Designing Automated, High-throughput, Continuous Cell Growth Experiments Using eVOLVER
In This Article
Summary
The eVOLVER framework enables high-throughput continuous microbial culture with high resolution and dynamic control over experimental parameters. This protocol demonstrates how to apply the system to conduct a complex fitness experiment, guiding users on programming automated control over many individual cultures, measuring, collecting, and interacting with experimental data in real-time.
Abstract
Continuous culture methods enable cells to be grown under quantitatively controlled environmental conditions, and are thus broadly useful for measuring fitness phenotypes and improving our understanding of how genotypes are shaped by selection. Extensive recent efforts to develop and apply niche continuous culture devices have revealed the benefits of conducting new forms of cell culture control. This includes defining custom selection pressures and increasing throughput for studies ranging from long-term experimental evolution to genome-wide library selections and synthetic gene circuit characterization. The eVOLVER platform was recently developed to meet this growing demand: a continuous culture platform with a high degree of scalability, flexibility, and automation. eVOLVER provides a single standardizing platform that can be (re)-configured and scaled with minimal effort to perform many different types of high-throughput or multi-dimensional growth selection experiments. Here, a protocol is presented to provide users of the eVOLVER framework a description for configuring the system to conduct a custom, large-scale continuous growth experiment. Specifically, the protocol guides users on how to program the system to multiplex two selection pressures — temperature and osmolarity — across many eVOLVER vials in order to quantify fitness landscapes of Saccharomyces cerevisiae mutants at fine resolution. We show how the device can be configured both programmatically, through its open-source web-based software, and physically, by arranging fluidic and hardware layouts. The process of physically setting up the device, programming the culture routine, monitoring and interacting with the experiment in real-time over the internet, sampling vials for subsequent offline analysis, and post experiment data analysis are detailed. This should serve as a starting point for researchers across diverse disciplines to apply eVOLVER in the design of their own complex and high-throughput cell growth experiments to study and manipulate biological systems.
Introduction
Continuous cell culture techniques, first developed nearly 70 years ago1,2, are enjoying a recent revival3,4. This is due to a confluence of factors. First, the development of high throughput -omics techniques, which have made it possible to read out and generate large numbers of genotypes5,6, has created a concomitant demand for experimental techniques that facilitate well-controlled cell growth and phenotyping. To this end, continuous culture represents a powerful experimental approach to capitalize on emergent genomic advances. By facilitating growth selections/experiments on cellular populations in precisely controlled (and dynamic) environmental conditions, continuous culture provides a means to rigorously map genotypes to phenotypes7,8, quantitatively characterize engineered strains and organisms9, and track adaptive genetic changes in laboratory evolution studies10,11,12.
Second, the recent emergence of accessible prototyping techniques, such as additive manufacturing and open-source hardware and software elements, has enabled a wider set of users to design and build their own cost-effective forms of continuous culture systems directly in the laboratory. All of this has led to an exciting array of do-it-yourself (DIY) devices that perform continuous culture functionalities, such as the chemostat13, turbidostat14, or morbidostat15. Unfortunately, though successful in addressing specific (niche) problems for which they were designed, these ad hoc solutions generally lack the ability to scale in throughput and/or experimental design complexity.
The eVOLVER system was designed with the goal of creating a single platform that can accommodate the growing experimental needs of continuous culture and match the speed and scale of emergent genomic techniques16 (Figure 1A). eVOLVER’s design implements common tenets underlying highly scalable technologies from other disciplines17, including standardized footprints, modular components, and open-source design principles. Thus, solutions for new niche applications can be designed without major modifications to the system. Comprised of highly modular and open-source wetware, hardware, electronics, and web-based software, eVOLVER is the first automated continuous culture system that can be cost-effectively and readily re-configured to carry out virtually any type of high-throughput growth experiment. Through modular and programmable Smart Sleeves which house all the sensors and actuators needed to control individual cultures, eVOLVER uniquely enables scaling of both throughput and individual control of culture conditions. Moreover, as a web-based platform, eVOLVER exchanges data and information with remote computers in real-time, permitting simultaneous monitoring of hundreds of individual cultures and automated culture perturbations through arbitrarily defined control algorithms.
In previous work16, eVOLVER’s robust performance was demonstrated in long-term experiments over hundreds of hours of operation, and its ability to cultivate various organisms, from E. coli and S. cerevisiae to undomesticated microbes. A series of distinct growth selection experiments were performed, in which programmatically defined multi-dimensional selection gradients were applied across an array of individual culture conditions and the resulting cellular fitness landscapes were quantified. Here, the objective is to provide eVOLVER users with a description of how to use the system to design and these types of experiments. As an illustrative example, methods quantifying the fitness landscape of S. cerevisiae mutants across a two-dimensional environmental gradient composed of temperature and osmotic stress are presented. The protocol guides users through configuring the eVOLVER framework for this experiment both programmatically, in using the software to set customized turbidity and temperature control routines for each of 16 parallel continuous cultures, and physically, through the fluidics layout to appropriately route medias of different salt concentrations. This protocol should serve as a general rubric for configuring eVOLVER to execute a wide variety of automated continuous culture experiments for diverse studies and disciplines.
Protocol
1. Preparing Media, Vials, and Inoculum
NOTE: This protocol assumes users have already calibrated the eVOLVER system and are using the Smart Sleeve configuration described in prior work16. Smart Sleeves are easily redesigned or modified, but setup details of volume and control parameters may differ for alternative configurations.
- The day before the experiment, prepare 2 mL overnight liquid cultures of S. cerevisiae BY4741 reference strain and variant strains in YPD media (Yeast Peptone Dextrose) at 30 °C in a shaking incubator set to at least 300 r.p.m.
- Transfer sterile YPD media into clean, autoclaved bottles fitted with tubing. Alternatively, media may be autoclaved directly in bottles pre-fitted with tubing.
NOTE: Bottles are fit with tubing by drilling a hole into the cap and running tubing through it with connectors to secure it in place. See Table of Materials. - Add stir bar to each vial and screw on a vial lid equipped with influx and efflux straws. Ensure that all components are clean by visual inspection.
- Place vials in an autoclavable rack, cover with foil, and autoclave on a gravity or vacuum cycle with a 20-min sterilization step.
2. Setting Up an Experiment
- In three large beakers, prepare 500 mL of 10% bleach, 200 mL of 10% bleach, and 300 mL of 70% ethanol.
- Using the switches on the eVOLVER device, turn on the 5 V power supply on the eVOLVER, wait for 5 s, then turn on the 12 V power supply.
- If running several vials from the same media bottle, connect multiple media input lines with tubing splitters. Custom tubing splitters can be constructed with Luer components.
- Submerge the media input lines in the first bleach beaker, and the media efflux lines in the second bleach beaker. Place the downstream end of the media input lines in the second beaker with the efflux lines. Place waste lines into waste carboy.
- Add 1-2 L of bleach into large empty waste carboy, which will sterilize waste generated during experiment. Consult with a safety coordinator to ensure proper disposal of waste.
- Using the touchscreen on the eVOLVER, navigate to the setup section, and run all pumps for 20 s to fill fluid lines with 10% bleach. While running, ensure by visual inspection that all lines have been filled and that pumps are operating normally. Allow bleach to sit in the lines for at least 30 min to sterilize.
- Run pumps again using the touchscreen app until lines are no longer submerged, pushing air through the lines to get as much of the bleach out as possible.
- Place media input lines in the ethanol beaker and repeat as in above, filling the lines with ethanol and flushing with air.
NOTE: As ethanol kills quickly, there is no need to wait 30 min for sterilization. - Attach media input lines to the media bottles with the Luer connectors and run the pumps until the media fully runs through the lines, flushing out any residual ethanol.
- Partially insert sterilized vials into the eVOLVER Smart Sleeves and hook up the lines to the proper positions, according to the color coding on the lines. Start by attaching input lines to the short influx straw, and then waste lines to the long efflux straw.
CAUTION: It is critical to check for loose connections or incorrectly routed lines. Failure to do so will cause overflows and potentially damage the Smart Sleeve. - Run all pumps in 10 s increments to fill the vials with media. Ensure by visual inspection efflux pumps are efficiently removing media through the efflux straws, to prevent overflows. If the efflux straws do not appear to be functioning efficiently, inspect the fluidic line connections and the peristaltic pump. Correct or replace parts as needed.
- Push vials down until fully encased by the Smart Sleeve.
- Set the initial conditions for experimental parameters using the eVOLVER touchscreen on the interactive setup page. For all vials, set the temperature to 30 °C and the stir to 10. This can be done for all vials at once by dragging a finger across the touchscreen to select all of the vials.
3. Configuring eVOLVER Software and Programming Algorithmic Culture Routines
- In the eVOLVER dashboard on a computer, navigate to the experiment manager page. Select the base experiment in the experiment navigator panel or an existing experiment as a starting point and click clone new.
NOTE: It is possible to use experiments from other groups by pasting the GitHub link to the repo where the experimental definition is saved into the experiment manager. - Specify the experiment name and the eVOLVER device that the experiment will be run on. Be sure that the desired calibration settings have been selected by modifying these fields on the page.
- Use the vial selector and parameter definition panes to set experimental conditions. For example, to set the temperature for vials 0-4 to 30 °C, select the vials in the vial selector, set the parameter definition to 30, and click Set. Repeat this for OD to set an upper and lower threshold for each vial.
- After experimental parameter definitions complete, save the experiment by clicking Save and click Start to run.
4. Initiating Experiment and Monitoring in Real-time
- Once the experiment is underway, navigate to the real-time data panel in the interactive dashboard. For each vial, check that OD values are holding at zero, and that temperatures are at or progressing towards programmed levels by browsing through the graphs.
- To prepare inoculum, measure the OD of overnight cultures, calculate the desired fold dilution assuming a vial volume of 25 mL, and pipette calculated volume of inoculum into vials through the sampling port. For example, to reach a desired starting OD of about 0.05 in the culture vial from overnight cultures that are at OD 2.0, 625 μL of cells should be added to each vial.
- Check the graphs on the dashboard to see that the OD graphs have been updated accordingly. An increase to near the desired starting OD should be seen in the graphs.
- Track different data types (such as OD, temperature, growth rate, generations, and media consumption) throughout the experiment by browsing through the corresponding graphs in the dashboard. These values may inform experimental decisions by the user (e.g. schedule timepoints) or even used in functions to perform automated feedback.
- To replace media bottles mid experiment, pause the experiment in the experiment manager panel to prevent pump events from occurring. Quickly unscrew the media line from the empty bottle and connect it to the new bottle. Avoid touching the ends of tubing to prevent contamination. Resume the experiment in the experiment manager.
5. Sampling Yeast Cells from eVOLVER Vials
- Prepare a cycloheximide solution to inhibit protein synthesis and fix yeast cells for flow cytometry analysis. In a 15 mL conical tube, add 12 mL of PBS and 12 μL of 20 mg/mL cycloheximide and vortex for 5 s.
- Using a multichannel pipette, aliquot 100 μL into each well of a 96-well round bottom plate.
- Wrap the plate in aluminum foil to block out light and keep at 4 °C until needed.
- To sample, pipette through the sampling port on the vial lid using extended length 200 μL tips.
- Pipette 100 μL from a vial into a well in the fixation plate, mixing 2-3x by pipetting up and down, then recover in foil and return the plate to 4 °C.
6. Experiment Break Down and Clean Up
- Prepare 1 L of 10% bleach and two 500 mL DI water solutions in large beakers.
- Stop the experiment by sending the stop command in the experiment manager panel. The data is still stored in the cloud-enabled database, and can still be viewed later in the real-time data panel of the dashboard, or downloaded for offline analysis.
- Remove vials from Smart Sleeves and place them in an autoclave rack to avoid overflows in subsequent steps.
- Unscrew the media input lines from the media bottles and place them into the beaker of 10% bleach. Run pumps from the eVOLVER user interface as in setup to sterilize lines and vials.
- Disconnect lines from vials and submerge lines in DI water. Run pumps to flush all bleach out of the system first with DI water, then with air.
- Turn off eVOLVER by first switching off the 12 V power supply, waiting 5 s, and then turning off the 5 V power supply.
- Soak stir bars and caps in 10% bleach, then rinse with DI water. Be sure to rinse the influx and efflux straws of each cap by running DI water through them. Scrub vials using test tube brush if film has formed on walls.
- Dispose of waste appropriately and rinse waste containers thoroughly.
CAUTION: Waste containers will contain a mixture of 10% bleach, sterilized cell culture waste, and trace amounts of ethanol. Consult a safety coordinator for proper handling and disposal.
7. Quantifying Fitness Using Flow Cytometry Analysis of Fractional Populations
- Analyze collected samples from section 5 using a flow cytometer equipped with the appropriate fluorescence channel and detectors16.
- Acquire at least 10,000 events per sample, and gate for intact cells using the forward and side scatter data.
- Gate based on the appropriate fluorescence channel (e.g. GFP) to determine the fractional distribution of each population.
- On a computer, save data to desired location from the Experiment Manager page by clicking the “Export” button.
- From the eVOLVER data files, determine the number of generations completed by each timepoint for each vial. In the eVOLVER code, generations are calculated by first segmenting the OD data between each dilution event, then each segment is fit with a basic exponential of the form
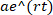 to each segment, yielding r, the growth rate16. The number of generations over a time period is then calculated using the following formula:
to each segment, yielding r, the growth rate16. The number of generations over a time period is then calculated using the following formula:
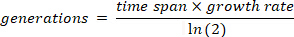
- Plot the log-ratio of the labelled and unlabeled populations from the flow cytometry data vs generation number from the above calculations at the time samples were taken. Identify the linear region and fit the slope according to the following equation. This slope is commonly defined as the competitive fitness18,19.
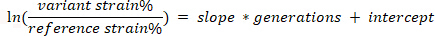
Representative Results
The protocol described here was used to construct fitness maps for genetic variants of S. cerevisiae across a two-dimensional stress gradient of temperature and salinity. Specifically, for each variant strain, we used eVOLVER to conduct pairwise competition experiments against a fluorescently-labeled reference strain (S. cerevisiae strain BY4741 with an integrated constitutive mNeonGreen reporter) in 16 different combinations of these stresses (Figure 1). For variants, two knockout strains were selected each predicted to be sensitive to one of the stress condition axes: ΔPRX1 which has been reported to have fitness defects in high temperatures20 and ΔPBS2 with fitness defects at high salt concentrations21. As a control variant, an unlabeled wild-type BY4741 strain was used, expected to perform similarly to the reference strain. In total, these three 72-hour competition experiments were conducted simultaneously in 48 vials across three eVOLVER (16-vial) devices all run from a single computer.
The eVOLVER interactive dashboard is used to navigate the large amount of data that is generated during each experiment (Figure 2A). Representative OD traces across all 16 vials of a single eVOLVER device are shown in Figure 2B. Each trace displays a characteristic sawtooth pattern from the turbidostat control algorithm, which uses feedback on OD to trigger dilutions and maintain cultures in a narrow density range (0.2-0.3 in this case). OD and temperature data are continuously measured, streamed to the cloud-enabled database, and updated in real-time in the eVOLVER interactive dashboard. From continuously streamed data, higher-order metrics (e.g. growth rate, cumulative generations) may be calculated and plotted in the dashboard (Figure 2C) and even used as feedback parameters in different selection schemes (e.g., morbidostat).
To generate fitness maps, we measure the rate at which the reference strain outcompetes the variant strain by incorporating both flow cytometry measurements and eVOLVER growth data. Specifically, population fractions are calculated as the log-ratio of variant strain over reference strain using flow cytometry data from each timepoint sample. For each culture and time point, the elapsed number of generations is interpolated from eVOLVER growth rate data in each dilution period. Plotting the log-ratios against generations, qualitative differences between conditions begin to emerge (Figure 3A). To calculate fitness, a slope is fit through the linear portion of each plot18,19, yielding a quantitative fitness value (with both sign and rate) that describes how the variant strain competes against the reference strain (Figure 3B). In this experiment, negative fitness values indicate that the variant strain is outcompeted by the reference strain. Constructing heatmaps from all the fitness calculations permits visualization of the two-dimensional fitness landscape, revealing subtle quantitative differences in performance between strains and conditions (Figure 3C).
From the fitness heatmaps, we see that each variant strain exhibits fitness defects that manifest in different ways. ΔPRX1 is primarily sensitive to high temperatures, but salt stress appears to have an additive effect, increasing the fitness defect. Conversely, ΔPBS2 shows fitness defects primarily in response to high salt concentrations, with minimal differences along the temperature axis. These results highlight the utility of high-resolution selection experiments particularly for deciphering interactions of multiple environmental stressors, which could have additive, synergistic, or epistatic effects.
Finally, it is important to note that in some cases, particularly for severe stress conditions, fitness calculations can lead to exaggerated or skewed results. For example, ΔPBS2 appears to show a steep positive slope in the 39 °C/1 M NaCl condition relative to the reference strain (Figure 3A). This is attributed to poor growth of both strains, which is reflected in the wild type data and limits the utility of this particular metric in this condition. An insufficient number of generations results in 1) poor separation of timepoints which interferes with slope fitting, and 2) sensitivity to stochastic effects originating from lag phase. While we did not elect to do so in this study, the real-time growth data collected by eVOLVER during the experiment could have been used to inform the decision to extend the experiment and collect additional timepoints for this condition with little effort. In follow-up experiments, starting ratios could also be modulated to extend the length of the linear region before saturation occurs.
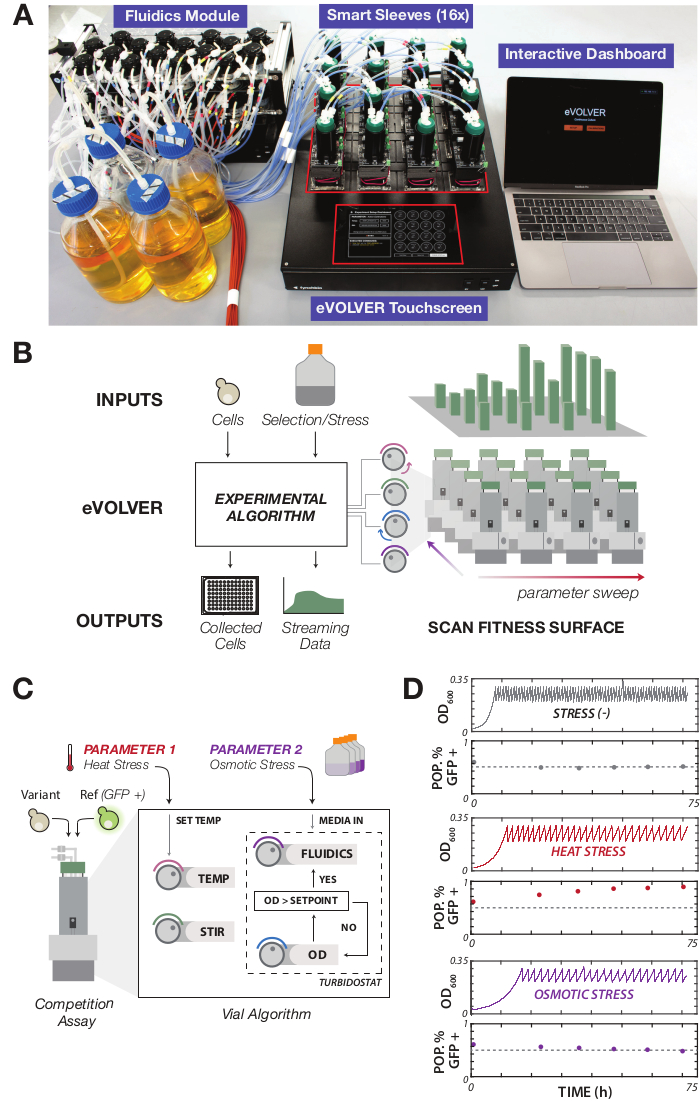
Figure 1: Overview of the eVOLVER framework and experimental design. (A) eVOLVER is an automated, continuous culture platform allowing multiparameter control of culture conditions. The platform consists of Smart Sleeves, which house the sensors and actuators for controlling culture conditions, a peristaltic pump array for flowing media and waste in and out of the cultures, a touchscreen for interacting manually with the device, and a computer used for interacting with the cloud infrastructure where data from the platform is streamed. (B) eVOLVER can be configured in multiple ways: physically, through cell and media inputs, and programmatically by adjusting the algorithms controlling Smart Sleeve culture vial modules. This can be utilized to program multidimensional selection/environmental gradients at high-resolution, with outputs consisting of physically collected cells at defined time points and real-time streaming of culture data. (C) Configuring eVOLVER to conduct a growth fitness experiment across a two-dimensional selection gradient, composed of temperature and osmotic stress. eVOLVER culture vials were seeded with equal proportions of a reference and variant yeast strain. A turbidostat routine was programmed to maintain all the cultures in exponential phase in a defined OD window (OD 0.2-0.3). Temperature and media conditions were varied across the Smart Sleeve array to form two independent stress gradients. Base media was composed of YPD (2% glucose) + 100 µg/mL carbenicillin + 25 µg/mL chloramphenicol as a precaution against bacterial contamination. Media compositions were varied by adding supplemental NaCl to 0 M, 0.6 M, 0.8 M, or 1.0 M. Vial temperatures were programmed to one of four values: 30 °C, 35 °C, 37 °C, or 39 °C. (D) Examples of raw output for culture OD and population fractions from three tested conditions. The competition experiment was maintained for 72 h, sampling at 24 h and subsequently every 12 h until the conclusion of the experiment. Please click here to view a larger version of this figure.
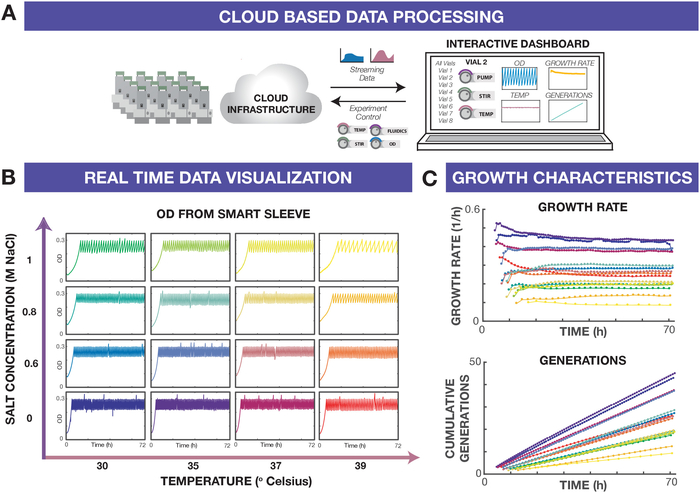
Figure 2: eVOLVER real-time data analysis dashboard. (A) eVOLVER data is processed and streamed in real time via cloud-based software to an interactive dashboard. Through the dashboard, users can define and initiate their own culture routines on a connected eVOLVER, monitor currently running experiments across all eVOLVER units, and vary experimental conditions manually on an individual vial or set of vials if needed. (B) OD traces for each vial across the matrix of conditions tested for the entire experiment duration. Traces can be viewed in real-time to ensure experiment is running as desired and used post-experiment for further analysis. (C) Growth rates and cell generations can be calculated from OD traces on the fly to track experiment progress. Please click here to view a larger version of this figure.
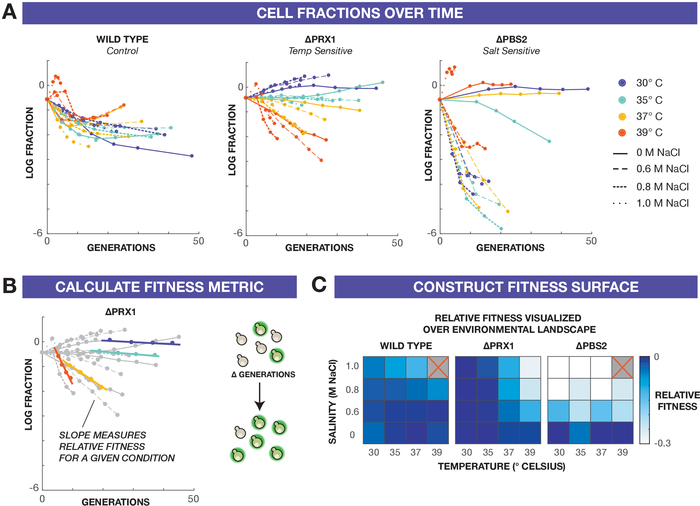
Figure 3: Construction of fitness surfaces. (A) Cell population fractions natural log (variant strain/reference strain) plotted against cell generations. Color indicates the temperature of selection while dashes distinguish salt concentrations. (B) To quantify relative fitness of the variant to the reference strain, a fitness metric is derived from the rate at which the cellular population fraction changes over generations. This metric is calculated from the linear region of the cellular fraction plots using a minimum of three points. (C) The relative fitness metrics are displayed in a heatmap to construct a fitness surface over the environmental stress gradients. The top right corner condition (39 °C/1.0 M NaCl) for the wild type and ΔPBS2 are not included in this analysis as the cultures went through insufficient generations (see Representative Results). Please click here to view a larger version of this figure.
Discussion
Growth selection is an indispensable tool in biology, broadly used to generate and characterize phenotypic differences between cellular populations. While batch cultures do permit growth selection in a limited way, continuous culture techniques dramatically expand the degree of control and predictability of these experiments, by exerting precise regulation over the form and dynamics of selection to generate repeatable, quantitative results22. Continuous culture has been employed to rigorously control selection for high-diversity libraries20,23,24,25, and to implement sophisticated adaptive regimes in experimental and directed evolution11,12,26,27. Continuous culture also enables precise characterization of cells across an array of quantitatively controlled conditions to better understand complex genetic systems and optimize engineered bioproduction strains9,14,28.
However, there is no universal protocol for continuous culture, as subtle changes to the selective conditions can lead to dramatic changes in biological outcomes4,29,30. Experimenters must be able to choose between selection regimes and adapt experimental protocols and equipment accordingly. In addition to offering a choice between control parameters, such systems would ideally be sophisticated enough to independently manage several parameters simultaneously in highly-parallel experiments that are needed to decipher interacting inputs in complex biological systems (e.g. epistasis). eVOLVER addresses this challenge by enabling users to arbitrarily program feedback control between culture conditions and fluidic functions in order to specify highly specialized environmental niches.
To overcome limitations in the current setup and expand or change control parameters, the Smart Sleeve could easily be redesigned to add new sensors or actuators. Additionally, reducing vial volume would decrease media expenditures, which can be significant in continuous culture. While the current design permits measurement and control of temperature, culture agitation, light induction, turbidity, and fluidics, other parameters must be measured externally by sampling from the vials. Current work includes incorporating the ability to monitor enzymatic activity via luciferase and regulate dissolved oxygen and pH directly in eVOLVER cultures. Additionally, while not demonstrated in this work, eVOLVER can interface with novel millifluidic multiplexing devices16 that draw on principles of large-scale integration (originating from electronics and adopted by microfluidics) in order to inexpensively enable more complex fluidic handling (e.g. multiplexed fluidic inputs, vial-to-vial transfers). These wetware modules may be designed and manufactured completely in the lab, allowing users to route fluids by programmatically actuating different combinations of valves in automated fluidic routines. This allows users to overcome the rigid fluidic designs traditionally used in continuous culture, but also to scale fluidic capabilities to high-throughput with a smaller number of costly control elements (e.g. peristaltic pumps). Lastly, we are hoping to incorporate an autosampling platform which will utilize these millifluidics and DIY components, overcoming the limitation of manual interaction during longer and larger experiments where sampling cultures would be cumbersome.
In addition to physical modifications to the platform, the web-based software opens new degrees of freedom by allowing users to write, edit, and share custom eVOLVER scripts, generating fully automated, feedback-enabled culture programs (e.g., turbidostat). Users may programmatically sweep across parameter ranges in subtle variations on the same selection scheme or connect control algorithms in novel combinations to specify any number of sophisticated selection schemes. Furthermore, the ability to easily monitor cultures in real-time transforms the way in which experiments are conducted. With real-time monitoring, users may 1) check for consistency between runs, a critical feature for bioproduction applications and high-throughput experiments, and 2) intervene during experiments if necessary, to troubleshoot challenging strains that exhibit poor growth or biofilm formation, or diagnose user errors (e.g., contamination). Finally, with multiple data streams being collected and interpreted in real-time for each individual culture, eVOLVER generates a high density of data, which may facilitate machine learning approaches for novel downstream analysis.
Beyond demonstrated uses for fitness characterization, library selection, and laboratory evolution, we view a number of related fields as ripe for implementation in eVOLVER with integrated fluidics. eVOLVER experiments with microbiome samples could assay community stability in controlled environments31,32, explore microbiota composition using culturomics techniques33, or dynamically mix species to interrogate ecological dynamics of colonization or invasion34,35. Numerous methods for continuous directed evolution of biomolecules could easily be implemented on the device as well26,36,37, greatly increasing accessibility and throughput of these systems. The ability to optimize growing conditions such as media composition, temperature, and strains in a dynamic, high throughput nature can aid in optimization efforts for industrial biomanufacturing applications9. We further envision vertically integrating eVOLVER with other analysis techniques such as microscopy and flow cytometry in a closed loop fashion, providing a fully automated system for growth and analysis of cellular cultures at both single cell and population levels. Moreover, with some hardware modifications to the Smart Sleeve such as sealing the vessel and controlling gas content, eVOLVER could potentially be adapted to support the growth of a wider range of cell types, such as suspension mammalian cells. It is also feasible to place the entire framework into an anaerobic chamber for anaerobic cell culture. Looking forward, we aim to build our software framework into a centralized cloud infrastructure and believe this would allow users to easily configure, analyze, and share their data remotely without needing to physically be present in the lab. Functioning as a data curator, the cloud infrastructure would also lend itself to large-scale meta analyses across experiments. We anticipate that eVOLVER and these future advances will greatly expand the scope of possible growth selection experiments by facilitating automation and innovation in continuous culture.
Disclosures
The authors, Brandon G. Wong and Ahmad S. Khalil, are co-founders of Fynch Bioscience Inc., which develops the eVOLVER platform used in this Article.
Acknowledgements
We thank B. Stafford for his assistance in design of the system, and H. Khalil, A. Soltanianzadeh, A. Sun, S. Pipe, and A. Cavale for help with construction of the system. We acknowledge the Electronics Design Facility (EDF), Engineering Product Innovation Center (EPIC), and the Software & Application Innovation Lab (SAIL) at the Hariri Institute for Computing at Boston University for their services. This work was supported by an NSF CAREER Award (MCB-1350949 to A.S.K.), and DARPA grants HR0011-15-C-0091 and HR0011-18-2-0014 (to A.S.K.). A.S.K. also acknowledges funding from the NIH Director’s New Innovator Award (1DP2AI131083-01), DARPA Young Faculty Award (D16AP00142), and NSF Expeditions in Computing (CCF-1522074).
Materials
| Name | Company | Catalog Number | Comments |
| 5 Gallon Plastic Hedpack with cap | Midwest Brewing and Winemaking Supplies | 45-56Y8-E2FR | For waste collection |
| a-D(+)-Glucose | Chem-Impex | 00805 | For YPD Medium |
| Attune NxT Autosampler | Thermo Fisher | Allows Flow Cytometer to run samples from 96 well plate | |
| Attune NxT Flow Cytometer | Thermo Fisher | Used to determine population fractions via single cell fluoresence | |
| Bacto Peptone | Fisher Scientific | DF0118-07-0 | For YPD Medium |
| Carbenicillin | Fisher Scientific | BP2648250 | For YPD Medium |
| Chemical-Resistant Barbed Tube Fitting Tee Connector, for 1/8" Tube ID, 250°F Maximum Temperature | McMaster- Carr | 5121K731 | For media input branching |
| Chloramphenicol | Fisher Scientific | BP904-100 | For YPD Medium |
| CLOROX GERMICIDAL Bleach 8.25 | Fisher Scientific | 50371500 | For Sterilization of fluidic lines |
| Custom eVOLVER vial lid | FynchBio | Lid has ports for sampling and fluidic input/output | |
| Cycloheximide | Fisher Scientific | ICN10018301 | For flow cytometry sampling plates |
| Ethanol, Anhydrous (Histological) | Fisher Scientific | A405P-4 | For sterilization of fluidic lines |
| eVOLVER Unit | FynchBio | ||
| Fisherbrand Extended-Length Tips (Lift Off Rack; 1 to 200 ul) | Fisher Scientific | 02-681-420 | For vial sampling |
| Fisherbrand Octagon Spinbar Magnetic Stirring Bars | Fisher Scientific | 14-513-57 | Diameter: 4.5 mm, Length, 12 mm |
| Fisherbrand Reusable Glass Media Bottles with Cap | Fisher Scientific | FB8002000 | Must be fitted with tubing |
| High-Temperature Silicone Rubber Tubing Semi-Clear White, Durometer 70A, 1/8" ID, 1/4" OD | McMaster- Carr | 51135K73 | For media bottles |
| Mac Mini | Apple | For running the experiment/collecting data | |
| Phosphate Buffered Saline (PBS) | Fisher Scientific | BP243820 | For flow cytometry sampling plates |
| Pipettes | Eppendorf | ||
| Plastic Quick-Turn Tube Coupling, Plugs, for 1/16" Barbed Tube ID, Polypropylene | McMaster- Carr | 51525K141 | For media bottles |
| Plastic Quick-Turn Tube Coupling, Plugs, for 5/32" Barbed Tube ID, Polypropylene | McMaster- Carr | 51525K144 | For media bottles |
| Plastic Quick-Turn Tube Coupling, Sockets, for 1/16" Barbed Tube ID, Polypropylene | McMaster- Carr | 51525K291 | For media bottles |
| Plastic Quick-Turn Tube Coupling, Sockets, for 5/32" Barbed Tube ID, Polypropylene | McMaster- Carr | 51525K294 | For media bottles |
| SCREW CAPS, OPEN TOP, WITH PTFE FACED SILICONE SEPTA, LAB-PAC, SEPTUM. Screw thread size: 24-400, GREEN | Chemglass | CG-4910-04 | Culture vials |
| Sodium Chloride (NaCl) | Fsher Scientific | S271-3 | For YPD Medium |
| SpectraMax M5 Multi-Mode Microplate Reader | Molecular Devices | For measuring OD600 of overnight cell cultures | |
| Vial Only, Sample, 40mL, Clear, 28x95mm, GPI 24-400 | Chemglass | CG-4902-08 | Culture vials |
| Yeast Extract | Fisher Scientific | BP1422-500 | For YPD Medium |
References
- Monod, J. The Growth of Bacterial Cultures. Annual Review of Microbiology. , (1949).
- Novick, A., Szilard, L. Experiments with the Chemostat on spontaneous mutations of bacteria. Proceedings of the National Academy of Sciences of the United States of America. 36 (12), 708-719 (1950).
- Bull, A. T. The renaissance of continuous culture in the post-genomics age. Journal of Industrial Microbiology and Biotechnology. 37 (10), 993-1021 (2010).
- Gresham, D., Dunham, M. J. The enduring utility of continuous culturing in experimental evolution. Genomics. 104 (6), 399-405 (2014).
- Lang, G. I., et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 500 (7464), 571-574 (2013).
- Maddamsetti, R., et al. Adaptation, Clonal Interference, and Frequency-Dependent Interactions in a Long-Term Evolution Experiment with Escherichia coli. Genetics. 200 (2), 619-631 (2015).
- Ishii, N., et al. Multiple High-Throughput Analyses Monitor the Response of E. coli to Perturbations. Science. 316 (5824), 593-597 (2007).
- Brauer, M. J., et al. Coordination of Growth Rate, Cell Cycle, Stress Response, and Metabolic Activity in Yeast. Molecular Biology of the Cell. 19 (1), 352-367 (2008).
- Moser, F., et al. Genetic circuit performance under conditions relevant for industrial bioreactors. ACS Synthetic Biology. 1 (11), 555-564 (2012).
- Kao, K. C., Sherlock, G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nature Genetics. 40 (12), 1499-1504 (2008).
- Toprak, E., Veres, A., Michel, J. -. B., Chait, R., Hartl, D. L., Kishony, R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nature Genetics. 44 (1), 101-105 (2011).
- Hope, E. A., Amorosi, C. J., Miller, A. W., Dang, K., Heil, C. S., Dunham, M. J. Experimental Evolution Reveals Favored Adaptive Routes to Cell Aggregation in Yeast. Genetics. 206 (2), 1153-1167 (2017).
- Miller, A. W., Befort, C., Kerr, E. O., Dunham, M. J. Design and Use of Multiplexed Chemostat Arrays. Journal of Visualized Experiments. (72), 2-7 (2013).
- Takahashi, C. N., Miller, A. W., Ekness, F., Dunham, M. J., Klavins, E. A low cost, customizable turbidostat for use in synthetic circuit characterization. ACS Synthetic Biology. 4 (1), 32-38 (2015).
- Toprak, E., et al. Building a morbidostat: an automated continuous-culture device for studying bacterial drug resistance under dynamically sustained drug inhibition. Nature Protocols. 8 (3), 555-567 (2013).
- Wong, B. G., Mancuso, C. P., Kiriakov, S., Bashor, C. J., Khalil, A. S. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nature Biotechnology. 36 (7), 614-623 (2018).
- Bondi, A. B. Characteristics of scalability and their impact on performance. Proceedings of the Second International Workshop on Software and Performance - WOSP ’00. , 195-203 (2000).
- Ziv, N., Brandt, N. J., Gresham, D. The Use of Chemostats in Microbial Systems Biology. Journal of Visualized Experiments. (80), e50168 (2013).
- Sanchez, M. R., et al. Differential paralog divergence modulates genome evolution across yeast species. PLOS Genetics. 13 (2), e1006585 (2017).
- Gibney, P. A., Lu, C., Caudy, A. A., Hess, D. C., Botstein, D. Yeast metabolic and signaling genes are required for heat-shock survival and have little overlap with the heat-induced genes. Proceedings of the National Academy of Sciences of the United States of America. 110 (46), E4393-E4402 (2013).
- Giaever, G., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418 (6896), 387-391 (2002).
- Piper, M. D. W., et al. Reproducibility of oligonucleotide microarray transcriptome analyses. An interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. Journal of Biological Chemistry. 277 (40), 37001-37008 (2002).
- Badarinarayana, V., et al. Selection analyses of insertional mutants using subgenic-resolution arrays. Nature Biotechnology. 19 (11), 1060-1065 (2001).
- Dai, J., Hyland, E. M., Yuan, D. S., Huang, H., Bader, J. S., Boeke, J. D. Probing Nucleosome Function: A Highly Versatile. Library of Synthetic Histone H3 and H4. 134 (6), 1066-1078 (2008).
- McGeachy, A. M., Meacham, Z. A., Ingolia, N. An Accessible Continuous-Culture Turbidostat for Pooled Analysis of Complex Libraries. bioRxiv. , 450536 (2018).
- Esvelt, K. M., Carlson, J. C., Liu, D. R. A system for the continuous directed evolution of biomolecules. Nature. 472 (7344), 499-503 (2011).
- Carlson, J. C., Badran, A. H., Guggiana-Nilo, D. A., Liu, D. R. Negative selection and stringency modulation in phage-assisted continuous evolution. Nature Chemical Biology. 10 (3), 216-222 (2014).
- Milias-Argeitis, A., Rullan, M., Aoki, S. K., Buchmann, P., Khammash, M. Automated optogenetic feedback control for precise and robust regulation of gene expression and cell growth. Nature Communications. 7 (May), 12546 (2016).
- Wenger, J. W., Piotrowski, J., Nagarajan, S., Chiotti, K., Sherlock, G., Rosenzweig, F. Hunger Artists: Yeast Adapted to Carbon Limitation Show Trade-Offs under Carbon Sufficiency. PLoS Genetics. 7 (8), e1002202 (2011).
- Yona, A. H., et al. Chromosomal duplication is a transient evolutionary solution to stress. Proceedings of the National Academy of Sciences of the United States of America. 109 (51), 21010-21015 (2012).
- Auchtung, J. M., Robinson, C. D., Britton, R. A. Cultivation of stable, reproducible microbial communities from different fecal donors using minibioreactor arrays (MBRAs). Microbiome. 3, 42 (2015).
- Goldford, J. E., et al. Emergent simplicity in microbial community assembly. Science. 361 (6401), 469-474 (2018).
- Lagier, J. -. C., Dubourg, G., Million, M., Cadoret, F., Fournier, P. Culturing the human microbiota and culturomics. Nature Reviews Microbiology. 16 (September), 540-550 (2018).
- Fukami, T. Historical Contingency in Community Assembly: Integrating Niches, Species Pools, and Priority Effects. Annual Review of Ecology, Evolution, and Systematics. 46 (1), 1-23 (2015).
- Friedman, J., Gore, J. Ecological systems biology: The dynamics of interacting populations. Current Opinion in Systems Biology. 1, 114-121 (2017).
- Crook, N., Abatemarco, J., Sun, J., Wagner, J. M., Schmitz, A., Alper, H. S. In vivo continuous evolution of genes and pathways in yeast. Nature Communications. 7, 13051 (2016).
- Ravikumar, A., et al. Scalable, Continuous Evolution of Genes at Mutation Rates above Genomic Error Thresholds. Cell. 175, (2018).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2025 MyJoVE Corporation. All rights reserved