Composition and Distribution Analysis of Bioaerosols Under Different Environmental Conditions
In This Article
Summary
Understanding the biological composition of environmental particulate matter is important for the study of its significant impacts on human health and disease spread. Here, we used three types of bioaerosol sampling methods and a biological analysis of airborne microbes to better explore airborne microbial communities under different environmental conditions.
Abstract
Variable microorganisms in particulate matter (PM) under different environmental conditions may have significant impacts on human health. In this study, we described a protocol for multiple analyses of the biological compositions in environmental PM. Five experiments are presented: (1) PM number monitoring by using a laser particle counter; (2) PM collection by using a cyclonic aerosol sampler; (3) PM collection by using a high-volume air sampler with filters; (4) culturable microbes collection by the Andersen six-stage sampler; and (5) detection of biological composition of environmental PM by bacterial 16SrDNA and fungal ITS region sequencing. We selected hazy days and a livestock farm as two typical examples of the application in this protocol. In this study, these two sampling methods, cyclonic aerosol sampler and filter sampler, showed different sampling efficiency. The cyclonic aerosol sampler performed much better in terms of collecting bacteria, while these two methods showed the same efficiency in collecting fungi. Filter samplers can work under low temperature conditions while cyclonic aerosol samplers have a sampling limitation for temperature. A solid impacting sampler, such as an Andersen six-stage sampler, can be used to sample bioaerosols directly into the culture medium, which increases the survival rate of culturable microorganisms. However, this method mainly relies on culture while more than 99% of microbes cannot be cultured. DNA extracted from the culturable bacteria collected by the Andersen six-stage sampler and samples collected by cyclonic aerosol sampler and filter sampler were detected by bacterial 16S rDNA and fungal ITS region sequencing.All the methods above may have wide application in many fields of study, such as environmental monitoring and airborne pathogen detection. From these results, we can conclude that these methods can be used under different conditions and may help other researchers further explore the health impacts of environmental bioaerosols.
Introduction
In natural environments, various kinds of microorganisms exist in bioaerosols, including fungi, bacteria, viruses and other microorganisms1. Airborne microorganisms, which can be emitted from some human activities such as animal feeding operations in livestock farms, are the important contents of the atmospheric environment2. These microorganisms may not only play important roles in the atmospheric environment but also have significant impacts on human health and diseases spread.
As an important way of spreading diseases, microbial aerosols have attracted wide attention throughout the world. In recent studies, many human diseases were found to be associated with the complex composition of environmental particulate matter (PM) in different locations, such as chemical factories, livestock farms and smoggy cities3,4. The biological composition of PM may contribute to some respiratory and cardiovascular diseases in PM-exposed humans5. Different body regions, such as the mucosa, skin, digestive tract and respiratory tract, can be potential targets of microbes attached to PM6,7. An increased risk of lung cancer might be caused by prolonged exposure to PM2.58.
Airborne bacteria have been surveyed at various locations in several countries around the world, including in subway stations, veterinary hospitals, slaughterhouses, composting facilities, tanneries, milk-processing facilities, coal mines, dental clinics and indoor environments9,10,11,12,13,14,15,16,17, generating a large number of reports about biological aerosols. Crowded places associated with campuses, livestock farms and large cities during hazy days are three particularly important public conditions for which we need to explore the connections between human health and the potential effects of PM exposure. Furthermore, during winter days in the northern cities of China, high PM2.5 values may affect human health. Although PM2.5 can generate toxic effects by targeting respiratory surfaces and dissolving into blood18, it is still not clear whether and how the microbes attached to PM2.5 could potentially affect human health19,20. Livestock farms are one of the main sources of PM and microbial aerosols in the air. The large numbers of pathogens, such as influenza virus and Brucella melitensis, that are carried by aerosols in the fields around livestock farms are important factors causing respiratory diseases in both livestock and poultry workers.
In this study, we explored multiple types of analyses of bioaerosols, including PM number monitoring, bioaerosol collection and biological composition analysis. Air samples were collected by a cyclonic aerosol sampler, a high-volume air sampler with filters and an Andersen six-stage sampler. Then, the samples collected by these three samplers were analyzed by biological analysis including bacterial 16S rDNA and fungal ITS sequencing to determine their biological compositions. Herein, we show representative results from the bioaerosol samples collected during Beijing hazy days and from livestock farms indicating that bioaerosols might have great impacts on human and animal health. The comparison between liquid and filter sampling methods were also explored in this study mainly based on the data from 16S DNA and fungal ITS sequencing.
Protocol
1. PM Number Monitoring
- Use an airborne laser particle counter (see Table of Materials) to determine the total PM number. Collect the PM by the air sampling port on the top of the airborne laser particle counter.
NOTE: This particle counter is an automation equipment and it can work independently when the program including sampling time, interval, collection times, etc. has been set on its touch screen.- Equip the instrument with a sensor to monitor temperature and relative humidity. Measure and record temperature and relative humidity data simultaneously every 5 min.
- In total, measure and record 6 different particle size classes (0.3-0.5 μm, 0.5-0.7 μm, 0.7-2.5 μm, 2.5-5 μm, 5-10 μm and > 10 μm) simultaneously every 5 min. Measure 4 other particle size classes (0.3-0.5 μm, 0.5-1 μm, 1-3 μm and ≥ 3 μm).
- Collect the air samples though the sampling hole on the top of the sampler and use the test modules inside the sampler to measure the particle size of each PM. Then the data is stored automatically. All the processes above can be performed automatically after the relative parameters, including sampling time and counting range, are set though the mini touch displayer of the airborne laser particle counter.
NOTE: This particle counter is an automation equipment and it has test modules that count the number of PM of different particle size classes.
- To assess experimental standard error, ensure that different particle size classes are counted with at least 15 replications. Ensure that readings are stored in the internal memory of the instrument and subsequently analyzed.
- Ensure that analysis of the primary data includes PM number concentration, the ANOVA test and the percentage of PM in each size class. For example, as shown in Figure 1A, the PM number concentrations in December (237.6 particles/cm3) were significantly higher than those in October (average: 110.2 particles/cm3) (ANOVA; P-value < 0.05). Figure 1B shows that the number of particles smaller than 3 μm accounted for more than 99% of the total number.
- Use the laser particle counter to monitor the PM number concentrations and store the data automatically. Use a USB flash disk to export data in the computer and perform the ANOVA test.
2. PM Collection by Cyclonic Aerosol Samplers
- Perform the PM sampling in an open area without any nearby major pollution sources at ∼2 m above the ground.
- Record the position of the sampling site. For example, the campus of Beijing Institute of Technology is the following: 39°57’51.0’’ N; 116°19’38.5’’ E.
- Use a cyclonic aerosol sampler to collect air samples. Use a sampler flow of 323 L/min, and a collection time of 6 h.
NOTE: The sampler flow rate is fixed and cannot be changed. The collection time is controlled by manual time-keeping as there is only a Start and Stop button on this sampler.- Use sterile water to wash the inside of the cyclonic aerosol sampler 3 times before collection, and use the automatic cleaning function 3 times by continuously pressing the collect button 3 times.
- Press the collect button to start sampling and press the pump button to stop sampling. Put the sampler on a shelf or floor with no one holding it in place.
- Preserve all samples in the dark at -20 °C until the subsequent analyses.
3. PM Collection by Filters
- Perform the PM sampling in an open area without any nearby major pollution sources at ∼2 m above the ground.
- Record the position of the sampling site. For example, the campus of Beijing Institute of Technology is as follows: 39°57’51.0’’ N; 116°19’38.5’’ E.
- Collect the PM samples on 20.32 × 25.4 cm2 filters (see Table of Materials) by using a high-volume air sampler (see Table of Materials) at a flow rate of 1,000 L/min. Set the flow rate and collection time by relative auto-programming.
- Preserve all sampled filters in the dark at -20 °C until the subsequent analyses.
4. Biological Composition Analysis
- For biological composition analysis, use each liquid sample from the cyclonic aerosol sampler or filter sample for DNA extraction by a multisource DNA extraction Kit (see Table of Materials) according to the manufacturer's protocols.
- For samples from the high-volume air sampler with filters, use 1/8 of each filter sample for DNA extraction. Put the filter into a 50 mL tube with the sample facing inward and the back facing towards the tube wall. Add 10 beads in the central sites towards the side of the filter containing the sample.
- Vortex the tube for 15 min at room temperature. Pipette the liquid in the tube into a clean 50 mL centrifuge tube to continue the DNA extraction process.
- Extract the DNA from the sample by the processes as follows.
- Centrifuge the bacteria sample at 2,000 x g for 5 min, remove the supernatant, and suspend bacteria in 2 mL of sterile PBS buffer.
- Collect the suspension of bacteria in a 2 mL centrifuge tube and then centrifuge at 2,000 x g for 5 min to discard the supernatant solution.
- Add 350 μL of sterile PBS buffer containing the suspended bacteria to the solution.
- Add 0.8 mL of RNase A.
- Add 150 μL of Buffer CL and 8 μL of proteinase K, and mix immediately by vortex shaking for 1 min. After brief centrifugation at 1,000 x g for 30 s (no residues on the wall), put the centrifugal tube in 56 °C water for 10 min.
- Add 350 μL of Buffer PD, and mix for 30 s and then centrifuge for 10 min at 12,000 x g.
- Place the DNA preparation tube in a 2 mL centrifuge tube, and transfer the mixture made in step 4.2.6 to the preparation tube. Then centrifuge for 1 min at 12,000 x g.
- Discard the filtrate, put back the contents of the preparation tube into the original 2 mL centrifuge tube, and add 50 μL of Buffer W1. Centrifuge the mixture for 1 min at 12,000 x g.
- Discard the filtrate, put back the contents of the preparation tube into the original 2 mL centrifuge tube, and add 700 μL of Buffer W2. Centrifuge the mixture for 1 min at 12,000 x g. Repeat this step to wash again with 700 μL of Buffer W2.
- Discard the waste liquid, and put back the contents of the preparation tube into the original 2 mL centrifuge tube. Centrifuge the mixture for 1 min at 12,000 x g.
- Put the contents of the DNA preparation tube into another clean 1.5 mL centrifuge tube, and add 100 μL of eluent or deionized water to the middle of the membrane in the preparation tube (deionized water or eluent was heated to 65 °C). Allow the mixture to stand at room temperature for 1 min, and Centrifuge the mixture for 1 min at 12,000 x g.
- Perform quantitative real-time polymerase chain reaction (Q-RT-PCR) to quantify the relative abundances of bacteria and fungi on the sampling filters.
- Use primers as follows: for bacterial 16S rDNA, 515F (5’- GTG CCA GCM GCC GCG GTA A-3’) and 806R (5’- GGA CTA CHV GGG TWT CTA AT-3’); and for the fungal ITS, ITS1 (5’-TCC GTA GGT GAA CCT GCG G-3’) and ITS1 (5’-GCT GCG TTC TTC ATC GAT GC-3’).
- Run the Q-RT-PCR assays on a Real-Time PCR System (see Table of Materials). RT-PCR condition: predegeneration, 95 °C, 10 min; degeneration, 95 °C, 15 s; anneal and extension, 60 °C, 1 min; 40 cycles from degeneration to anneal and extension.
- For bacterial and fungal community structure analysis, amplify the V1–V3 region of the bacterial 16S rDNA and the ITS region of the fungal rRNA operon by PCR.
- Use the primers as follows: for bacteria, V1-9F (5′-CCT ATC CCC TGT GTG CCT TGG CAG TCT CAG ACG AGT TTG ATC MTG GCT CAG-3′) and V3-541R (5′-CCA TCT CAT CCC TGC GTG TCT CCG ACT CAG-barcode-ACW TTA CCG CGG CTG CTG G-3′); and for fungi, ITS-3F (5’-CCT ATC CCC TGT GTG CCT TGG CAG TCT CAG CAC ATC GAT GAA GAA CGC AGC-3’) and ITS-4R (5’-CCA TCT CAT CCC TGC GTG TCT CCG ACT CAG-barcode-GCT CCT CCG CTT ATT GAT ATG C-3’).
- Ensure that the 20 μL mixture of PCRs included 2 μL of 2.5 mM dNTPs, 4 μL of 5× FastPfu Buffer, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA (see Table of Materials).
- Use the PCR program as follows: 94 °C for 5 min; followed by 10 cycles of 94 °C for 30 s, 55-60 °C for 45 s, and 72 °C for 90 s; 20 cycles of 94 °C for 30 s, 55 °C for 45 s and 72 °C for 90 s; and a final extension at 72 °C for 5 min.
- Perform DNA purification, DNA quantification and pyrosequencing as described in a previous study 21.
5. Bioaerosol Sampling and Cultivation
- Use an international standard Andersen six-stage sampler (see Table of Materials) to sample culturable airborne bacteria and fungi at a flow rate of 28.3 L/min. Use a sampling time of 35 min. Put the culture plate in each stage of the sampler. Sufficiently disinfect the sampler with 75% ethyl alcohol after each sampling.
NOTE: The sampler flow rate is fixed and the collection time is controlled by manual time-keeping.- Sample the six stages with the Andersen six-stage sampler, which is defined by the aerodynamic diameters of the airborne particles, including stage VI (0.65–1.1 μm), stage V (1.1–2.1 μm), stage IV (2.1–3.3 μm), stage III (3.3–4.7 μm), stage II (4.7–7.0 μm) and stage I (≥7.0 μm).
- Collect the bacterial particles and deposit onto the culture plate in each stage containing Soybean-Casein Digest Agar (see Table of Materials). There is a container with many air intakes on the top in each stage of the sampler.
- Culture the airborne bacteria collection plates at 37 °C for 24-48 h; then, count the colony numbers of bacteria on the sample dish for each stage.
- To prevent the particles collected by the Andersen six-stage sampler from overlapping, correct the number of colonies at each sampler level by the following formula:
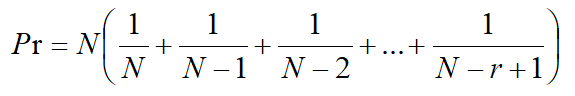
where Pr: corrected number of colonies at each level,
N: number of sampling holes of the sampler at all levels (400), and
r: the actual count of colonies. - Calculate the unit of colonies per m3 of air as follows:
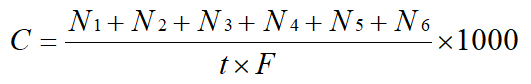
where C: concentration (CFU/m3),
N1-N6: the corrected number of colonies at each level,
T: sampling time (min), and
F: sampling flow rate (28.3 L/min). - Use methods from Steps 5.1 - 5.3 to study the bioaerosol in livestock farm, including four types of piggeries.
NOTE: The livestock farm is located in Changchun, China. The central position 2 m above the ground in each piggery was selected as sampling location.) After culture of bacteria, treat all colonies in the plates as described in Steps 6.1 - 6.2.
6. Identification of Culturable Bacteria
- After 48 h or 72 h of culturing, place the bacteria into a 2 mL centrifuge tube. Extract the DNA of these bacteria or fungi by using a multisource DNA extraction Kit (see Table of Materials). The DNA extraction process is described in section 4.2.
- Use 200 μL DNA extraction sample for 16S rDNA sequencing. Use the same processes as described in Steps 4.2, 4.3 and 4.4 on biological composition analysis.
Representative Results
In this study, we performed an assessment of the overall PM distribution and a comprehensive analysis of the bioaerosols in a dairy farm from September to December. Many environmental factors contribute to the distribution of aerosol particles. We studied the concentration and size distributions of PM in a cow house by using a TSI laser particle counter. As shown in Figure 1A, the concentration of aerosol particles was highest in December and lowest in October, which might be caused by changes in temperature and humidity (Table 1). The concentration of inhalable aerosol particles (0.3-3.0 µm) accounted for more than 99% of the total particle concentration (Figure 1B), and the particles in this range could reach the deep respiratory tract, causing serious hazards for humans and animals.
Biological composition analysis of samples can be performed by DNA extraction, bacterial 16SrDNA and fungal ITS region sequencing instead of microorganism culture. From the biological analysis of bioaerosol samples collected by using a cyclonic aerosol sampler or a high-volume air sampler with filters, we can preliminarily compare the efficiencies of these two methods in collecting bacteria and fungi. Figure 2 showed the analysis results of the bioaerosol samples collected during Beijing hazy days in the campus of Beijing Institute of Technology in December 20, 2016. For bacteria collection, the results indicated that the cyclonic aerosol sampler collected many more genera than the high-volume air sampler with filters (Figure 2A). For fungi collection, these samplers showed equal collection efficiencies and almost the same genus abundances (Figure 2B). From the results presented in Figure 2, we were able to measure the different collection efficiencies of these two methods for bacteria and fungi. For bacteria collection, the cyclonic aerosol sampler performed much better than the high-volume air sampler with filters because the samples from the former showed a higher genus abundance (Figure 2A). However, the fungal sequencing analysis of the two samples from different sampling methods showed nearly identical community structures (Figure 2B).
We studied airborne culturable bacteria by using an Andersen six-stage sampler. As shown in Figure 3, the colony numbers of culturable bacteria for stage I-VI particles was reduced. Stage I particles (particle size > 8.2 µm) had the highest numbers of culturable bacteria colonies. The percentage of stage I colonies in four different types of piggeries including farrowing house, pregnant sow house, fattening house and weaning house was 33%, 30%, 26% and 34%, respectively. The percentage of stage II colonies in four different types of piggeries was 20%, 22%, 19% and 20% respectively. The percentage of stage III colonies in four different types of piggeries was 18%, 18%, 18% and 19% respectively. The percentage of stage IV colonies in four different types of piggeries was 17%, 16%, 16% and 16% respectively. The percentage of stage V colonies in four different types of piggeries was 10%, 10%, 14% and 6% respectively. Stage VI particles (particle size < 1.0 µm) had the lowest numbers of culturable bacteria colonies. The percentage of stage VI colonies in the four different types of piggeries was 3%, 5%, 6% and 5%, respectively.
The air samples were collected in four different types of piggeries by using an Andersen six-stage sampler and then cultured under suitable conditions. The whole-genome DNA of the culturable bacteria collected from each particle stage was extracted and detected by bacterial 16S rDNA and fungal ITS region sequencing. A total of 91 genera and 158 species of bacteria were identified in the culturable bacteria in piggeries. The culturable bacteria community structures in four different types of piggeries, including farrowing house, pregnant sow house, fattening house and weaning house are shown in Figure 4 with data from stage I to stage VI. The content of different predominant bacterial genera is not the same among different piggeries.
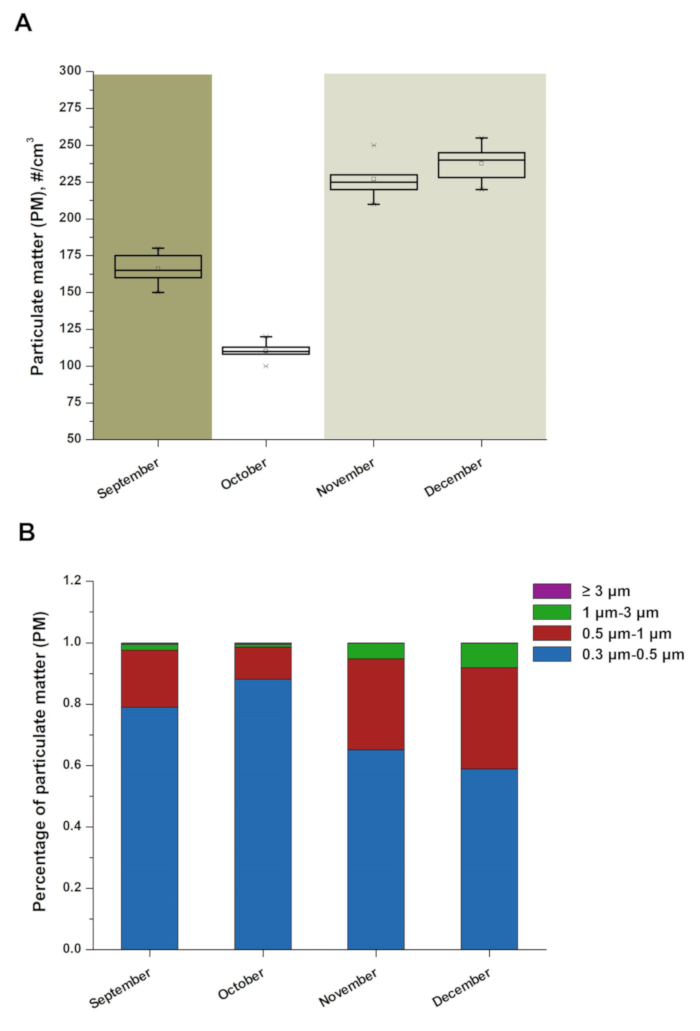
Figure 1: The concentration and size distribution of PM in four different months. (A) Boxplot of PM number during the study period. Each boxplot includes the Maximum, minimum, median, two quartiles and abnormal value of the database. (B) Average size distribution maps of PM. There were eighty cows in the house from September to December. The percentage of PM (≥ 3 µm) in four months (September, October, November and December) was 0.005, 0.005, 0.002 and 0.002 respectively. Please click here to view a larger version of this figure.
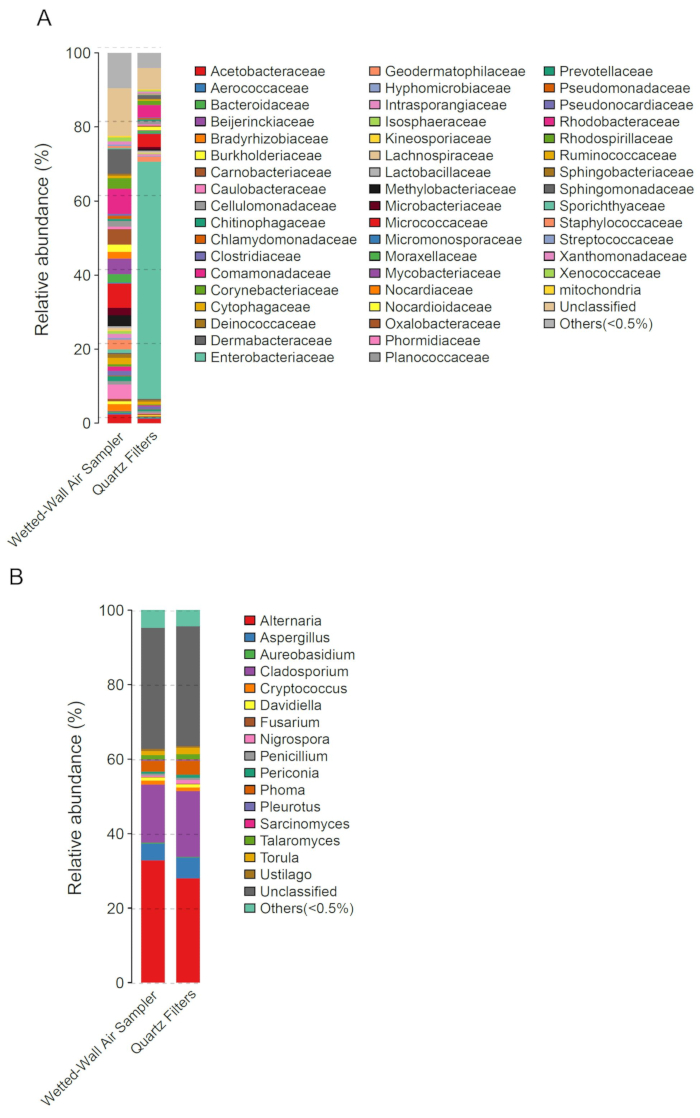
Figure 2: Biological analysis of bioaerosol samples collected by two samplers. (A) and (B) show the abundances of bacterial or fungal genera in bioaerosol samples obtained with different collection methods. Wetted-Wall Air Sampler represents the cyclonic aerosol sampler. Quartz Filters represents the high-volume air sampler with filters. Please click here to view a larger version of this figure.
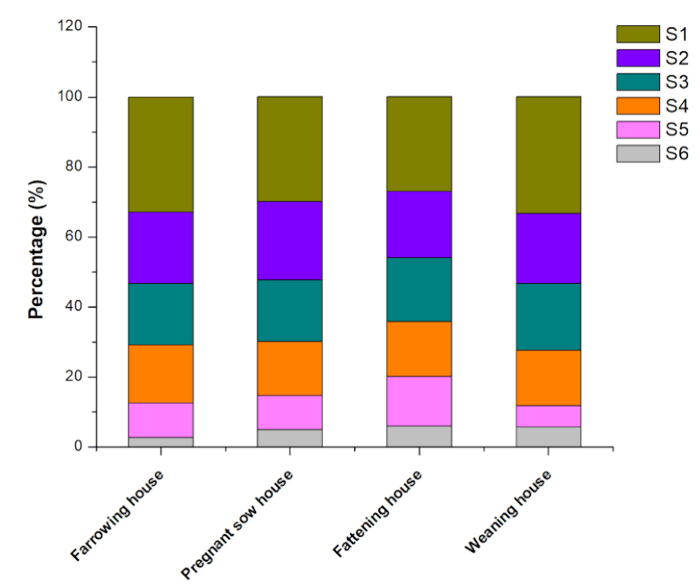
Figure 3: Average hierarchical distribution maps of culturable bacteria in four types of piggeries. The four types of piggeries are farrowing house, pregnant sow house, fattening house and weaning house. S1 to S6 represent the six particle stages (I to VI) collected by the Andersen six-stage sampler. The stages were defined by the aerodynamic diameters of the airborne particles, including stage VI (0.65-1.1 µm), stage V (1.1-2.1 µm), stage IV (2.1-3.3 µm), stage III (3.3-4.7 µm), stage II (4.7-7.0 µm) and stage I (7.0 µm). Please click here to view a larger version of this figure.
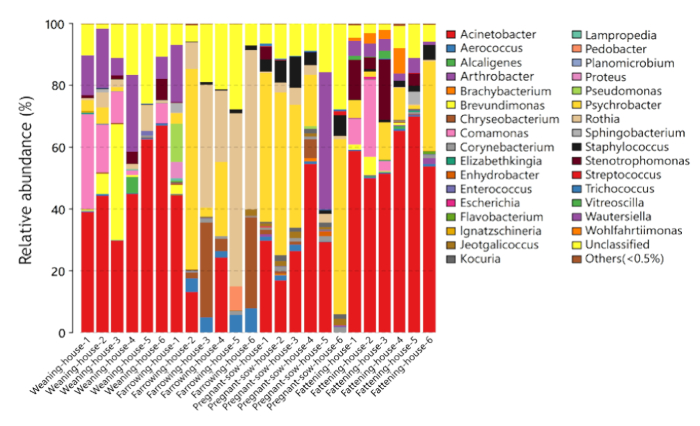
Figure 4: Bacterial community structures with different abundances in the air samples. The air samples were collected in four different types of piggeries by using an Andersen six-stage sampler and then cultured under suitable conditions. The whole-genome DNA of the culturable bacteria in each particle stage collected by the Andersen six-stage sampler was extracted and detected by bacterial 16S rDNA sequencing. The numbers 1 to 6 in each type of piggery represent particle stages I to VI measured by the Andersen six-stage sampler. The text on the right side includes the genus name for each bacterium. Please click here to view a larger version of this figure.
Discussion
In this study, we provided some representative results that were obtained during hazy days and in livestock farms. The results from bioaerosol samples taken during Beijing hazy days facilitated a better understanding of the biological compositions of the biological composition of PM without PM present during Beijing hazy days. The results from samples taken from livestock farms will also provide basic data for environmental air quality control in piggeries and a theoretical foundation and technical support for healthy breeding and safe production in livestock farms. Many environmental factors, such as temperature, humidity and wind speed, might contribute to the distribution of aerosol particles in the cow house (Figure 1). Previous studies have revealed that the PM in livestock farms mainly comes from the fodder, feces, fur and feather, which may be related to the animal activities. The environmental factors can affect not only animal activities, but also the aggregation and diffusion of PM in a relatively closed cow house. Therefore, we detect different concentrations and size distributions of PM in four different months. Besides, the sanitary condition, feeding method and animal activity were different between the four types of piggeries, which might also affect their community structure of culturable bacteria in the air (Figure 4).
However, in this study, we mainly focused on the available methods that we could use to study the composition and distribution of bioaerosols under different environmental conditions. Compared with other microbial samples, those of airborne microorganisms have very low concentrations and are mixed with a large number of impurities, such as inorganic dust particles, which introduce certain difficulties during the collection and detection of such microorganisms21. Therefore, appropriate methods of collection and detection should be selected for microbial aerosols. The collection of microbial aerosol samples is generally performed by using the precipitation method or specialized equipment to collect the microorganisms in liquid, semi-solid or solid sampling medium22,23,24. Then, some corresponding technical treatment and specific testing and analysis are subsequently carried out25. The sampling medium should keep microorganisms intact to reduce the error associated with detection and analysis26. However, different aerosol microorganism samplers have different effects on the integrity of samples due to their different sampling principles and media. People have designed many kinds of bioaerosol samplers that use different sampling principles, such as inertial impaction, filtration resistance, and electrostatic precipitation27.
Impacting samplers can push airborne PM into the sampling medium at high speed by using extraction equipment. There are two types of impacting samplers: solid and liquid.Solid impacting samplers can be used to sample bioaerosols at a low concentration and can be almost impervious to air flow28. Aerosol particles of different sizes can be preliminarily screened, and microbes can be sampled directly into the culture medium, which increases the survival rate of culturable microorganisms10. Due to inertial impact, microbial aerosol particles easily collide at the same site, and colonies may overlap easily after culture. At present, the most common solid impacting sampler is the Andersen-6 microbial aerosol sampler. In this study, we used an Andersen six-stage sampler to study the culturable bacteria distributed in airborne PM of different sizes.
Cyclonic aerosol samplers use cyclones to spiral air at high speed into a cylinder or cone. Bioaerosol particles can be separated from the airflow by centrifugal force, which means that microbes can bump into the inner wall of the sampler and then be collected by the sampling buffer. This method is convenient and can be used for long sampling times in large flow and sampling operations. However, this method cannot be implemented at low temperatures because the operation is dependent on liquid. The cyclonic aerosol sampler used in this study is a cyclonic wetted-wall aerosol sampler that can extract and transfer airborne pathogens and particles from sampled air into a small volume of water for analysis29.
Filter samplers can work under low temperature conditions and can sample particles above a certain size. However, they have a great impact on microbial activity and are prone to damage, which will greatly influence subsequent studies on sampling activity. In this study, bioaerosol samples from Beijing hazy days were collected by using both a high-volume air sampler with filters and a cyclonic aerosol sampler. The aim of this experiment was to analyses the biological composition of PM without the separation and culturing of airborne microbes. Therefore, these two sampling methods were suitable for this study. For the cyclonic aerosol sampler method, airborne microbes at a low concentration can be easily extracted into the running buffer and then can be analyzed conveniently without the additional treatment that is commonly used for filter samples. In this study, these two sampling methods, cyclonic aerosol sampler and filter sampler, showed different sampling efficiency. The additional treatment that is commonly used for filter samples, such as recovering the sample from the filter, is one of the main differences between these two methods. Besides, the air samples were collected directly into the running buffer by cyclonic aerosol sampler while the other method collected samples on filters. The characteristics of different type of sampling method might contribute to this different sampling efficiency. We can assume that the cyclonic aerosol sampler is the better choice for microorganism collection, and this assumption needs further confirmation.
In this study, bacterial 16S rDNA and fungal ITS region sequencing were used to perform the biological analysis of bioaerosols. 16SrDNA sequencing is the determination of 16S rDNA segments in the microbial genome30. 16S rDNA widely exists in prokaryotes with high conservation and specificity, which makes it useful for the identification of microbial species31. Whole-genome sequencing requires only the extraction of genomic DNA and subsequent sequencing. In addition to producing a large amount of data, this process also allows for a more comprehensive analysis of microbial community structure. Besides, metagenomics can also be used in this field of study by providing more information in the future. Handelsman et al. first proposed the concept of the metagenome in a 1998 paper on microbes in soil32. In subsequent studies, the concept of the metagenome was gradually accepted, and much research was carried out on the microbes included in the human gut, ocean and soil33,34,35. With the support of high-throughput sequencing technology, metagenomics has developed rapidly, and it plays an increasingly important role in the study of pathogen detection. Traditional microbial research methods mainly rely on culture for separation and purification. However, many studies cannot be carried out because more than 99% of microbes cannot be cultured. In contrast to traditional methods, metagenomics can take the genetic information of all the microbes in the environment as a whole without the need to separate individual organisms36. A comprehensive analysis of all the resulting microorganisms can be conducted directly.
In summary, this study showed several detection, sampling and analysis methods that could be used for studies on the biological composition of environmental PM, including PM monitoring; PM sampling by an Andersen six-stage sampler, a high-volume air sampler with filters or cyclonic aerosol sampler; and subsequent biological analysis based on DNA sequencing. In practice, these methods can be used under different environmental conditions, such as many types of livestock farms. Our protocols and results may help other researchers all over the world further explore the health impacts of fungal and bacterial bioaerosols in the environment.
Acknowledgements
Financial support for this study came from the National Natural Science Foundation of China (No. 41775148). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Materials
| Name | Company | Catalog Number | Comments |
| airborne laser particle counter | TSI Inc, MN, USA | model 9306 | |
| Andersen six-stage sampler | Tisch Inc, USA | TE-20-600 | |
| AxyPrep multisource DNA Miniprep Kit | Axygen, NY, USA | AP-MN-MIS-GDNA-50G | |
| FastPfu Polymerase | TransGen Inc., Beijing, China | AP221-01 | |
| High-volume air sampler | Beijing HuaRui HeAn Technology Co., Ltd., China | HH02-LS120 | |
| Real-Time PCR System | Thermo Fisher Scientific, USA | Applied Biosystems® 7500 | |
| Soybean-Casein Digest Agar | Becton, Dickinson and company, MD, USA | 211043 | |
| Tissuquartz filters | Pall, NY, USA | 7204 | |
| Wetted-Wall Air Sampler | Research International, Inc. 17161 Beaton Road SE Monroe, Washington 98272-1034 USA | SASS 2300 |
References
- Jones, A. M., Harrison, R. M. The effects of meteorological factors on atmospheric bioaerosol concentrations--a review. The Science of the total environment. 326 (1-3), 151-180 (2004).
- Ko, G., et al. Investigation of bioaerosols released from swine farms using conventional and alternative waste treatment and management technologies. Environmental science & technology. 42 (23), 8849-8857 (2008).
- Seagrave, J., et al. Lung toxicity of ambient particulate matter from southeastern U.S. sites with different contributing sources: relationships between composition and effects. Environmental health perspectives. 114 (9), 1387-1393 (2006).
- Janssen, N. A., et al. Black carbon as an additional indicator of the adverse health effects of airborne particles compared with PM10 and PM2.5. Environmental health perspectives. 119 (12), 1691-1699 (2011).
- Langrish, J. P., et al. Reducing personal exposure to particulate air pollution improves cardiovascular health in patients with coronary heart disease. Environmental health perspectives. 120 (3), 367-372 (2012).
- Haas, D., et al. The concentrations of culturable microorganisms in relation to particulate matter in urban air. Atmospheric Environment. 65 (Supplement C), 215-222 (2013).
- Stahlhofen, W., Gebhart, J., Heyder, J. Experimental determination of the regional deposition of aerosol particles in the human respiratory tract. American Industrial Hygiene Association. 41 (6), 385-398 (1980).
- Abba, E. J., Unnikrishnan, S., Kumar, R., Yeole, B., Chowdhury, Z. Fine aerosol and PAH carcinogenicity estimation in outdoor environment of Mumbai City, India. International journal of environmental health research. 22 (2), 134-149 (2012).
- Dybwad, M., Skogan, G., Martha Blatny, J. Temporal Variability of the Bioaerosol Background at a Subway Station. Concentration Level, Size Distribution, and Diversity of Airborne Bacteria. 80, (2013).
- Harper, T. A., et al. Bioaerosol sampling for airborne bacteria in a small animal veterinary teaching hospital. Infection ecology & epidemiology. 3, (2013).
- Hall, R. J., et al. Metagenomic detection of viruses in aerosol samples from workers in animal slaughterhouses. PloS one. 8 (8), e72226 (2013).
- Wery, N. Bioaerosols from composting facilities--a review. Frontiers in cellular and infection microbiology. 4, 42 (2014).
- Skora, J., Gutarowska, B., Stepien, L., Otlewska, A., Pielech-Przybylska, K. The evaluation of microbial contamination in the working environment of tanneries. Medycyna pracy. 65 (1), 15-32 (2014).
- Brandl, H., et al. Distribution and identification of culturable airborne microorganisms in a Swiss milk processing facility. Journal of dairy science. 97 (1), 240-246 (2014).
- Wei, M., Yu, Z., Zhang, H. Molecular characterization of microbial communities in bioaerosols of a coal mine by 454 pyrosequencing and real-time PCR. Journal of environmental sciences. 30, 241-251 (2015).
- Polednik, B. Aerosol and bioaerosol particles in a dental office. Environmental research. 134, 405-409 (2014).
- Veillette, M., et al. Microbial contents of vacuum cleaner bag dust and emitted bioaerosols and their implications for human exposure indoors. Applied and environmental microbiology. 79 (20), 6331-6336 (2013).
- Cao, C., et al. Inhalable microorganisms in Beijing's PM2.5 and PM10 pollutants during a severe smog event. Environmental science & technology. 48 (3), 1499-1507 (2014).
- Lippmann, M., Chen, L. C. Health effects of concentrated ambient air particulate matter (CAPs) and its components. Critical reviews in toxicology. 39 (10), 865-913 (2009).
- Kunzli, N., et al. Comparison of oxidative properties, light absorbance, total and elemental mass concentration of ambient PM2.5 collected at 20 European sites. Environmental health perspectives. 114 (5), 684-690 (2006).
- Wei, K., et al. Ambient bioaerosol particle dynamics observed during haze and sunny days in Beijing. The Science of the total environment. 550, 751-759 (2016).
- Nehme, B., Letourneau, V., Forster, R. J., Veillette, M., Duchaine, C. Culture-independent approach of the bacterial bioaerosol diversity in the standard swine confinement buildings, and assessment of the seasonal effect. Environmental microbiology. 10 (3), 665-675 (2008).
- Riemenschneider, L., et al. Characterization of reaerosolization from impingers in an effort to improve airborne virus sampling. Journal of applied microbiology. 108 (1), 315-324 (2010).
- Mehta, S. K., Bell-Robinson, D. M., Groves, T. O., Stetzenbach, L. D., Pierson, D. L. Evaluation of portable air samplers for monitoring airborne culturable bacteria. AIHAJ : a journal for the science of occupational and environmental health and safety. 61 (6), 850-854 (2000).
- Sun, Z., Mu, Y., Liu, Y., Shao, L. A comparison study on airborne particles during haze days and non-haze days in Beijing. The Science of the total environment. , 1-8 (2013).
- Lednicky, J., et al. . Highly efficient collection of infectious pandemic Influenza H1N1 virus (2009) through laminar-flow water based condensation. 50, (2016).
- Verreault, D., Moineau, S., Duchaine, C. Methods for sampling of airborne viruses. Microbiology and molecular biology reviews : MMBR. 72 (3), 413-444 (2008).
- Dybwad, M., Skogan, G., Blatny, J. M. Temporal variability of the bioaerosol background at a subway station: concentration level, size distribution, and diversity of airborne bacteria. Applied and environmental microbiology. 80 (1), 257-270 (2014).
- Hietala, S. K., Hullinger, P. J., Crossley, B. M., Kinde, H., Ardans, A. A. Environmental air sampling to detect exotic Newcastle disease virus in two California commercial poultry flocks. Journal of veterinary diagnostic investigation: official publication of the American Association of Veterinary Laboratory Diagnosticians, Inc. 17 (2), 198-200 (2005).
- Logares, R., et al. Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environmental microbiology. 16 (9), 2659-2671 (2014).
- Kogawa, M., Hosokawa, M., Nishikawa, Y., Mori, K., Takeyama, H. Obtaining high-quality draft genomes- from uncultured microbes by cleaning and co-assembly of single-cell amplified genomes. Scientific reports. 8 (1), 2059 (2018).
- Handelsman, J., Rondon, M. R., Brady, S. F., Clardy, J., Goodman, R. M. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chemistry & biology. 5 (10), R245-R249 (1998).
- Moon, C. D., Young, W. Metagenomic insights into the roles of Proteobacteria in the gastrointestinal microbiomes of healthy dogs and cats. MicrobiologyOpen. , e00677 (2018).
- Ribicic, D., et al. Microbial community and metagenome dynamics during biodegradation of dispersed oil reveals potential key-players in cold Norwegian seawater. Marine pollution bulletin. 129 (1), 370-378 (2018).
- Vera-Gargallo, B., Navarro-Sampedro, L., Carballo, M., Ventosa, A. Metagenome Sequencing of Prokaryotic Microbiota from Two Hypersaline Soils of the Odiel Salt Marshes in Huelva, Southwestern Spain. Genome announcements. 6 (9), (2018).
- Lloyd-Price, J., et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 550 (7674), 61-66 (2017).
Explore More Articles
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved