A Luciferase-fluorescent Reporter Influenza Virus for Live Imaging and Quantification of Viral Infection
In This Article
Summary
Influenza A viruses (IAVs) are contagious respiratory pathogens that cause annual epidemics and occasional pandemics. Here, we describe a protocol to track viral infections in vivo using a novel recombinant luciferase and fluorescence-expressing bi-reporter IAV (BIRFLU). This approach provides researchers with an excellent tool to study IAV in vivo.
Abstract
Influenza A viruses (IAVs) cause human respiratory disease that is associated with significant health and economic consequences. As with other viruses, studying IAV requires the use of laborious secondary approaches to detect the presence of the virus in infected cells and/or in animal models of infection. This limitation has been recently circumvented with the generation of recombinant IAVs expressing easily traceable fluorescent or bioluminescent (luciferase) reporter proteins. However, researchers have been forced to select fluorescent or luciferase reporter genes due to the restricted capacity of the IAV genome for including foreign sequences. To overcome this limitation, we have generated a recombinant replication-competent bi-reporter IAV (BIRFLU) stably expressing both a fluorescent and a luciferase reporter gene to easily track IAV infections in vitro and in vivo. To this end, the viral non-structural (NS) and hemagglutinin (HA) viral segments of influenza A/Puerto Rico/8/34 H1N1 (PR8) were modified to encode the fluorescent Venus and the bioluminescent Nanoluc luciferase proteins, respectively. Here, we describe the use of BIRFLU in a mouse model of IAV infection and the detection of both reporter genes using an in vivo imaging system. Notably, we have observed a good correlation between the expressions of both reporters and viral replication. The combination of cutting-edge techniques in molecular biology, animal research and imaging technologies, provides researchers the unique opportunity to use this tool for influenza research, including the study of virus-host interactions and dynamics of viral infections. Importantly, the feasibility to genetically alter the viral genome to express two foreign genes from different viral segments opens up opportunities to use this approach for: (i) the development of novel IAV vaccines, (ii) the generation of recombinant IAVs that can be used as vaccine vectors for the treatment of other human pathogen infections.
Introduction
Influenza A virus (IAV) is an enveloped single-stranded negative-sense segmented RNA virus of the Orthomyxoviridae family1,2,3. The World Health Organization (WHO) estimates 3-5 million annual influenza cases and over 250,000 deaths from influenza worldwide4,5,6. Groups that are particularly vulnerable to influenza include the elderly, immunocompromised individuals, and children7,8,9,10,11. Although vaccines are available and represent the most common and effective intervention against viral infection, IAV is able to rapidly evolve and escape preexisting immunity3,12,13,14,15. The re-emergence of a pandemic H1N1 strain in 2009 and the emergence of pathogenic IAV reiterates the constant threat to human public health worldwide4,16.
During an epidemic or pandemic, it is crucial to rapidly determine the pathogenicity and transmissibility of newly isolated viruses. Currently available techniques to detect the virus are time-consuming and sometimes require the use of laborious approaches, which can delay the completion of these analyses17,18,19,20. Moreover, present viral assays are difficult to scale up, which could be necessary during the event of an outbreak. Finally, the use of validated animal models of infection, such as mice, guinea pigs and ferrets are routinely used and are vital in studying influenza infections, immune responses, and the efficacy of new vaccines and/or antivirals. However, these models are restrictive due to the inability to observe viral dynamics in real time; this limits the studies to static imaging of viral infections21,22,23,24,25. Animals used in these assays are also euthanized in order to determine viral load, thus increasing the number of animals required to complete these studies26. To circumvent all these limitations, many researchers rely on the use of recombinant replication-competent, reporter-expressing IAVs, which are capable of accelerating virological assays and detecting viral load and dissemination in vivo in real-time26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41. Importantly, these reporter-expressing IAVs are able to replicate similarly to wild-type (WT) IAVs in cell culture and in animal models of infection33,42.
Fluorescent and bioluminescent proteins are two reporter systems commonly used by researchers due to their sensitivity, stability and ease of use. In addition, there is tremendous support and advancement in fluorescent and bioluminescent protein detection technologies43,44,45,46,47,48. Fluorescent proteins and luciferase have different properties that allow them to glow, specifically differing in how excited states are generated and how emittance is detected43,44,45,46,47,48. Fluorescent proteins are first excited by absorbing energy, which is subsequently released as light at a different wavelength as the molecules decrease to a lower energy state43. On the other hand, bioluminescence is derived from a chemical exothermic reaction that involves a substrate, oxygen, and sometimes ATP in order to produce light45. Due to the varying properties of these two types of reporter proteins, one maybe more advantageous than the other depending on the study of interest. While fluorescent proteins are widely used to observe cellular localization28,41, their in vivo signals have inadequate intensity and are often obscured by autofluorescence in live tissues49. Therefore, researchers rely on luciferases to evaluate viral dynamics in live organisms, although fluorescent proteins can be preferred for ex vivo studies50,51,52,53. Unlike fluorescent proteins, luciferases are more convenient for in vivo studies and more applicable in non-invasive approaches26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,54. Ultimately, based on the type of study, researchers must choose between the use of either a fluorescent or a luciferase reporter protein as their readout, which subjects their study to a trade-off of functionalities and sensitivities, and severely restricts the usefulness of the recombinant reporter viruses. Moreover, there are concerns regarding the correlation among the expression of different reporter genes using fluorescence or luciferase systems and viral replication or dissemination, which might jeopardize the interpretation of the data obtained with reporter-expressing IAVs.
We have overcome this limitation by generating a recombinant replication-competent bi-reporter IAV (BIRFLU) that encodes for both a fluorescent and a luciferase protein in the same viral genome55 (Figure 1). Here, NanoLuc luciferase (Nluc), a small and bright bioluminescent protein48, was inserted upstream of the hemagglutinin (HA) sequence in the viral HA segment of influenza A/Puerto Rico/08/1934 H1N1 (PR8)24,33,40,55,56,57. In addition, Venus, a frequently used monomeric fluorescent protein, was inserted into the non-structural (NS) viral segment32,33,36,41,55. Since BIRFLU encodes for both fluorescent and luciferase reporter genes, either reporter protein signal can be used as readout to determine viral replication and dissemination in vitro or in vivo55. Additional information regarding the generation and in vitro or in vivo characterization of BIRFLU can be found in our recent publication55. BIRFLU can be used to test the effectiveness of antiviral drugs or neutralizing antibodies via a novel fluorescent- and bioluminescent-based microneutralization assay55. Moreover, BIRFLU can also be used to evaluate viral dynamics in a mouse model of infection55. In this manuscript, we describe the procedures to characterize BIRFLU55 in vitro and how to study BIRFLU infection in mice using in vivo luminescence imaging systems for the detection of Nluc in vivo or of Venus ex vivo.
The combination of cutting-edge techniques in molecular biology, animal research and imaging technologies, brings researchers the unique opportunity to use BIRFLU for IAV research, including the study of virus-host interactions, dynamics of viral infection; the development of novel vaccine approaches for the therapeutic treatment of IAV infections or the potential use of IAV as a vaccine vector for the treatment of other pathogen infections.
Protocol
All protocols involving mice have been approved by the Institutional Animal Care and Use Committee (IACUC) and the Institutional Biosafety Committee (IBC) at the University of Rochester, School of Medicine and Dentistry. All the experiments performed in animals follow the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council58. The Vivarium and Division of Laboratory Animal Medicine facilities at the School of Medicine and Dentistry at University of Rochester is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AALAC) International and conform to federal and state laws and National Institutes of Health (NIH) policy. Proper Personal Protection Equipment (PPE) is required when working with mice. Similar policies and requirements should be implemented when performing experiments outlined within this manuscript at each institution.
1. Use of Small Vertebrate Animals
- Purchase five to seven-week-old female BALB/c mice and maintain them in the animal care facility under specific pathogen-free conditions. Upon mice arrival to the determined facilities, allow period of rest for 4–5 days to allow animals acclimate to their new environment.
- Following IACUC protocols, place a maximum of 5 mice per cage.
NOTE: At experiment conclusion, in order to ensure that the animal is dead, mice were euthanized using two approved procedures, taking into consideration that the second must be a physical method. In this study, after mice infection with BIRFLU and in vivo imaging, animals are euthanized with a lethal dose of 2,2,2-tribromoethanol (TBE), and by cutting the hepatic vein as the physical secondary method as we have previously shown23.
2. Biosafety
NOTE: In this manuscript, BIRFLU was generated in the backbone of influenza A/Puerto Rico/08/34 H1N1 (PR8), which is a common mouse-adapted laboratory IAV strain23,32,33,56. The virus was generated using previously described plasmid-based reverse genetics approaches and a complete description of the generation, and in vitro and in vivo characterization of BIRFLU can be found in our recent publication55. All procedures that involve IAV infections (in vitro or in vivo) were performed in a biological safety cabinet under biosafety level (BSL)-2 conditions.
CAUTION: An appropriate biosafety level should be determined in accordance with a biosafety risk assessment. Additional information on performing biosafety risk assessments and establishing effective biosafety containment should be consulted with the institution where the experiments will be performed.
- Clean the biosafety cabinets with 70% ethanol or chlorine dioxide disinfectant before and after performing all the experimental procedures. For mice work, sterilize all dissection material (scissors, dissecting forceps, etc.) and the Dounce homogenizer before and after their use.
- Discard all biological material produced during the procedures under proper IBC and IACUC guidelines.
3. In Vitro Characterization of BIRFLU (Figure 2)
NOTE: Refer to Table 1 for all buffer and media compositions.
- Analyze protein expression by fluorescence (Figure 2A) and indirect immunofluorescence (Figure 2B)
- One day before infection, seed 24-well plates with Madin-Darby Canine Kidney (MDCK) epithelial cells (1 x 105 cells/well, triplicates) in tissue culture media and maintain the plates in a 37 °C incubator with 5% CO2. Prepare enough wells to evaluate all the chosen antibodies in both mock-infected and BIRFLU-infected cells.
NOTE: We recommend visualizing the cells under a light microscope to confirm a monolayer before starting the viral infection. A 90% confluency monolayer of MDCK cells by the time of infection is recommended. - Prepare dilutions of WT or BIRFLU IAVs in infection media and infect the seeded MDCK cells with a multiplicity of infection (MOI) of 0.1 plaque-forming units (PFU) per cell in a final volume of 0.25 mL/well of infection media.
- Remove the tissue culture medium from step 3.1.1 and wash the MDCK cells twice with 1x phosphate-buffered saline (PBS). Add the virus dilutions from 3.1.2 to the MDCK cells and place the plates on a rocking platform for 1 h at room temperature to allow viral adsorption.
NOTE: Mock-infected cells are incubated only with infection media in the absence of virus. - After viral adsorption (step 3.1.3), remove the viral inoculum by aspiration and add 1 mL of post-infection media containing 1 μg/mL of TPCK-treated trypsin to each well. Incubate cells for 18 h in a 5% CO2 humidified incubator at 33 °C.
- After 18 h of infection, remove the tissue culture supernatant from the 24-well plates (3.1.4). Fix and permeabilize the cells with 0.25 mL/well of fixation/permeabilization solution for 20 min at room temperature.
NOTE: Prepare the fixation/permeabilization solution in a fume hood to prevent formaldehyde exposure. - Remove the fixation/permeabilization solution from step 3.1.5, wash the cells twice with 1x PBS, and incubate the cells with 0.25 mL/well of blocking solution for 1 h at room temperature.
NOTE: Proceed to the next step or store cells in blocking solution at 4 °C overnight. - Remove the blocking solution (step 3.1.6) and add 0.25 mL/well (1 µg/mL) of a mouse monoclonal antibody (MAb) against the IAV nucleoprotein, NP (HB-65) or a 1:1,000 dilution of a rabbit polyclonal antibody (pAb) against NLuc (see the Table of Materials), both diluted in antibody dilution solution (1x PBS, 2.5% BSA), and incubate the cells for 1 h at 37 °C.
- Remove the primary antibody from step 3.1.7, wash the cells three times with 1x PBS and incubate them with 0.25 mL/well of a Texas Red-conjugated anti-mouse or anti-rabbit secondary antibodies (see the Table of Materials) diluted 1:200 in antibody dilution solution.
NOTE: Cell nuclei can be stained at the same time by adding 0.5 µg/mL of 4’,6’-diamidino-2-phenylindole (DAPI) to the same secondary antibody solution. Incubate the cells for 1 h at 37 °C in the dark. Other secondary antibodies conjugated with other fluorophores can be used. - Remove the secondary antibody and DAPI from step 3.1.8, wash the cells three times with 1x PBS. After washing, leave the cells in 0.25 mL/well of 1x PBS.
- Place the plate on the stage of a fluorescence microscope to detect Venus and Nluc reporter expression, and NP from infected cells using the proper fluorescent filters. Capture images using a fluorescence microscope (20x magnification) and merge them using an image editing software (Figure 2A,B).
- One day before infection, seed 24-well plates with Madin-Darby Canine Kidney (MDCK) epithelial cells (1 x 105 cells/well, triplicates) in tissue culture media and maintain the plates in a 37 °C incubator with 5% CO2. Prepare enough wells to evaluate all the chosen antibodies in both mock-infected and BIRFLU-infected cells.
- Analyze the Nluc activity (Figure 2C) and viral replication (Figure 2D).
- One day before infection, seed 12-well plates with MDCK cells (2 x 105cells/well, triplicates) using tissue culture media in a 37 °C incubator with 5% CO2 to reach approximately 90% confluency by the time of infection.
NOTE: Before infection, check the cells under a microscope to verify a monolayer of MDCK cells. - The day of infection prepare dilutions of the WT and BIRFLU viruses in infection media to infect the MDCK cells (step 3.2.1.) in triplicate with a MOI of 0.001 PFU in 0.5 mL/well. Remove the tissue culture medium and wash the MDCK cells twice with 1x PBS.
- Add the virus dilutions to the MDCK monolayers and allow viral adsorption at room temperature for 1 h on a rocking platform. After viral adsorption, remove the virus inoculum and add 1.5 mL of post-infection media containing 1 μg/mL of TPCK-treated trypsin, to each well. Incubate infected cells in a 5% CO2 humidified incubator at 33 °C and collect supernatants at the indicated time points indicated in step 3.2.4.
- At 24, 48, 72, and 96 h post infection (p.i.) collect 150 µL of tissue culture supernatant from each well and store the samples in a microcentrifuge tube at -80 °C to perform the Nluc assays and viral titrations.
- To perform the Nluc activity assay, follow the manufacturer’s indications. First, thaw the tissue culture supernatant stored at -80 °C (step 3.2.4) on ice.
NOTE: Refer to the recommendations from the manufacturer and optimize the assay if required. - Prepare the luciferase assay solution (see the Table of Materials) by diluting the Nluc substrate 1:50 with the dilution buffer. In a white flat-bottom 96-well microplate for luciferase assays, mix 25 µL of luciferase assay solution with 10 to 25 µL of the collected tissue culture supernatant samples. Incubate the mix for 2–3 min before measuring the luminescence using a luminometer (Figure 2C).
NOTE: Presence of infectious viral particles in the tissue culture supernatants is determined by fluorescence immune-focus assay (Venus) or indirect immunofluorescence (viral antigen staining) as previously described23,32,33,56. - A day before viral titrations, seed MDCK cells in a 96-well plate (2 x 104 cells/well, triplicates) with tissue culture media and allow cells to reach 90% confluence by the time of infection in an incubator set at 37 °C with 5% CO2.
- Use the tissue culture supernatant samples that were thawed on ice (step 3.2.4.). Add 90 μL of infection media to each of the wells in a new 96-well plate, then add 10 μL of the thawed tissue culture supernatant (step 3.2.4) to the first well (row A). Use a multichannel pipet to mix and transfer 10 μL from row A to row B. Change the tips between dilutions and mix well.
NOTE: This procedure should be repeated until the last row (H). We recommend performing viral titrations in triplicates for accurate measure of viral titers. - Remove the tissue culture medium from the MDCK cells in the 96-well plates (step 3.2.7) and wash twice with 1x PBS. Add 50 µL of the supernatant dilutions of the replica 96-well plate containing the virus dilutions (step 3.2.8) to each well in the 96-well plate containing MDCK cells.
- Incubate the 96-well plate on a rocking platform for 1 h at room temperature to allow viral adsorption. After viral adsorption, remove the inoculum and add 100 μL/well of post-infection media containing 1 μg/mL of TPCK-treated trypsin. Incubate infected cells for 12 h in a 33 °C incubator with 5% CO2.
- Since BIRFLU expresses Venus, the number of infected cells can be directly counted using a fluorescence microscope. Alternatively, viral titers can be determined by indirect immunofluorescence as previously described using an antibody against a viral protein23,56,57,59. For the latter, proceed to fix/permeabilize the cells and stain using the anti-NP mAb HB-65 as described in section 4.4.
- Determine the viral titers by counting the number of fluorescent-forming units (FFU)/mL using the formula: ((number of FFU) x 20 x 1/dilution (Figure 2D).
- One day before infection, seed 12-well plates with MDCK cells (2 x 105cells/well, triplicates) using tissue culture media in a 37 °C incubator with 5% CO2 to reach approximately 90% confluency by the time of infection.
4. In Vivo Characterization of BIRFLU (Figure 3 and Figure 4)
- Mouse infection
NOTE: Intranasal infection of mice was done as previously described23. For a more detailed protocol of IAV infection in vivo using the mouse model of infection, we recommend viewing the video associated with the previous publication23. This section only summarizes the steps required for mouse infection with BIRFLU.- Inspect the mice to evaluate their health and overall physical appearance. Prepare the dilution of BIRFLU in 1x PBS to inoculate mice with 1 x 106 PFU of BIRFLU in a total volume of 30 μL/mouse. Maintain the virus on ice until mouse inoculation.
NOTE: Mock-infected (1x PBS) mice will be needed as internal control for imaging. Place mock-infected mice in a different cage than BIRFLU-infected animals. - Anesthetize five-to-seven-week-old female BALB/c mice intraperitoneally with 240–250 mg/kg of tribromoethanol (TBE) by inserting the needle into the caudal 2/3 of the right side of the abdomen. Then, return the mouse to the cage and wait for around 5 min. Check that the mouse is anesthetized before virus inoculation by the absence of the toe-pinch reflex.
NOTE: Injectable sedatives are recommended over inhaled sedatives for influenza infections in vivo, as the latter can modify the behavior and uptake of the virus by the airway epithelium. Moreover, they can also affect local immune responses of the lung and interfere with the in vivo imaging of infected mice. Finally, inhaled sedatives have reduced duration of effect, which would be a problem for in vivo imaging of the infected animals. - Inoculate the mice intranasally with the 30 μL of the prepared BIRFLU dilution. Check that the mice are breathing properly before returning them to the cage.
- Inspect the mice to evaluate their health and overall physical appearance. Prepare the dilution of BIRFLU in 1x PBS to inoculate mice with 1 x 106 PFU of BIRFLU in a total volume of 30 μL/mouse. Maintain the virus on ice until mouse inoculation.
- Bioluminescence monitoring of mice infected with BIRFLU (Figure 4A)
NOTE: In this manuscript reporter expression of Nluc or Venus and viral replication in the lungs of mice infected with BIRFLU are determined at 3 days post-infection. However, the nature of bioluminescence imaging allows repeated monitoring of Nluc expression in individual BIRFLU-infected animals without the need to euthanize them for each experimental time-point. An in vivo imaging system with an isoflurane anesthesia manifold is required to perform the described experimental procedures. See the Table of Material for details regarding the imaging instrument and image software used for the image acquisition and data analysis.- Shave the mice chest to improve the bioluminescence signal. Open the imaging software and press Initialize. Next, set the parameters that will be used, including setting the imaging mode to bioluminescence, auto saving, exposure time to auto, open filter, etc.
- Once the machine is fully initialized, turn on the isoflurane anesthesia system. Place animals in the anesthesia chamber. Mice are simultaneously and lightly anesthetized with a mixture of oxygen gas and vaporized 1–2% isoflurane.
- Once mice are anesthetized, administer the Nluc substrate (see the Table of Materials) diluted 1:10 in 1x PBS (final volume 100 µL/mouse) via retro-orbital route using a syringe with a 22 G needle.
- Immediately after Nluc reagent administration, place the animals in the imaging instrument with their chests facing up and snout inside the manifold cone to keep animal anesthetized during imaging. Immediately after closing the imager door, click Acquire in the software program (Figure 4A, top).
- After imaging, return the mice to their cages monitoring them until they have fully recovered and turn off the isoflurane vaporizer. Then, proceed with the ex vivo imaging of mice lungs to evaluate Venus reporter gene expression (section 4.3).
- Use the imaging software tools to analyze the acquired bioluminescence data. Utilize the tool ROI (region of interest) to designate the specific signal and perform flux measurements in the region of interest (usually around the chest) (Figure 4A, bottom). Although ROI shape is irrelevant, larger ROIs are generally preferred to capture the entire signal diffusion area.
- Click Measure. Assess the bioluminescence in photons since it provides an absolute photon emission measurement that is comparable to output measurements provided by different parameters or imaging instruments.
- Fluorescence analysis in mice infected with BIRFLU (Figure 3 and Figure 4B)
- Collect the mouse lungs after in vivo imaging, as previously described23.
- Briefly, euthanize mice with a lethal dose of TBE (500 mg/kg). Disinfect the incision site with 70% ethanol. With a scalpel, make an incision from the sternum to the base of the abdomen and then cut from the bottom of the incision to the sides with scissors. Next, cut the hepatic vein (second physical euthanasia method) to bleed the animal.
NOTE: To avoid high background signal during imaging, it is important to minimize the amount of blood in the lungs (see below).
- Briefly, euthanize mice with a lethal dose of TBE (500 mg/kg). Disinfect the incision site with 70% ethanol. With a scalpel, make an incision from the sternum to the base of the abdomen and then cut from the bottom of the incision to the sides with scissors. Next, cut the hepatic vein (second physical euthanasia method) to bleed the animal.
- Place the mouse in dorsal recumbency, and use the scissors to cut the pleura and open the rib cage. Subsequently, remove the lungs by snipping the end of the trachea with scissors while gently holding the lungs with forceps.
- Place the lungs in a six-well plate with 2 mL of 1x PBS and wash the lungs three times with 1x PBS.
NOTE: To avoid contamination between samples, clean and disinfect the dissecting tools between each animal.
- Place the lungs in a six-well plate with 2 mL of 1x PBS and wash the lungs three times with 1x PBS.
- Start the image acquisition software by click initialize and set the parameters for imaging, including setting imaging mode to fluorescence, auto saving, exposure time to auto, excitation (500 nm) and emission (540 nm) filters.
- When the machine is initialized, place the lungs in a black background tray, ensuring that the tissues are separated from one another, introduce the tray into the imager, and click Acquire on the imaging system program after closing the imager door (Figure 4B, top).
- After imaging, remove the tissues immediately and store them on ice (4 °C) if the samples are processed the same day; or on a tube and dry ice to freeze them quickly, before storing them at -80 °C, if the samples will be processed later on a different day.
- For image processing, select the ROI tool and draw ROIs around each of the individual lungs. Click Measure. Then, using the resulting average radiant efficiency measurements, subtract the values from the mock-infected mice (Figure 4B, bottom).
NOTE: With BIRFLU, there is a good correlation of the levels of Nluc and Venus expression, and signal distribution. Therefore, it is important to analyze Nluc (whole mouse) and Venus (excised lungs) expression from the same animal, and maintain the same orientation.
- Collect the mouse lungs after in vivo imaging, as previously described23.
- Evaluation of BIRFLU replication in mice lungs (Figure 3 and Figure 4C)
- Homogenize mice lungs for viral titration by plaque assay as previously described23.
- Place the lungs into a Dounce homogenizer with 1 mL of chilled infection media. Homogenize the lungs with the pestle for 1 min at room temperature until it has fully disintegrated and store the samples in a sterile tube at 4 °C.
- Centrifuge the samples at 300 x g for 10 min and collect the supernatant in a sterile tube. Store the samples on ice if they will be used on the same day or freeze them at -80 °C to evaluate viral titers at a later time.
NOTE: A new Dounce homogenizer should be used for each lung sample. - To perform the plaque assay, the day prior to performing viral titration, seed six-well plates with MDCK cells (5 x 105 cells/well) in tissue culture media. Incubate the cells overnight at 37 °C with 5% CO2 to reach 90% confluence next day. Before infection, confirm that the cells form a monolayer under a light microscope.
- Prepare 1:10 serial dilutions of the supernatant from the homogenized samples (step 4.4.1) in infection media. Prepare microcentrifuge tubes with 540 µL of infection media, add 60 µL from the homogenized lungs sample to the first tube, mix by pipetting up and down, and then using a new tip, transfer 60 µL to the next tube. Repeat this serial dilution process until the last dilution.
NOTE: In our experience 7 dilutions are sufficient to determine viral titers by plaque assay from the lungs of infected mice. - Wash the MDCK cells (step 4.4.3) twice with 1x PBS and transfer 500 µL of the serially diluted samples to each well in the six-well plate. Place the plates on a rocking platform and allow viral absorption to occur for 1 h at room temperature.
- After 1 h of viral absorption, remove the viral inoculum, add 2 mL/well of overlay medium containing 1 μg/mL of TPCK-treated trypsin, and then incubate the cells at 33 °C under 5% CO2 for 3 days.
- Fix infected cells (step 4.4.6) with 1 mL/well of 4% formaldehyde diluted in 1x PBS at room temperature for 2 h, then carefully remove the overlay medium and add 1 mL of 1x PBS to each well. For visualization of Venus, image the six-well plates with an imaging system for the detection of fluorescence.
NOTE: Viral plaques can be colored using an image editing software (see the Table of Materials). - To evaluate Nluc expression, visualize viral plaques by immunostaining using an anti-Nluc pAb. For this, remove the 1x PBS, permeabilize cells using 0.5 mL/well of permeabilization solution for 15 min at room temperature.
- Remove the permeabilization solution, wash the cells three times with 1x PBS and block with 0.5 mL/well of blocking solution for 1 h at room temperature.
- Remove the blocking solution from step 4.4.9 and incubate the cells with 0.5 mL/well of the pAb anti-Nluc diluted 1:1,000 in dilution solution for 1 h at 37 °C.
- Use ABC Alkaline Phosphatase kit and DAB Peroxidase Substrate Kit following manufacturer’s recommendation for visualization of Nluc-expressing plaques.
- Briefly, wash cells from 4.4.10 three times with 1x PBS and incubate them with 0.5 mL/well of biotinylated anti-rabbit secondary antibody for 1 h at 37 °C.
- Remove the secondary antibody, wash the cells three times with 1x PBS, and incubate with the ABC solution for 1 h at 37 °C. Wash the cells three times with 1x PBS and visualize the viral plaques with the DAB HRP substrate Kit. Scan the immunostained plaques using a conventional scanner.
NOTE: Other similar kits for immunostaining could be used.
- Stain the viral plaques with crystal violet solution for 1 h at room temperature. Discard the crystal violet, wash the plates three times with water, allow plates to dry, and scan the plates again.
- To determine viral titers, count the plaques revealed after crystal violet staining. Scan the plaques using a conventional scanner. Calculate virus titer as plaque forming units (PFU) per mL (PFU/mL).
- To assess BIRFLU stability in vivo, calculate the percentage of reporter-expressing viruses by counting the number of crystal violet-stained plaques (number of infectious viruses, step 4.4.12) and compare with the number of Venus- and Nluc-expressing plaques (step 4.4.7 and step 4.4.11, respectively).
- Centrifuge the samples at 300 x g for 10 min and collect the supernatant in a sterile tube. Store the samples on ice if they will be used on the same day or freeze them at -80 °C to evaluate viral titers at a later time.
Representative Results
Generation and characterization of BIRFLU in vitro (Figure 1 and Figure 2)
A recombinant replication-competent IAV expressing two different reporter genes (BIRFLU) was constructed using state of the art molecular biology and plasmid-based reverse genetics techniques (Figure 1). Here, we chose to use Nluc due to several advantages over other luciferases, including its small size, ATP-independence, greater intensity, and optimized substrate48,60. Nluc was cloned into the HA segment of IAV PR8 followed by the porcine teschovirus (PTV) 2A cleavage site (2A) in front of the open reading frame (ORF) of HA (Figure 1). The ORF of HA included silent mutations to remove the original packing signals and avoid any possible recombination. The complete HA packaging signal was added in front of Nluc to allow proper incorporation of the modified HA segment into the virion and Nluc and HA expression from the same viral RNA segment (Figure 1). In addition, the fluorescent protein Venus was cloned into a modified IAV PR8 NS segment which encodes the two viral proteins NS1 and NEP from a single transcript32,36,41,54,57. To that end, Venus was fused to the C-terminal of NS1 and the entire NEP ORF was cloned downstream of the PTV 2A cleavage site that was placed between the NS1-Venus and NEP sequences (Figure 1). Ultimately, these two modified HA and NS viral plasmid constructs were used in combination with the rest of the IAV PR8 reverse genetics plasmids to generate BIRFLU (Figure 1). The in vitro and in vivo characterization of BIRFLU has been described previously55.
In Figure 2, we characterized in vitro BIRFLU by determining Venus, Nluc, and NP expression levels using fluorescence and indirect immunofluorescence approaches (Figure 2A,B). Confluent monolayers of MDCK cells were either mock-infected or infected (MOI 0.1) with WT or BIRFLU PR8 viruses and, at 18 h post-infection, Venus expression was directly evaluated using fluorescence microscopy (Figure 2A,B). Nluc (Figure 2A) and NP (Figure 2B) expression were visualized by indirect immunofluorescence using antibodies specific for each protein. As anticipated, Venus and Nluc expression were detected only in cells infected by BIRFLU and not in cells infected by WT PR8 virus. In addition, indirect immunofluorescence microscopy revealed NP expression in both WT and BIRFLU PR8-infected cells. No expression of Venus, Nluc or NP were detected, as expected, in mock-infected cells (Figure A,B).
To evaluate Nluc expression levels in vitro, MDCK cells were infected (MOI 0.001) with WT or BIRFLU PR8 viruses and Nluc activity in tissue culture supernatants was assessed at 24, 48, 72 and 96 h post-infection (Figure 2C). Only Nluc activity was detected in tissue culture supernatants of MDCK cells infected with BIRFLU (Figure 2C). Nluc activity in the tissue culture supernatants was detected as early as 24 h post-infection with higher expression levels at 96 h post-infection, most probably because the cytopathic effect (CPE) induced during viral infection release the Nluc protein retained into the cell. To evaluate the fitness of BIRFLU in cultured cells, growth kinetics of WT and BIRFLU PR8 viruses were also evaluated (Figure 2D) and the presence of infectious virus in tissue culture supernatants was determined by immune-focus assay (Figure 2D). Notably, BIRFLU replication kinetics were comparable to those of WT PR8 virus, although BIRFLU replication was slightly delayed and did not reach same viral titers as WT PR8. However, BIRFLU was able to reach titers of 5 x 107 PFU/mL (Figure 2D), indicating that expression of two reporter genes in the viral genome does not significantly interfere with BIRFLU replication in MDCK cells.
Tracking BIRFLU infection in mice (Figure 3 and Figure 4)
Figure 3 is a schematic flow chart for the assessment of BIRFLU dynamics in a mouse model of IAV infection. Five-to-seven-week-old female BALB/C mice were either mock-infected with 1x PBS or infected with 1 x 106 PFU of BIRFLU intranasally. At 3 days post-infection, mice were anesthetized with isoflurane and then injected with Nluc substrate retro-orbitally. All mice were immediately placed in the IVIS instrument and Nluc signal was assessed in vivo using the IVIS. Next, mice were euthanized and lungs were harvested. Excised lungs were then analyzed ex vivo using the in vivo imager to determine fluorescence intensity via Venus expression. Lastly, mice lungs were homogenized, and viral titers and stability were determined by plaque assay. Plaques were assessed by the direct fluorescence of Venus, by immunostaining using antibodies specific for Nluc and by crystal violet staining.
Previously described replication-competent reporter-expressing IAVs express a single reporter gene, most frequently either a fluorescent or a bioluminescent protein, as surrogate for viral infection and replication. However, BIRFLU, is able to express both types of reporter genes upon viral infection. To assess the correlation between bioluminescence (in vivo imaging) and fluorescence (ex vivo imaging) after BIRFLU infection, five-to-seven-week-old female BALB/c mice were mock-infected with 1x PBS or inoculated with BIRFLU (106 PFU) intranasally. Nluc activity (Figure 4A) was evaluated by administration of Nluc substrate injected retro-orbitally at 3 days post-infection using an in vivo imaging instrument. We chose to evaluate bioluminescence at day 3 because previous studies indicated that IAV replication, including PR8, peaks between days 2 and 4 post-infection24,54. Bioluminescence was monitored (Figure 4A, top) and used to calculate the average total flux (Flux (log10 p/s) (Figure 4A, bottom). As predicted, mice inoculated with BIRFLU displayed high bioluminescence activity but no signal was detected in mock-infected mice. Thereafter, the lungs of infected mice were harvested and Venus expression was assessed using ex vivo imaging (Figure 4B, top). Moreover, the fluorescence average radiant efficiency was calculated (Figure 4B, bottom). The excised mice lungs were also homogenized to determine the viral titers and the genetic stability of BIRFLU in vivo (Figure 4C,D). Genetic stability of BIRFLU was analyzed through plaque assay using the viruses isolated from mice lungs and fluorescent microscopy (Venus, top), immunostaining (Nluc, middle) and crystal violet staining (bottom). BIRFLU recovered from mice lungs were able to form plaques and stably expresses both reporter genes (Figure 4C). Notably, we observed a good correlation between bioluminescence and fluorescence signals with viral replication.
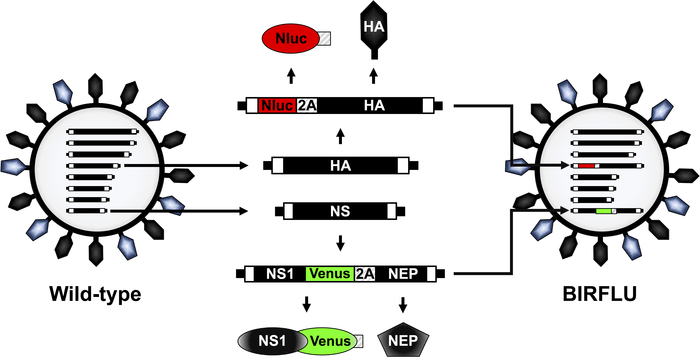
Figure 1: Schematic representation of IAV PR8 WT and BIRFLU virion structure and genome segments. IAV are surrounded by a lipid bilayer containing the two major viral glycoproteins hemagglutinin (HA; black) and neuraminidase (NA; blue). IAV contain eight single-stranded, negative-sense, RNA segments (PB2, PB1, PA, HA, NP, NA, M, and NS). Each viral segment contains non-coding regions (NCR) at the 3’ and 5’ ends (black boxes). Also, at the 3’ and 5’ end of the viral (v)RNAs are the packaging signals, responsible for the efficient encapsidation of vRNAs into nascent virions (white boxes). IAV PR8 HA and NS viral segments and products are indicated in black. Sequences of Nluc, Venus, and PTV 2A are indicated in red, green and striped boxes, respectively. The schematic representation of the modified HA and NS segments expressing Nluc and Venus, respectively, in BIRFLU are also indicated. This figure has been adapted from Nogales et al.55. Please click here to view a larger version of this figure.
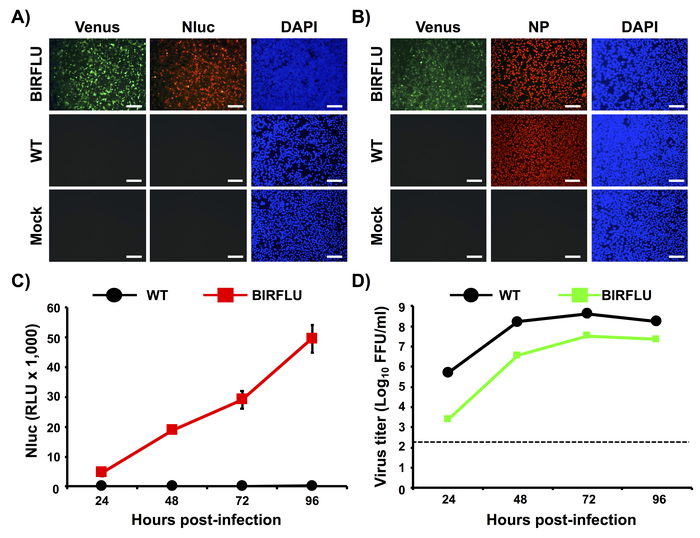
Figure 2: In vitro characterization of BIRFLU. (A, B) Analysis of protein expression by direct fluorescence and immunofluorescence. MDCK cells were mock-infected or infected (MOI 0.1) with PR8 WT or BIRFLU viruses. Infected cells were fixed at 18 h post-infection to directly visualize Venus expression by direct fluorescent microscopy and to visualize Nluc (A) and viral NP (B) expression using specific antibodies and indirect immunofluorescence. Nuclei were stained with DAPI. Representative images (20x magnification) are shown. Scale bars = 100 μm. (C, D) Growth kinetics of PR8 WT and BIRFLU. Nluc activity (C) and viral titers (D) in tissue culture supernatants from MDCK cells infected (MOI 0.001) with WT and BIRFLU PR8 viruses were assessed at the indicated times post-infection. Data represent the means ± SD of triplicates. Viral titers were determined by immune-focus assay (FFU/mL). Dotted line denotes the limit of detection (200 FFU/ml). This figure has been adapted from Nogales et al.55. Please click here to view a larger version of this figure.
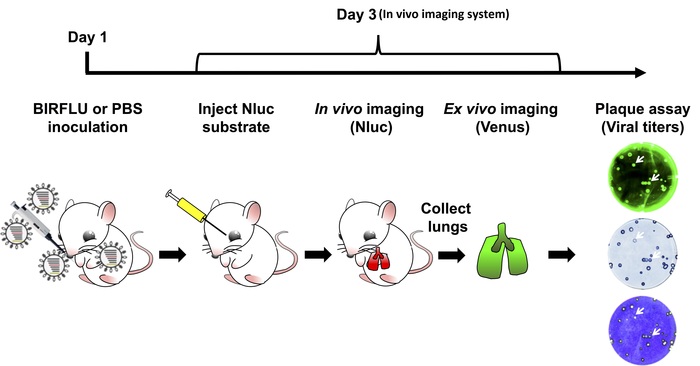
Figure 3: Schematic representation for the study of BIRFLU in mice. Expression of Nluc and Venus reporter genes was evaluated in mice infected with 1 x 106 PFU of BIRFLU using in vivo or ex vivo imaging. Briefly, on day 1, 5 to 7 week-old female BALB/c mice were mock-infected (1x PBS) or inoculated with 1 x 106 PFU of BIRFLU intranasally. At day 3 post-infection, mice were mildly anesthetized using isoflurane and Nluc substrate was injected retro-orbitally. Nluc signal was directly assessed using in vivo imaging. Immediately after imaging, mice were euthanized and expression of Venus in whole excised lungs was analyzed using ex vivo imaging. Recovered mice lungs were homogenized to evaluate viral replication and stability by plaque assay. Arrows indicate correlation between fluorescence (Venus), immunostaining (Nluc) and crystal violet staining. Please click here to view a larger version of this figure.
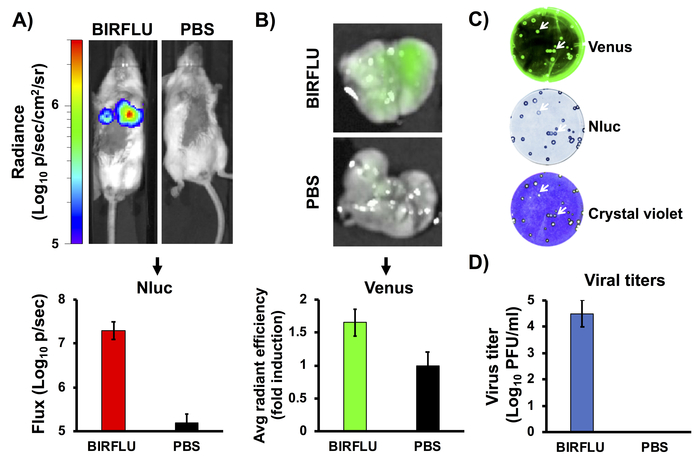
Figure 4: In vivo bioluminescence and fluorescence expression. Female five-to-seven-week-old BALB/c mice were mock-infected (1x PBS) or inoculated with 1 x 106 PFU of BIRFLU intranasally. At day 3 post-infection, Nluc activity (A) in the whole mouse was determined. Representative images of a single mouse showing radiance scale (p/sec/cm2/sr). Bioluminescence radiance values were quantitated and the average total flux is shown (Flux (Log10 p/s). After Nluc imaging, lungs were harvested for ex vivo imaging (B). Representative pictures from whole lungs are shown. To quantify Venus expression, mean values of regions of interest (ROIs) were normalized to lung auto-fluorescence from mock-infected mice and fold changes were calculated. To analyze the genetic stability of BIRFLU in vivo, viruses recovered from mice lungs were analyzed by plaque assay using fluorescent microscopy (Venus, top), immunostaining (Nluc, middle) and crystal violet staining (bottom) (C). Representative images from one mouse are shown. To evaluate virus replication, whole lungs were homogenized after imaging and used to infect MDCK cells and determine viral titers by plaque assay (PFU/mL) (D). Arrows indicate correlation between fluorescence (Venus), immunostaining (Nluc), and crystal violet staining. Bars represent the mean ± SD of lung virus titers. This figure has been adapted from Logales et al.55. Please click here to view a larger version of this figure.
| Tissue culture media and solutions | Composition | Storage | Use |
| Tissue culture media: Dulbecco’s modified Eagle’s medium (DMEM), 10 % Fetal Bovine Serum (FBS), 1% Penicillin-Streptomycin-L-glutamine (PSG) (DMEM 10 % FBS 1% PSG). | 445 ml DMEM, 50 mL of FBS and 5 mL of 100x PSG. | 4 °C | Maintenance of MDCK cells |
| Post-infection media: DMEM 0.3% Bovine Albumin (BA), 1% PSG (DMEM 0.3 % BA 1% PSG). | 491 ml DMEM, 4.2 ml of 35 % BA and 5 ml of 100x PSG | 4 °C | Maintenance of MDCK cells after viral infection |
| 10x Phosphate buffered saline (PBS) | 80 g of NaCl, 2 g of KCl, 11.5 g of Na2HPO4.7H2O, 2 g of KH2PO4. Add ddH2O up to 1 L. Adjust pH to 7.3 | Room temperature | To prepare 1x PBS |
| 1x PBS | Dilute 10x PBS with ddH2O | Room temperature | Wash cells |
| Infection media: 1x PBS, 0.3% BA, 1% Penicillin-Streptomycin (PS) (PBS/BA/PS). | 487 mL 1x PBS sterile, 4.2 mL of 35% BA and 5 ml of 100x 1% PS(100 U/mL) | 4 °C | Viral infections |
| Fixation/permeabilization solution: 4% formaldehyde, 0.5% triton X-100 diluted in 1x PBS. | 400 mL neutral buffered formalin 10%, 5 ml of Triton X-100 and 595 mL of 1x PBS | Room temperature | Fix and permeabilization of MDCK cells. |
| Blocking solution: 2.5% Bovine Serum Albumin (BSA) in 1x PBS. | 2.5 g of BSA in 97.5 mL of 1x PBS | 4 °C | Blocking solution for immunofluorescence and plaque assays. |
| Antibody dilution solution (1% BSA in 1x PBS) | 1 g of BSA in 99 mL of 1x PBS | 4 °C | Dilution of primary and secondary antibodies. |
| 0.1% crystal violet solution | 1 g of crystal violet in 400 mL of methanol. Add 600 ml of ddH2O | Room temperature | Staining of MDCK cells in plaque assays. |
| Tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin | Prepare a 1,000x stock solution at 1 mg/mL in ddH2O | -20 °C | For viral infections. |
Table 1: Tissue culture media and solutions.
Discussion
Researchers have relied on recombinant reporter-expressing viruses as vital molecular tools to understand and expand upon the current understanding of viral replication and pathogenesis26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,54. The most commonly favored reporter genes are luciferases and fluorescent proteins, mainly due to the technological advancements in their identification, development of improved variants, and detection by imaging technologies43,44,45,46,47,48. Recombinant reporter viruses are often used to accelerate virological assays, study the dynamics of viruses in vitro and in vivo, and to test the effectiveness of currently approved or new vaccine and therapeutic approaches26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,54. Unfortunately, in the case of IAV, past studies were limited to the expression of a single reporter gene, which hinders the type of study that could be conducted 26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,54. To avoid this limitation, we have generated a replication-competent bi-reporter IAV that expresses a Nluc luciferase and a Venus fluorescent protein (BIRFLU).
In this report, we describe the in vitro characterization of BIRFLU and the experimental approaches to use BIRFLU to track viral infection in vivo using a mouse model of IAV infection. BIRFLU Nluc and Venus expression correlated with viral titers. In addition, BIRFLU remained stable and continued to express both reporter genes after being recovered from the lungs of infected mice. This approach provides researchers with an excellent opportunity to study IAV in cultured cells and in animal models, including the identification and develop of new therapeutic alternatives for the treatment of IAV infections.
Although BIRFLU has been generated using the backbone of PR8, other recombinant IAV using different type, subtype or viral strain backbones could be generated using the same experimental approach. Likewise, in this report we described the experimental procedures for the use of BIRFLU in a mouse model of IAV. However, BIRFLU could be a valuable technology to evaluate IAV infection in other animal models.
Acknowledgements
Research on influenza virus in LM-S laboratory is partially funded by The New York Influenza Center of Excellence (NYICE) (NIH 272201400005C), a member of the NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract No. HHSN272201400005C (NYICE) and by the Department of Defense (DoD) Peer Reviewed Medical Research Program (PRMRP) grant W81XWH-18-1-0460.
Materials
| Name | Company | Catalog Number | Comments |
| 12-well Cell Culture Plate | Greiner Bio-one | 665102 | |
| 24-well Cell Culture Plate | Greiner Bio-one | 662160 | |
| 6-well Cell Culture Plate | Greiner Bio-one | 657160 | |
| 96-well Cell Culture Plate | Greiner Bio-one | 655-180 | |
| Adobe Photoshop CS4 | Adobe | This software is used in 3.1.10 and 4.4.7 | |
| Bovin Albumin solution (BA) | Sigma-Aldrich | A7409 | Store at 4° C |
| Bovin Serum Albumin (BSA) | Sigma-Aldrich | A9647 | Store at 4 °C |
| Cell Culture dishes 100mm | Greiner Bio-one | 664-160 | |
| ChemiDoc MP Imaging System | BioRad | This instrument is used in 4.4.7 | |
| Crystal Violet | Thermo Fisher Scientific | C581-100 | Store at Room temperature |
| Dounce Tissue Grinders | Thomas Scientific | 7722-7 | |
| Dulbecco’s modified Eagle’s medium (DMEM) | Corning Cellgro | 15-013-CV | Store at 4 °C |
| Fetal Bovine Serum (FBS) | Seradigm | 1500-050 | Store at -20 °C |
| Five- to seven-week-old female BALB/c mice | National Cancer Institute (NCI) | 555 | |
| Isoflurane | Baxter | 1001936040 | Store at Room temperature |
| IVIS Spectrum | PerkinElmer | 124262 | This instrument is used for in vivo imaging (4.2 and 4.3) |
| IX81 Motorized Inverted Microscope | Olympus | Olympus IX81 | |
| Living Image 4.7.2 software | PerkinElmer | This instrument is used for in vivo imaging (4.2 and 4.3) | |
| Lumicount | Packard | This instrument is used for quantifying luciferase activity (3.2.6) | |
| Madin-Darby Canine Kidney (MDCK) epithelial cells | ATCC | CCL-34 | |
| Monoclonal Antibody anti-NP Influenza A Virus HB-65 | ATCC | H16-L10-4R5 | Store at -20 °C |
| Nano-Glo Luciferase Assay Reagent | Promega | N1110 | This reagent is used to measure Nluc activity. Store at -20 °C |
| Neutral Buffered Formalin 10% | EMD | 65346-85 | Store at RT |
| Nunc MicroWell 96-Well Microplates | Thermo Fisher Scientific | 269620 | |
| Penicillin/Streptomycin (PS) 100x | Corning | 30-00-CI | Store at -20 °C |
| Penicillin/Streptomycin/L-Glutamine (PSG) 100x | Corning | 30-009-CI | Store at -20 °C |
| Retiga 20000R Fast1394 Camera | Qimaging | Retiga 2000R | |
| Scanner | HP | ||
| Texas Red-conjugated anti-mouse -rabbit secondary antibodies | Jackson | 715-075-150 | Store at -20 °C |
| Tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin | Sigma-Aldrich | T8802 | Store at -20 °C |
| Triton X-100 | J.T.Baker | X198-07 | Store at RT |
| Vmax Kinetic plate reader | Molecular Devices |
References
- Palese, P., Shaw, M. L., Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A. Orthomyxoviridae: The Viruses and Their Replication. Fields Virology. , (2007).
- Martinez-Sobrido, L., Peersen, O., Nogales, A. Temperature Sensitive Mutations in Influenza A Viral Ribonucleoprotein Complex Responsible for the Attenuation of the Live Attenuated Influenza Vaccine. Viruses. 10 (10), (2018).
- Nogales, A., Martinez-Sobrido, L. Reverse Genetics Approaches for the Development of Influenza Vaccines. International Journal of Molecular Sciences. 18 (1), (2016).
- Neumann, G., Noda, T., Kawaoka, Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 459 (7249), 931-939 (2009).
- Louie, J. K., et al. A review of adult mortality due to 2009 pandemic (H1N1) influenza A in California. PLoS One. 6 (4), e18221 (2011).
- Barr, I. G., et al. Epidemiological, antigenic and genetic characteristics of seasonal influenza A(H1N1), A(H3N2) and B influenza viruses: basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009-2010 Northern Hemisphere season. Vaccine. 28 (5), 1156-1167 (2010).
- Simonsen, L., et al. Impact of influenza vaccination on seasonal mortality in the US elderly population. A.M.A. archives of internal medicine. 165 (3), 265-272 (2005).
- McLean, H. Q., Peterson, S. H., King, J. P., Meece, J. K., Belongia, E. A. School absenteeism among school-aged children with medically attended acute viral respiratory illness during three influenza seasons, 2012-2013 through 2014-2015. Influenza and Other Respiratory Viruses. 11 (3), 220-229 (2017).
- Principi, N., Esposito, S. Protection of children against influenza: Emerging problems. Human Vaccines and Immunotherapeutics. , 1-8 (2017).
- Falsey, A. R., Treanor, J. J., Tornieporth, N., Capellan, J., Randomized Gorse, G. J. double-blind controlled phase 3 trial comparing the immunogenicity of high-dose and standard-dose influenza vaccine in adults 65 years of age and older. Journal of Infectious Diseases. 200 (2), 172-180 (2009).
- Fuller, J. D., et al. Influenza vaccination of human immunodeficiency virus (HIV)-infected adults: impact on plasma levels of HIV type 1 RNA and determinants of antibody response. Clinical Infectious Diseases. 28 (3), 541-547 (1999).
- Carrat, F., Flahault, A. Influenza vaccine: the challenge of antigenic drift. Vaccine. 25 (39-40), 6852-6862 (2007).
- Doherty, P. C., Kelso, A. Toward a broadly protective influenza vaccine. Journal of Clinical Investigation. 118 (10), 3273-3275 (2008).
- Fiore, A. E., et al. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Recommendations and Reports. 59 (RR-8), 1-62 (2010).
- Pica, N., Palese, P. Toward a universal influenza virus vaccine: prospects and challenges. Annual Review of Medicine. 64, 189-202 (2013).
- To, K. K., Chan, J. F., Chen, H., Li, L., Yuen, K. Y. The emergence of influenza A H7N9 in human beings 16 years after influenza A H5N1: a tale of two cities. Lancet Infectious Diseases. 13 (9), 809-821 (2013).
- Baker, S. F., Nogales, A., Santiago, F. W., Topham, D. J., Martinez-Sobrido, L. Competitive detection of influenza neutralizing antibodies using a novel bivalent fluorescence-based microneutralization assay (BiFMA). Vaccine. 33 (30), 3562-3570 (2015).
- He, W., Mullarkey, C. E., Miller, M. S. Measuring the neutralization potency of influenza A virus hemagglutinin stalk/stem-binding antibodies in polyclonal preparations by microneutralization assay. Methods. , (2015).
- Kayali, G., et al. Testing human sera for antibodies against avian influenza viruses: horse RBC hemagglutination inhibition vs. microneutralization assays. Journal of Clinical Virology. 43 (1), 73-78 (2008).
- Stephenson, I., et al. Reproducibility of serologic assays for influenza virus A (H5N1). Emerging Infectious Diseases. 15 (8), 1252-1259 (2009).
- Maher, J. A., DeStefano, J. The ferret: an animal model to study influenza virus. Lab Animal (NY). 33 (9), 50-53 (2004).
- Webster, R. G., Cox, N., Stoehr, K. . WHO/CDS/CSR/NCS/2002.5 Rev. 1. , (2002).
- Rodriguez, L., Nogales, A., Martínez-Sobrido, L. Influenza A Virus Studies in a Mouse Model of Infection. Journal of Visualized Experiments. (127), (2017).
- Cox, A., Baker, S. F., Nogales, A., Martínez-Sobrido, L., Dewhurst, S. Development of a mouse-adapted live attenuated influenza virus that permits in vivo analysis of enhancements to the safety of live attenuated influenza virus vaccine. Journal of Virology. 89 (6), 3421-3426 (2015).
- Steel, J., Lowen, A. C., Mubareka, S., Palese, P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathogens. 5 (1), e1000252 (2009).
- Tran, V., Moser, L. A., Poole, D. S., Mehle, A. Highly sensitive real-time in vivo imaging of an influenza reporter virus reveals dynamics of replication and spread. Journal of Virology. 87 (24), 13321-13329 (2013).
- Fukuyama, S., et al. Multi-spectral fluorescent reporter influenza viruses (Color-flu) as powerful tools for in vivo studies. Nature Communications. 6, 6600 (2015).
- Manicassamy, B., et al. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proceedings of the National Academy of Sciences U S A. 107 (25), 11531-11536 (2010).
- Perez, J. T., Garcia-Sastre, A., Manicassamy, B. Insertion of a GFP reporter gene in influenza virus. Current Protocols in Microbiology. , (2013).
- Reuther, P., et al. Generation of a variety of stable Influenza A reporter viruses by genetic engineering of the NS gene segment. Scientific Reports. 5, 11346 (2015).
- Tran, V., et al. Multi-Modal Imaging with a Toolbox of Influenza A Reporter Viruses. Viruses. 7 (10), 5319-5327 (2015).
- Breen, M., Nogales, A., Baker, S. F., Perez, D. R., Martinez-Sobrido, L. Replication-Competent Influenza A and B Viruses Expressing a Fluorescent Dynamic Timer Protein for In Vitro and In Vivo Studies. PLoS One. 11 (1), e0147723 (2016).
- Nogales, A., Baker, S. F., Martinez-Sobrido, L. Replication-competent influenza A viruses expressing a red fluorescent protein. Virology. , 206-216 (2014).
- Nogales, A., et al. Replication-competent fluorescent-expressing influenza B virus. Virus Research. 213, 69-81 (2015).
- Avilov, S. V., et al. Replication-competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. Journal of Virology. 86 (3), 1433-1448 (2012).
- Breen, M., Nogales, A., Baker, S. F., Martínez-Sobrido, L. Replication-Competent Influenza A Viruses Expressing Reporter Genes. Viruses. 8 (7), (2016).
- Eckert, N., et al. Influenza A virus encoding secreted Gaussia luciferase as useful tool to analyze viral replication and its inhibition by antiviral compounds and cellular proteins. PLoS One. 9 (5), e97695 (2014).
- Karlsson, E. A., et al. Visualizing real-time influenza virus infection, transmission and protection in ferrets. Nature Communications. 6, 6378 (2015).
- Czako, R., et al. In Vivo Imaging of Influenza Virus Infection in Immunized Mice. MBio. 8 (3), (2017).
- Harding, A. T., Heaton, B. E., Dumm, R. E., Heaton, N. S. Rationally Designed Influenza Virus Vaccines That Are Antigenically Stable during Growth in Eggs. MBio. 8 (3), (2017).
- DiPiazza, A., et al. Pandemic 2009 H1N1 Influenza Venus reporter virus reveals broad diversity of MHC class II-positive antigen-bearing cells following infection in vivo. Scientific Reports. 7 (1), 10857 (2017).
- Yan, D., et al. Replication-Competent Influenza Virus and Respiratory Syncytial Virus Luciferase Reporter Strains Engineered for Co-Infections Identify Antiviral Compounds in Combination Screens. Biochemistry. 54 (36), 5589-5604 (2015).
- Shaner, N. C., Patterson, G. H., Davidson, M. W. Advances in fluorescent protein technology. Journal of Cell Science. 120 (Pt 24), 4247-4260 (2007).
- Shaner, N. C., Steinbach, P. A., Tsien, R. Y. A guide to choosing fluorescent proteins. Nature Methods. 2 (12), 905-909 (2005).
- Kelkar, M., De, A. Bioluminescence based in vivo screening technologies. Current Opinion in Pharmacology. 12 (5), 592-600 (2012).
- Welsh, D. K., Noguchi, T. Cellular bioluminescence imaging. Cold Spring Harbor Protocols. 2012 (8), (2012).
- Zhao, H., et al. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. Journal of Biomedical Optics. 10 (4), 41210 (2005).
- Stacer, A. C., et al. NanoLuc reporter for dual luciferase imaging in living animals. Molecular Imaging. 12 (7), 1-13 (2013).
- Vintersten, K., et al. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 40 (4), 241-246 (2004).
- Stacer, A. C., et al. NanoLuc reporter for dual luciferase imaging in living animals. Mol Imaging. 12 (7), 1-13 (2013).
- Schoggins, J. W., et al. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proc Natl Acad Sci U S A. 109 (36), 14610-14615 (2012).
- Pan, W., et al. Visualizing influenza virus infection in living mice. Nat Commun. 4, 2369 (2013).
- Heaton, N. S., et al. In vivo bioluminescent imaging of influenza a virus infection and characterization of novel cross-protective monoclonal antibodies. J Virol. 87 (15), 8272-8281 (2013).
- Nogales, A., Baker, S. F., Martínez-Sobrido, L. Replication-competent influenza A viruses expressing a red fluorescent protein. Virology. 476, 206-216 (2015).
- Nogales, A., et al. A novel fluorescent and bioluminescent Bi-Reporter influenza A virus (BIRFLU) to evaluate viral infections. Journal of Virology. , (2019).
- Nogales, A., et al. Influenza A Virus Attenuation by Codon Deoptimization of the NS Gene for Vaccine Development. Journal of Virology. 88 (18), 10525-10540 (2014).
- Nogales, A., DeDiego, M. L., Topham, D. J., Martinez-Sobrido, L. Rearrangement of Influenza Virus Spliced Segments for the Development of Live-Attenuated Vaccines. Journal of Virology. 90 (14), 6291-6302 (2016).
- National Research Council (U.S.). Committee for the Update of the Guide for the Care and Use of Laboratory Animals., Institute for Laboratory Animal Research (U.S.) & National Academies Press (U.S.). . Guide for the care and use of laboratory animals. , (2011).
- Nogales, A., Martinez-Sobrido, L., Chiem, K., Topham, D. J., DeDiego, M. L. Functional Evolution of the 2009 Pandemic H1N1 Influenza Virus NS1 and PA in Humans. Journal of Virology. 92 (19), (2018).
- Hall, M. P., et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chemical Biology. 7 (11), 1848-1857 (2012).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved