Visualization of Candida albicans in the Murine Gastrointestinal Tract Using Fluorescent In Situ Hybridization
In This Article
Summary
The purpose of this protocol is to visualize Candida albicans cell shape and localization in the mammalian gastrointestinal tract.
Abstract
Candida albicans is a fungal component of the gut microbiota in humans and many other mammals. Although C. albicans does not cause symptoms in most colonized hosts, the commensal reservoir does serve as a repository for infectious disease, and the presence of high fungal titers in the gut is associated with inflammatory bowel disease. Here, we describe a method to visualize C. albicans cell morphology and localization in a mouse model of stable gastrointestinal colonization. Colonization is established using a single dose of C. albicans in animals that have been treated with oral antibiotics. Segments of gut tissue are fixed in a manner that preserves the architecture of luminal contents (microorganisms and mucus) as well as the host mucosa. Finally, fluorescent in situ hybridization is performed using probes against fungal rRNA to stain for C. albicans and hyphae. A key advantage of this protocol is that it allows for simultaneous observation of C. albicans cell morphology and its spatial association with host structures during gastrointestinal colonization.
Introduction
Candida albicans is a fungal commensal as well as an opportunistic human pathogen. This yeast lacks a defined environmental niche and instead propagates within the gastrointestinal (GI) tract, skin, and genitourinary tract of humans and other mammals1. Whereas early research on C. albicans focused primarily on its virulence potential, several recent reports suggest that commensally propagating organisms within the gut may play important roles in normal health, including the immune development of the host2,3,4. To facilitate investigations of C. albicans commensalism within the mammalian gut, we developed a mouse model of stable GI colonization and a fluorescence in situ hybridization (FISH)-based method to visualize fungal yeast cells and hyphae within the intestinal lumen.
With some exceptions5, laboratory-bred mice typically exhibit resistance to fungal colonization of the digestive tract. Colonization resistance is thought to be mediated by specific bacterial species; however, this can be overcome by treatment of the animals with antibiotics6,7 or use of a chemically-defined diet that presumably alters bacterial species composition8,9. Similarly, in humans, the use of broad-spectrum antibiotics has been associated with C. albicans overgrowth and dissemination10. Our murine colonization model uses broad-spectrum antibiotics to establish C. albicans colonization of immunocompetent, conventionally reared mice. Penicillin and streptomycin are provided in the animals’ drinking water for one week prior to gavage with 108 colony forming units (CFUs) of C. albicans. As long as the antibiotic-infused water is continued, C. albicans will propagate through the GI tract, reaching fecal titers of 106−108 CFUs/g. Despite the high level of fungal colonization, animals remain healthy and gain weight at the same rate as uninfected controls. This model has been used successfully to screen for and characterize multiple C. albicans commensalism factors11,12.
Like other members of the fungal kingdom, C. albicans is capable of enormous morphological plasticity13. Under in vitro conditions, it has been shown to transition among at least six unicellular yeast cell types, as well as multicellular hyphae and pseudohyphae. The yeast-to-hypha transition is one of its best-characterized virulence attributes, and hyphae and pseudohyphae predominate in most mammalian disease models, as well as in infected human tissues. To determine C. albicans localization and cell morphology within the murine digestive tract, we developed a FISH technique for staining yeasts and hyphae in fixed histological sections. The probes consist of fluorescently labeled DNA oligonucleotides that hybridize to fungal 23S ribosomal RNA (rRNA), which is distributed throughout the fungal cytoplasm. Because host tissues are fixed in a manner that preserves the three-dimensional architecture of the gut, including the host mucosa, digestive material, the bacterial microbiota, and mucus within the gut lumen, this technique permits the localization of fungal cells with respect to these landmarks when stained. The FISH technique compares favorably to traditional histological stains for fungi, such as Periodic Acid Schiff (PAS) or Gomori's Methenamine-Silver (GMS), as well as commercially available antifungal antibodies, because these reagents are not specific for C. albicans. Moreover, standard fixatives remove the mucus layer and disrupt other contents of the gut lumen14,15.
In this article, we provide detailed instructions for establishing high-grade C. albicans colonization of the mouse GI tract, for dissection of the digestive tract from euthanized animals, for tissue fixation in a manner that preserves luminal architecture, and for detection of C. albicans within host tissues using FISH. In addition to wild type and mutant strains of C. albicans, the gavage technique can be used to deliver other microorganisms. The fixation technique would be useful for any study in which preservation of gut contents is desired. The FISH procedure can be completed within a day and can be used to localize multiple fungal species by using multiple, differentially labeled probes.
Protocol
The steps described below have been approved by the UCSF Institutional Animal Care and Use Committee (IACUC).
1. Gastrointestinal Colonization of Mice with C. albicans Using Oral Gavage
- Provide housing for 18−21 g male or female mice (8−10 weeks old) as approved by the local IACUC. Use autoclaved food and water starting on the first day.
NOTE: The use of sterilized cages, bedding, food, and water reduces the risk of contamination with antibiotic-resistant environmental bacteria that can potentially displace C. albicans from the gut. Further, depending on the experimental question, individual housing of animals should be considered because mice are coprophagic and gut contents will be shared among cohoused animals. - The following day, replace the autoclaved drinking water with autoclaved water containing 5% glucose, 1,500 U/mL penicillin G, and 2 mg/mL streptomycin.
- For convenience, prepare 200x antibiotic stock solutions (Table 1) in advance, filter sterilize, and freeze at -20 °C.
- Maintain animals on antibiotics for one week.
- On days 3 and 6 after antibiotics have been started, assess each animal’s feces for contamination with antibiotic-resistant aerobic bacteria. Holding an animal with one hand, place an uncapped microfuge tube near the anus and collect at least one fecal pellet.
- Resuspend each fecal pellet in 1 mL of sterile water and plate 100 µL on Luria broth (LB), yeast extract peptone dextrose (YEPD), or brain heart infusion (BHI) plus blood agar plates. Incubate the plates overnight in a standard incubator (not anaerobic) at 37 °C and assess for bacterial growth.
NOTE: Plates should exhibit minimal growth (0−20 colonies). If >20 colonies of bacteria are recovered from animals treated with penicillin and streptomycin, then it is unlikely that high-level colonization with C. albicans will be established and the experiment should be aborted.
- Three days prior to gavage, streak C. albicans from frozen glycerol stocks to YEPD + 2% agar plates and incubate at 30 °C for 2 days.
- One day before gavage, inoculate one colony of each C. albicans strain to be tested into 5 mL of liquid YEPD. Incubate at 30 °C overnight with shaking.
- The morning of gavage, dilute the saturated culture of C. albicans to an optical density at 600 nm (OD600) of 0.1 in 100 mL of YEPD prewarmed to 30 °C. Shake in an orbital shaker at 200 rpm at 30 °C for between 4 h and 5 h until OD600 reaches 1 (mid-log growth).
- Transfer each 100 mL culture to two 50 mL conical tubes and centrifuge at 3,000 x g for 5 min at room temperature (RT).
NOTE: C. albicans propagates relatively slowly at RT. If adapting this protocol for another species, the inocula may need to be kept on ice to prevent continued growth. - Discard the supernatant, resuspend the pelleted cells in 20 mL of sterile saline per 50 mL of culture, and pool the duplicate resuspended pellets into single conical tubes. Centrifuge at 3,000 x g for 5 min.
- Discard the supernatant and resuspend in 20 mL of sterile saline. Vortex briefly and remove 20 µL for OD600 measurement. Dilute 1:50 in sterile normal saline. Centrifuge the remaining resuspended cells at 3,000 x g for 5 min.
- Discard the supernatant. For an inoculum size of 1 x 108 CFU, resuspend the pelleted cells to approximately 5 x 108 CFUs/mL in sterile normal saline based on the estimated concentration determined from OD600 measurement.
NOTE: Typically, an OD600 of 1 corresponds to ~1 x 107 yeast cells/mL. This value may vary by spectrophotometer and should be verified. If a change in concentration is desired for infections with a different microorganism, plan for an inoculum volume of ≤10 µL/g mouse to minimize discomfort to the animal. - Verify the concentration of the inoculum by counting a 1:1,000 dilution with a hemocytometer. Adjust the volume of cells to be gavaged based on the concentration determined using the hemocytometer.
- Using an animal feeding needle (Figure 1A), gavage each animal with the calculated volume of a given inoculum. See steps 1.13.1−1.13.8 for the gavage procedure.
NOTE: Training for gavage should be obtained from an experienced practitioner, and the procedure should be mastered in advance. A successful procedure for a single animal typically takes no longer than 1 min.- Attach an autoclaved animal-feeding needle to a 1 mL syringe and fill the syringe with one gavage volume, removing any air bubbles to avoid an inaccurate dose. Place on a sterile surface.
- Secure the animal for gavage. Hold the animal at the base of the tail with the dominant hand and slide nondominant hand up the animal’s back to the loose skin just behind the ears.
- Using the forefinger and thumb of the nondominant hand, gather the loose skin together tightly; this is the “scruff”. Pick up the animal by its scruff and loop small finger of the same (nondominant) hand around the tail to immobilize animal against the palm of the hand. Gently extend the animal’s head so that its head and body (and therefore neck and esophagus) are in a straight line.
NOTE: Attempting a gavage while the animal’s body is bent or twisted may result in complications such as esophageal perforation and death of the animal. - Pick up the syringe with the dominant hand and hold perpendicular to the animal, with the curved needle pointing down (Figure 1B). Using the ball of the needle, gently hook an inside corner of the mouth, gently advance the needle along the side of the mouth to the back of the throat, and finally move the needle to the center.
NOTE: Introducing the needle along a lateral path helps to avoid resistance from the animal’s teeth and tongue. - Once the ball of the needle is at the back of throat, tilt the needle up so that it is parallel with the animal’s body line (Figure 1C).
NOTE: At this point, the animal will exhibit a gag reflex. This is a good sign that indicates that the needle is positioned correctly, and the gagging motion helps to guide the needle down the esophagus. If there is no gag reflex, the needle may be in the trachea rather than the esophagus. Gently withdraw the needle and proceed back to step 1.13.1. - Allow the animal to swallow the inoculum until only a few millimeters remain visible (Figure 1D). Advance the needle smoothly.
NOTE: If the needle does not advance, avoid using force, which can damage the esophagus. Sometimes advancing the last half centimeter will require an extra-large gag from the animal. If the needle seems to be stuck, slowly withdraw it and try again from step 1.13.4. - Test that the ball of needle is in the correct location (stomach) by gently advancing the plunger. If there is no resistance, continue delivering the inoculum. Once the inoculum has been administered, slowly withdraw the needle.
- Replace the animal in its cage and continue to monitor the animal for 5−10 min for signs of labored breathing or distress. Verify that the animal resumes normal activity soon after gavage.
- If the same inoculum will be used with the next animal, clean the needle with an alcohol wipe. Proceed back to step 1.13.1. If a different inoculum will be used, use a new needle and proceed back to step 1.13.1.
NOTE: It is important to inspect animals the day after gavage. Animals exhibiting signs of distress (e.g., hunched posture, labored breathing) should be humanely euthanized, as they have likely suffered esophageal rupture, damage to the lungs, or another internal injury from which recovery is unlikely.
- Following gavage of the animals, plate dilutions of the inocula for dose verification.
- Prepare 10-fold serial dilutions up to 1:105. Plate one gavage volume of the 1:105 dilution onto a 100 mm plate of Sabouraud dextrose agar.
- Incubate the plate for 2 days at 30 °C. Count CFUs to confirm inoculum dose.
2. Preparation of Gastrointestinal Tissues for Histology
NOTE: This section of the protocol is adapted from Johansson and Hansson16.
- Prepare 500 mL of methacarn solution (60% methanol, 30% chloroform, 10% glacial acetic acid) in a large screw cap plastic jar for every 2 animals dissected.
CAUTION: Methanol and chloroform are harmful if inhaled and should be manipulated in a chemical hood. Glacial acetic acid can be corrosive and should be handled with the proper personal protective equipment. Methanol, chloroform, and glacial acetic acid should be discarded as hazardous waste. - At the experimental endpoint, euthanize animals humanely according to a protocol approved by the local IACUC.
NOTE: In antibiotic-treated animals, C. albicans colonization typically ranges from ~106 CFUs/g on day 5 up to ~5 x 107 CFUs/g by day 25. - Sterilize dissection tools with 10% bleach and rinse with 70% ethanol.
- Secure each animal to a dissection surface, with ventral side facing up. Use 30 G needles to secure the limbs to the dissection surface (Figure 2A). Spray the animal with 70% ethanol to clean the fur and minimize sticking to the tools.
- Using blunt-ended forceps, pinch a section of abdominal skin with nondominant hand. Using scissors with dominant hand, incise the skin and underlying fascia near the base of the pelvis. Extend the incision in a U shape along each side of the peritoneal cavity and up to the rib cage. Lift the skin out of the way.
- Using a pencil, prelabel histology cassettes for the GI segments to be assessed. For example, for each animal, label separate cassettes as “stomach,” “proximal small intestines,” “mid small intestines,” “distal small intestines,” “cecum,” and “large intestines.” Place a foam pad on the bottom of each cassette (Figure 2A).
NOTE: Figure 2B outlines suggested sections to place in cassettes. - Using blunt-ended forceps, gently extract the cecum from the peritoneal cavity. Use scissors to sever connections between the cecum and the small and large intestines.
NOTE: Cecal tissue is very delicate and easy to rupture. Take care when manipulating. - Place the entire cecum into a histology cassette so that it is untwisted and lays flat (Figure 2C). Place a second foam pad on top and close the cassette. Place the cassette into a screw cap jar containing methacarn.
- Excise a section of the large intestines that contains 1−2 fecal pellets, with a length less than that of the cassette.
- Place the tissue section into the cassette, keeping the tissue flat and untwisted. Overlay with a second foam pad and close the cassette. Place the cassette into methacarn.
- For each section of the small intestines to be sampled, excise a 1−2 cm segment that contains some digestive material.
- Place the segment into a histology cassette, keeping the tissue flat and untwisted. Overlay with a second foam pad and close the cassette. Place the cassette into methacarn.
- For the stomach, sever the connections to the esophagus and small intestines. Place in a cassette, keeping the tissue flat and untwisted. Overlay with a second foam pad and close the cassette. Place the cassette into methacarn.
- Leave the cassettes in methacarn solution at RT for at least 3 h and less than 2 weeks.
NOTE: There are several points during which the fixed GI segments can be transferred to a commercial histology service. These tissues can be difficult to section without disruption of luminal contents, and optimal results will be obtained by an experienced technician. If the commercial service is willing to perform washes prior to embedding the tissues in paraffin, the cassettes can be sent at this stage along with instructions for step 2.16. - Begin melting enough paraffin wax to cover all tissue cassettes in a large beaker in a preheated 70 °C hybridization oven.
- Following fixation, remove the foam pads and return each intact tissue section back into its cassette. Wash the tissues with the following solutions at RT with shaking: twice for 35 min in 100% methanol, twice for 25 min in 100% ethanol, twice for 20 min in xylene.
CAUTION: Xylene is flammable, toxic, and can cause damage if inhaled. Use in a chemical hood with proper protection. Dispose of xylene via hazardous waste procedures. - Pat the cassettes dry on a paper towel and place in premelted paraffin wax for 2 h at 70 °C in a hybridization oven. Ensure that the cassettes are filled with paraffin and no bubbles remain.
NOTE: This step allows the wax to infiltrate the GI segment and to stabilize the tissue and contents. - Remove the cassettes from wax, allow the excess wax to drain, and store at RT until they are embedded in wax blocks. Discard the wax containing trace amounts of xylenes as hazardous waste.
NOTE: The Noble Lab uses a commercial histology service for embedding and sectioning of tissues. For standard visualization of C. albicans yeasts and hyphae, 4 µm sections are used. To evaluate connectivity of hyphal segments, which typically extend through multiple planes, 8 µm sections can be used. Depending on the ability of FISH probes to penetrate into the specimen, it may be possible to use even thicker sections.
3. Validation of Newly Designed Probes
NOTE: The following protocol for validating C. albicans probes was adapted from Swidsinski et al.17.
- Streak C. albicans SC5314 and closely related comparator species (e.g., Candida dubliniensis, Candida tropicalis) to YEPD + 2% agar plates. Incubate for 1−2 days at 30 °C.
- Inoculate 5 mL of liquid YEPD medium with a single colony.
- Pipette 1 mL of the suspended cells to a microcentrifuge tube and centrifuge at 3,000 x g for 5 min. Discard supernatant.
- Resuspend the pellet in 100 µL of phosphate-buffered saline (PBS).
- Pipette 5−10 µL of resuspended cells and spot onto a glass slide. Spread into a larger circle and allow to dry.
NOTE: If cells are not adherent, use poly-L-lysine coated slides. - Circle the cell spot with a PAP pen and cover with 50 µL of 2.5% paraformaldehyde (PFA) in PBS. Incubate at RT for 10 min.
CAUTION: PFA is flammable and can cause health problems if inhaled or in contact with skin. Items contaminated with PFA should be discarded as hazardous waste. - Wash 3x with PBS. Tap liquid off onto paper towel and cover with 100 µL of PBS. Repeat twice. Discard final wash.
- Prepare slides for multiday storage. If performing FISH on the same day, proceed to step 3.9 without the steps in this section.
NOTE: It is preferable to perform the hybridization immediately but if short on time follow step 3.8 before storing. Slides can be stored for several days at 4 °C.- Cover the spot with 60% ethanol. Incubate at RT for 3 min. Discard the solution.
- Cover the spot with 80% ethanol. Incubate at RT for 3 min. Discard the solution.
- Cover the spot with 100% ethanol. Incubate at RT for 3 min. Discard the solution. Proceed to step 3.9.
- Place the slides uncovered in a hybridization oven at 50 °C for 1 h. If step 3.8 was followed, store the slides at 4 °C. If not, proceed to step 4.2 to perform the hybridization protocol.
4. FISH Sstaining in GI Tract Tissues
- Dewax paraffin-embedded histological sections.
- In a hybridization oven, prewarm a Coplin jar filled with xylene to 60 °C.
- Insert the slides into the xylene-filled jar. Place at 60 °C for 10 min. Discard the xylene wash. Keep the slides in the jar and pour off the xylene into a waste container.
- Pour fresh RT xylene into the jar and incubate at 60 °C for 10 min. Discard the second xylene wash.
- Fill the jar with 100% ethanol and incubate at RT for 5 min. Remove the slides from the jar and air dry.
- Set the hybridization oven to 50 °C for steps in section 4.2.
- Stain C. albicans with the FISH probe.
- Prepare 1 mL of fresh hybridization solution (20 mM Tris–HCl, pH 7.4, 0.9 M NaCl, 0.1% sodium dodecyl sulfate [SDS], 1% formamide in RNase-free water). See Table 1 for sample calculations.
- Mix together 50 µL of hybridization solution with 1 µL of C. albicans probe (5’ Cy3- ACAGCAGAAGCCGTGCC 3’; 50 µg/mL in RNase-free water) for a final probe amount of 0.5 µg per slide.
NOTE: Many laboratory-bred mice do not contain any fungi among their microbiota. In this case, it is possible to substitute a pan-fungal probe (5’ Cy3-CTCTGGCTTCACCCTATTC 3’) that recognizes 23S rRNA of most fungal species, not just C. albicans. In the authors’ experience, the panfungal probe yields brighter staining than the C. albicans-specific probe. - Protect the solution from light and prewarm to 50 °C.
- Using a PAP pen, draw a circle around tissue section from section 4.1.
- Pipette 50 μL of probe-containing hybridization solution onto the fixed tissue and gently spread over the tissue section with a pipette tip. Overlay the liquid with a hybridization coverslip making sure to prevent bubbles.
- Seal slides in a watertight hybridization chamber and incubate the sections for 3 h at 50 °C in the dark in a hybridization oven.
- Meanwhile, prepare 50 mL of FISH washing solution (20 mM Tris-HCl, pH 7.4, 0.9 M NaCl in RNase-free water). See Table 1 for sample calculations.
- Move the washing solution to 50 °C to prewarm.
- After 3 h, add the washing solution to a Coplin jar. Then, carefully remove the coverslip from the slide with forceps and place the slide in the washing solution.
- Cover the jar with aluminum foil and incubate at 50 °C for 20 min.
- During this incubation, prepare mucin-nuclei staining solution (20 ng/mL 4′,6-diamidino-2-phenylindole [DAPI], 1.6 µg/mL fluorescein isothiocyanate [FITC]-lectin in PBS). See Table 1 for sample calculations. For stomach and large intestines, use FITC-Ulex europaeus agglutinin 1 (UEA-1). For small intestines and cecum, use a combination of FITC-UEA-1 and FITC-wheat germ agglutinin (WGA).
NOTE: Lectins from different species recognize different sugar modifications of glycoproteins such as mucin. The lectin combinations recommended here were determined empirically for optimal staining of mucus in different GI compartments. - Wash the slides by pouring off the FISH washing solution and refilling the jar with RT PBS. Pour off the PBS immediately and refill with PBS. Pour off the PBS for a total of 2 washes.
- Wick away the PBS with a delicate task wiper tissue and circle the intestinal tissue section with a PAP pen. Place the slides in an opaque plastic container and add 50 µL of mucin-nuclei staining solution to the tissue section. Cover the container with aluminum foil to protect from light. Incubate at 4 °C for 45 min.
- Place the slides in a Coplin jar and wash quickly twice in PBS at RT.
- Tap away the excess PBS on a paper towel, and then wick away the final drops of PBS with a tissue.
- Place one drop of mounting medium on each section and overlay with a glass coverslip, preventing bubbles. Let the mounting medium spread under the entire coverslip. For immediate imaging, anchor the coverslip in place with nail polish at the corners. For long term storage, seal around the coverslip with nail polish.
- Image the slides using a fluorescent microscope.
Representative Results
Following the instructions provided, the outcome of this technique will be fluorescent labelling of nuclei in the host epithelium, mucus, and C. albicans in sectioned GI tissues. Wild type Candida albicans cells should appear as either round, unicellular yeast cells or highly elongated, sometimes branching, multicellular hyphae (cell-cell divisions will not be apparent in the hyphae). Mutants of C. albicans may additionally appear as smaller, slightly elongated “gray” yeasts or as larger, slightly elongated “GUT” (gastrointestinal-induced transition) or “opaque” yeasts.
Figure 1 depicts the feeding needle used for oral gavage, as well as key positions of the needle during the gavage procedure. Figure 2 provides a typical setup used for organ dissection and the appearance of GI segments of the mouse.
Figure 3 displays FISH staining of different C. albicans cell types following propagation in vitro and within the mouse model. Figure 3A shows fluorescence and phase images of fixed, permeabilized, in vitro-propagated hyphae. Hypha formation was induced with liquid Lee’s medium18 with 2% N-acetylglucosamine, pH 6.8 at 37 °C. Figure 3B shows fluorescence and phase images of fixed, permeabilized, in vitro-propagated yeast cells. Figure 3C,D shows fixed, permeabilized, in vitro-propagated GUT cells and opaque cells, respectively. GUT and opaque cell types are morphologically indistinguishable except when visualized by scanning electron microscopy. Please note that FISH staining of in vitro-propagated cells will be suboptimal if the cells are not well permeabilized. An example of weak and diffuse staining is shown in Figure 3C. For best results, cell fixation and permeabilization conditions should be determined empirically for each cell type and species to be investigated. Fortunately, the recommended protocol for visualizing C. albicans in sectioned tissues produces more consistent permeabilization.
Figure 3E-G depict C. albicans in mouse large intestines, stained with the C. albicans-specific probe (Figure 3E) or the panfungal probe (Figure 3F,G). C. albicans appears red in these images, host cell nuclei are blue, and the mucus layer is green. Both round yeasts and highly elongated hyphae (white arrowheads) occur in the large intestines. Please note that staining is brighter with the panfungal probe. Figure 3G depicts a ume6 mutant in the same compartment, stained with the panfungal probe. Ume6 encodes a transcription factor that is required for hypha formation under in vitro conditions19. Interestingly, the yeast-locked phenotype displayed by this mutant under in vitro conditions is not recapitulated within the gut, suggesting that a redundant factor must be activated within the host12. Figure 4 depicts the appearance of wild type C. albicans in different segments of the murine GI tract, including the regions immediately adjacent to the host mucosa and more central regions of the gut lumen.
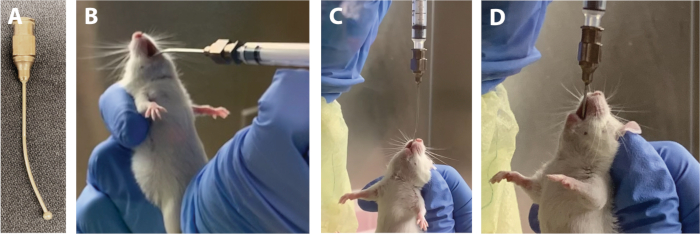
Figure 1: Key feeding needle positions during gavage. (A) Feeding needle. (B) Feeding needle ball inserted to the side of the tongue. (C) Feeding needle in the upright position with insertion in the esophagus. (D) Feeding needle inserted into the stomach with a few millimeters visible above the nose. Please click here to view a larger version of this figure.

Figure 2: Example dissection layout. (A) Dissection pad and tools. (B) Excised GI tract. Proximal (esophagus) to distal (anus) sections: I. Stomach. II. Proximal small intestines. III. Medial small intestines. IV. Distal small intestines. V. Cecum. VI. Large intestine. Pink dashed border boxes indicate the tissues to be fixed. (C) Tissues positioned in cassettes. Roman numerals are the same as in panel B. Please click here to view a larger version of this figure.
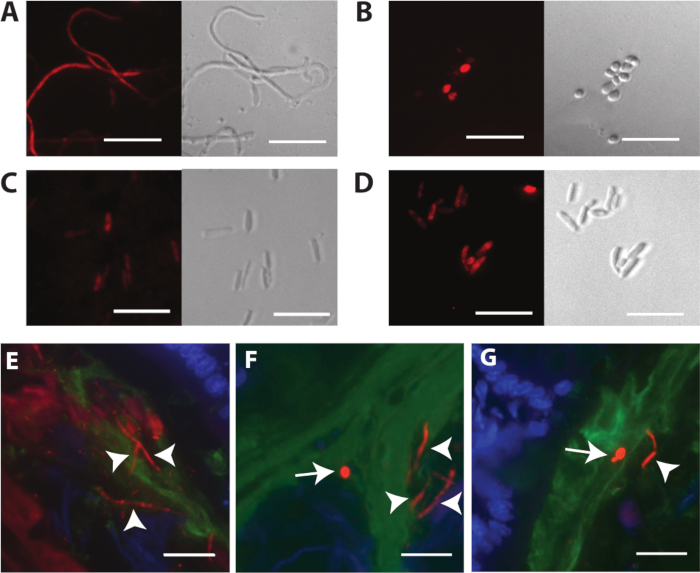
Figure 3: Representative images of FISH-stained C. albicans. (A) FISH of C. albicans hyphae grown in liquid Lee’s media18 with 2% N-acetylglucosamine, pH 6.8. (B) FISH of C. albicans round yeast cells grown on YEPD + 2% agar plates. (C) FISH of C. albicans GUT cells (ySN1045) grown on YEPD + 2% agar plates. (D) FISH of C. albicans opaque cells grown on YEPD + 2% agar plates. (E) Wild type C. albicans (ySN250) in large intestines, stained with the C. albicans-specific probe. (F) Wild type C. albicans in large intestines, stained with the panfungal probe. (G) Ume6 mutant (ySN1479) in large intestines, stained with the panfungal probe: red is C. albicans, blue is host epithelial nuclei, and green is mucin. Arrowheads indicate hyphae. Arrows indicate yeast. Scale bars = 20 µm. Images F and G are adapted from Witchley et al.12. Please click here to view a larger version of this figure.
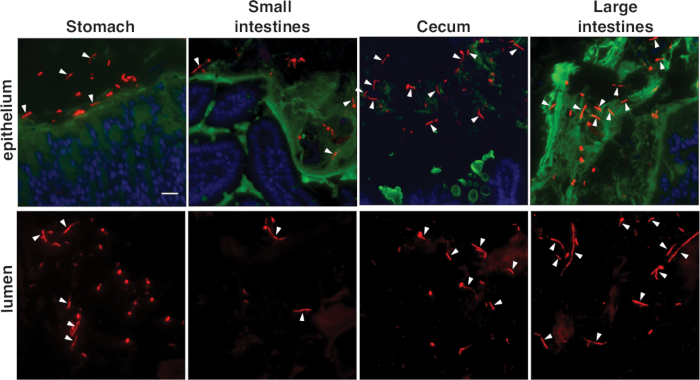
Figure 4: Appearance of FISH-stained C. albicans in different gut compartments. Wild type C. albicans is shown in the indicated segments of the mouse GI tract. Within each compartment, images depict the area adjacent to the host mucosa and the central region of the gut lumen. Staining was performed with the panfungal probe. Scale bar = 20 µm. Images are adapted from Witchley et al.12. Please click here to view a larger version of this figure.
| Antibiotic stocks (200x) | |||
| Penicillin G | 181 mg/mL | ||
| Streptomycin | 400 mg/µL | ||
| Methacarn | |||
| Stock | Final concentration | Volume | Units |
| Methanol | 60% | 300 | mL |
| Chloroform | 30% | 150 | mL |
| Glacial acetic acid | 10% | 50 | mL |
| Hybridization solution | |||
| Stock | Final concentration | Volume | Units |
| 1 M Tris-HCl, pH 7.4 | 20 mM | 20 | µL |
| 5 M NaCl | 0.9 M | 180 | µL |
| 10% SDS | 0.10% | 10 | µL |
| 100% formamide | 1.00% | 10 | µL (kept in small aliquots at -20 °C between uses) |
| RNase-free water | 780 | µL | |
| FISH washing solution | |||
| 1 M Tris-HCl, pH 7.4 | 20 mM | 1 | mL |
| 5 M NaCl | 0.9 M | 9 | mL |
| RNase-free water | 40 | mL | |
| Mucin-nuclei staining solution | |||
| DAPI, 10 µg/mL | 20 ng/mL | 2 | µL |
| lectin, 40 µg/mL | 1.6 µg/mL | 40 | µL |
| PBS, pH 7.4 | 958 | µL | |
Table 1: Typical volumes and concentrations of solutions used in this protocol.
Discussion
The method described here allows for visualization of C. albicans yeasts and hyphae in the GI tracts of commensally colonized mice of any sex or strain. The FISH probe hybridizes to 23S rRNA, which is distributed throughout the fungal cytoplasm. Our method was adapted from a previously reported protocol for visualizing gut bacteria20. Because C. albicans changes its morphology within the host, the method is useful to monitor fungal cell shape as well as localization. For example, we have used this method to disprove the hypothesis that yeasts predominate throughout the digestive tract, and to expose discrepancies between the in vivo and in vitro phenotypes of certain “filamentation-defective” mutants12.
Several mouse models exist for C. albicans commensal colonization. The microbiota of laboratory mice and humans are distinct and, in mice, the use of antibiotics or a specialized diet is required to establish stable colonization. Treatment with antibiotics also enhances C. albicans colonization of humans and is a major risk factor for disseminated disease10. The antibiotics used in this study are relatively inexpensive and yield reliable decreases in the bacterial microbiota. Note that antibiotics are used to decrease the burden of antagonistic bacterial species, not to eliminate all bacteria from the animals. If researchers wish to study C. albicans-host interactions in the absence of bacteria or to avoid the use of antibiotics or a special diet, germ-free animals can be substituted for conventionally reared animals; however, gnotobiotic mice exhibit certain immune and anatomic abnormalities and therefore may not be suitable for all purposes.
Several steps are important for successful FISH staining: The recommended fixation method is critical to preserve the structural integrity of GI tissues, particularly the fragile contents of the gut lumen, such as the mucus layer and the three-dimensional organization of fungi and bacteria16. Please note that many commonly used fixation solutions that contain water are highly damaging to the luminal architecture. The fixatives and solutions for post-fixation washes recommended in this protocol do not contain water, and it is important to avoid contamination with water. Another critical step is the hybridization step, where it is important to avoid evaporation of the hybridization solution during incubation of slides in the hybridization oven. We suggest placing the slides in a watertight container to avoid this problem. Alternatively, one can use watertight hybridization chambers such as those originally used for hybridization of microarrays.
A C. albicans-specific FISH probe is described in this protocol. However, because many laboratory-reared mice do not contain C. albicans or other fungi as part of their natural gut microbiota, a panfungal probe may specifically stain C. albicans in experimentally colonized animals. Substitution of a panfungal probe may be desirable because of its superior hybridization characteristics and higher signal-to-noise ratio. If the panfungal probe is used as a pseudospecific probe for C. albicans, it is important to document a lack of staining in uninfected animals (i.e., treated with antibiotics but not C. albicans). Staining of a noncolonized control animal is also useful to assess background staining that may result from adherence of FISH probes to food particulates. Overall, the use of FISH probes offers enhanced specificity over most other methods of staining fungi, such as Calcofluor white (which stains chitin, a cell wall component that may vary between cell types), GMS, or commercially available antifungal antibodies. Moreover, FISH staining allows for costaining of multiple organisms using differentially labeled FISH probes to species-specific rRNAs.
One caveat of this technique is that certain C. albicans yeast cell types appear very similar by FISH (for example, the opaque and GUT cell types). To distinguish among these cell types, it would be useful to develop hybridization probes to cell type-specific fungal mRNAs. Nevertheless, in its current form, the FISH technique has already revealed surprising discrepancies between the in vivo and in vitro behaviors of C. albicans mutants evaluated under both conditions12, indicating complex interactions in the natural commensal environment that are not adequately mimicked by existing in vitro assays. Further studies of different C. albicans stains in additional wild type and mutant hosts are likely to yield additional insights into fungal-host interactions. In particular, it will be instructive to profile C. albicans in models of inflammatory bowel disease, which is associated with high titers of C. albicans in humans21, and other models of C. albicans overgrowth and disease. The FISH technique may also be combined with immunohistochemistry to stain specific host cells. Overall, the method presented here provides a reasonably quick and reliable means to determine C. albicans localization and morphology within the mammalian GI tract.
Acknowledgements
The authors would like to thank Carolina Tropini, Katharine Ng, Justin Sonnenburg, and KC Huang for guidance in developing the FISH technique. Teresa O’Meara provided helpful comments on the manuscript, and Miriam Levy assisted with photography. This work was supported by NIH grants R01AI108992, R01DK113788, and a Burroughs Welcome Award in the Pathogenesis of Infectious Disease.
Materials
| Name | Company | Catalog Number | Comments |
| 1 mL syringe | BD | 309659 | can be substituted from any vendor |
| BHI blood agar | can be substituted from any vendor | ||
| C. albicans FISH probe | IDT DNA Technologies | custom order | |
| chamber for hybridization incubation | watertight chamber meant to reduce evaporation | ||
| chloroform | Sigma | C2432 | >=99.5% |
| DAPI | Roche | 10236276001 | can be substituted from any vendor |
| delicate task wiper tissues | Kimberley-Clark | 34256CT | Kimwipes |
| D-glucose | Sigma | G7021-5KG | can be substituted from any vendor |
| ethanol | Sigma | E7023 | molecular biology grade |
| feeding needles | Cadence, Inc. | 7910 | metal; 20 X1-1/2" W/2-1/4; can be autoclaved to sterilize |
| FITC-UEA-1 | Sigma | L9006-1MG | other fluorophores available |
| FITC-WGA | Sigma | L4895-2MG | other fluorophores available |
| Foam pads | Fisher | 22038221 | Order foam pads that will fit within cassettes |
| formamide | Sigma | 47671 | molecular biology grade |
| glacial acetic acid | Macron Fine Chemicals | MK881746 | ACS reagent, >=99.5% |
| glass Coplin jar | Fisher | 08-815 | hold up to 10 slides back to back |
| Histology cassettes | Simport | M512 | Deep cassettes so cecum is not squished |
| hybridization coverslips | Sigma | GBL712222 | RNase-free |
| hybridization oven | can be substituted from any vendor | ||
| LB | can be substituted from any vendor | ||
| Lee's media | prepared as described in Lee et al. 1975 | ||
| methanol | Sigma | 179337 | ACS reagent, >=99.8% |
| mice | Charles River Laboratories | 028 | adult BALB/c; 18-21 grams (8-10 weeks) |
| paraformaldehyde | Fisher | 50-980-487 | 16% solution |
| parrafin wax | Sigma | P3558-1KG | Paraplast for tissue embedding |
| PBS, pH 7.4 | UCSF Cell Culture Facility Media Production Unit | CCFAL003 | calcium, magnesium-free; can be substituted from any vendor |
| penicillin G | Sigma | PENNA-100MU | |
| Sabouraud dextrose agar | can be substituted from any vendor | ||
| saline | Baxter | 2F7123 | sterile, can be substituted from any vendor |
| sodium chloride (NaCl) | Sigma | S3014 | can be substituted from any vendor; maintain RNase-free |
| sodium dodecyl sulfate (SDS) | Sigma | L3771 | can be substituted from any vendor; maintain RNase-free |
| streptomycin | Sigma | S9137-100G | |
| super PAP pen | Life Technologies | 8899 | can be substituted from any vendor |
| Tris-HCl | Sigma | T3253 | can be substituted from any vendor; maintain RNase-free |
| Vectashield | Vector Laboratories | H-1000 | does not contain DAPI |
| xylenes | Sigma | 214736 | reagent grade |
| YEPD | can be substituted from any vendor |
References
- Noble, S. M., Gianetti, B. A., Witchley, J. N. Candida Albicans Cell-Type Switching and Functional Plasticity in the Mammalian Host. Nature Reviews in Microbiology. 15 (2), 96-108 (2017).
- Bacher, P., et al. Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans. Cell. 176 (6), 1340-1355 (2019).
- Shao, T. Y., et al. Commensal Candida albicans Positively Calibrates Systemic Th17 Immunological Responses. Cell Host and Microbe. 25 (3), 404-417 (2019).
- Jiang, T. T., et al. Commensal Fungi Recapitulate the Protective Benefits of Intestinal Bacteria. Cell Host and Microbe. 22 (6), 809-816 (2017).
- Iliev, I. D., et al. Interactions Between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science. 336 (6086), 1314-1317 (2012).
- Fan, D., et al. Activation of HIF-1 alpha and LL-37 by Commensal Bacteria Inhibits Candida Albicans Colonization. Nature Medicine. 21 (7), 808-814 (2015).
- Koh, A. Y., Kohler, J. R., Coggshall, K. T., Van Rooijen, N., Pier, G. B. Mucosal Damage and Neutropenia Are Required for Candida Albicans Dissemination. PLoS Pathogens. 4 (2), e35 (2008).
- Yamaguchi, N., et al. Gastric Colonization of Candida Albicans Differs in Mice Fed Commercial and Purified Diets. Journal of Nutrition. 135 (1), 109-115 (2005).
- Kadosh, D., et al. Effect of Antifungal Treatment in a Diet-Based Murine Model of Disseminated Candidiasis Acquired via the Gastrointestinal Tract. Antimicrobial Agents and Chemotherapy. 60 (11), 6703-6708 (2016).
- Perlroth, J., Choi, B., Spellberg, B. Nosocomial Fungal Infections: Epidemiology, Diagnosis, and Treatment. Medical Mycology. 45 (4), 321-346 (2007).
- Pande, K., Chen, C., Noble, S. M. Passage Through the Mammalian Gut Triggers a Phenotypic Switch That Promotes Candida Albicans Commensalism. Nature Genetics. 45 (9), 1088-1091 (2013).
- Witchley, J. N., et al. Candida Albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection. Cell Host and Microbe. 25 (3), 432-443 (2019).
- Noble, S. M., Gianetti, B. A., Witchley, J. N. Candida Albicans Cell-Type Switching and Functional Plasticity in the Mammalian Host. Nature Reviews in Microbiology. 15 (2), 96-108 (2017).
- Balish, E., Filutowicz, H., Oberley, T. D. Correlates of Cell-Mediated Immunity in Candida Albicans-Colonized Gnotobiotic Mice. Infection and Immunity. 58 (1), 107-113 (1990).
- Guarner, J., Brandt, M. E. Histopathologic Diagnosis of Fungal Infections in the 21st Century. Clinical Microbiology Reviews. 24 (2), (2011).
- Johansson, M. E., Hansson, G. C. Preservation of Mucus in Histological Sections, Immunostaining of Mucins in Fixed Tissue, and Localization of Bacteria with FISH. Methods in Molecular Biology. 842, 229-235 (2012).
- Swidsinski, A., Weber, J., Loening-Baucke, V., Hale, L. P., Lochs, H. Spatial Organization and Composition of the Mucosal Flora in Patients with Inflammatory Bowel Disease. Journal of Clinical Microbiology. 43 (7), 3380-3389 (2005).
- Lee, K. L., Buckley, H. R., Campbell, C. C. An Amino Acid Liquid Synthetic Medium for the Development of Mycelial and Yeast Forms of Candida Albicans. Sabouraudia. 13 (2), 148-153 (1975).
- Banerjee, M., et al. UME6, a Novel Filament-Specific Regulator of Candida Albicans Hyphal Extension and Virulence. Molecular Biology of the Cell. 19 (4), 1354-1365 (2008).
- Earle, K. A., et al. Quantitative Imaging of Gut Microbiota Spatial Organization. Cell Host and Microbe. 18 (4), 478-488 (2015).
- Sokol, H., et al. Fungal Microbiota Dysbiosis in IBD. Gut. 66 (6), 1039-1048 (2017).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved