnology tool kits for diverse biomedical and biotechnology applications. International Journal of Nanomedicine. 11, 3287-3303 (2016).
- Van Saun, R. J. Nutritional requirements and assessing nutritional status in camelids. Veterinary Clinics of North America: Food Animal Practice. 25 (2), 265-279 (2009).
- Jones, M., Boileau, M. Camelid herd health. Veterinary Clinics of North America: Food Animal Practice. 25 (2), 239-263 (2009).
- Daley, L. P., Gagliardo, L. F., Duffy, M. S., Smith, M. C., Appleton, J. A. Application of monoclonal antibodies in functional and comparative investigations of heavy-chain immunoglobulins in new world camelids. Clinical and Diagnostic Laboratory Immunology. 12 (3), 380-386 (2005).
- Romao, E., et al. Identification of Useful Nanobodies by Phage Display of Immune Single Domain Libraries Derived from Camelid Heavy Chain Antibodies. Current Pharmaceutical Design. 22 (43), 6500-6518 (2016).
- Henry, K. A., et al. Isolation of TGF-beta-neutralizing single-domain antibodies of predetermined epitope specificity using next-generation DNA sequencing. Protein Engineering, Design and Selection. 29 (10), 439-443 (2016).
- Burg, J. S., et al. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science. 347 (6226), 1113-1117 (2015).
- Wesolowski, J., et al. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Medical Microbiology and Immunology. 198 (3), 157-174 (2009).
- Harmsen, M. M., De Haard, H. J. Properties, production, and applications of camelid single-domain antibody fragments. Applied Microbiology and Biotechnology. 77 (1), 13-22 (2007).
- McMahon, C., et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nature Structural & Molecular Biology. 25 (3), 289-296 (2018).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved