Detection of Tissue-resident Bacteria in Bladder Biopsies by 16S rRNA Fluorescence In Situ Hybridization
In This Article
Summary
This protocol is for the unbiased detection of tissue-associated bacteria in patient biopsies by 16S rRNA in situ hybridization and confocal microscopy.
Abstract
Visualization of the interaction of bacteria with host mucosal surfaces and tissues can provide valuable insight into mechanisms of pathogenesis. While visualization of bacterial pathogens in animal models of infection can rely on bacterial strains engineered to express fluorescent proteins such as GFP, visualization of bacteria within the mucosa of biopsies or tissue obtained from human patients requires an unbiased method. Here, we describe an efficient method for the detection of tissue-associated bacteria in human biopsy sections. This method utilizes fluorescent in situ hybridization (FISH) with a fluorescently labeled universal oligonucleotide probe for 16S rRNA to label tissue-associated bacteria within bladder biopsy sections acquired from patients suffering from recurrent urinary tract infection. Through use of a universal 16S rRNA probe, bacteria can be detected without prior knowledge of species, genera, or biochemical characteristics, such as lipopolysaccharide (LPS), that would be required for detection by immunofluorescence experiments. We describe a complete protocol for 16S rRNA FISH from biopsy fixation to imaging by confocal microscopy. This protocol can be adapted for use in almost any type of tissue and represents a powerful tool for the unbiased visualization of clinically-relevant bacterial-host interactions in patient tissue. Furthermore, using species or genera-specific probes, this protocol can be adapted for the detection of specific bacterial pathogens within patient tissue.
Introduction
The urinary tract, consisting of the urethra, bladder, ureters and kidneys is constantly exposed to bacteria that comprise the urinary microbiome as well as invading uropathogens, like uropathogenic E. coli (UPEC), from the gastrointestinal tract1,2. A layer of hydrated mucus consisting of glycosaminoglycans and an impermeable plaque of glycosylated uroplakin proteins expressed on the surface of the superficial cells form a barrier that routinely protects the bladder epithelium from invasion by adherent bacteria3,4. During urinary tract infection (UTI), these barriers are disturbed or destroyed, facilitating attachment to and invasion of the bladder epithelium by uropathogenic bacteria5,6. Work in murine models has revealed that many uropathogenic bacteria including UPEC, Klebsiella pneumoniae, and Enterococcus faecalis can form replicative intracellular communities (IBCs) within the cytoplasm of superficial cells and quiescent intracellular reservoirs (QIRs) within transitional epithelial cells7,8,9. Although UPEC has been identified within shed epithelial cells from human UTI patients, the interaction of uropathogens with the bladder mucosa in humans had not been previously visualized10.
We adapted a common technique, fluorescence in situ hybridization (FISH), to detect bacteria within the mucosa of bladder biopsies obtained from postmenopausal patients undergoing cystoscopy with electrofulguration of trigonitis (CEFT) for the advanced management of antibiotic-refractory recurrent UTI11. Using a universal probe for 16S rRNA, we were able to objectively detect bacterial species associated with the bladder mucosa of recurrent UTI patients and determine their position within the bladder wall12. The universal 16S rRNA nucleotide probe was previously designed to target a conserved region of the bacterial 16S rRNA13, which corresponds to positions 388-355 of the E. coli 16s rRNA. The 16S rRNA and scramble probe sequences have been previously validated and published for use in the mouse gastrointestinal tract14,15. The sequences and properties of the probes are described in Table 1. It is essential to use two sequential sections in this protocol, one for the 16S rRNA probe and one for the scramble probe, to be able to distinguish between true and background signal as the bladder epithelium, collagen and elastin exhibit autofluorescence16. In this protocol, the 16S rRNA and scramble probes were designed with fluorescent Alexa Fluor 488 labels on both the 3' and 5' termini via N-hydroxysuccinimide (NHS) ester linkages to increase fluorescent signal.
Although this protocol was developed for use on human bladder biopsy sections, it can readily be adapted for use on paraffin-embedded sections from any tissue where bacteria are believed to reside. Unlike immunohistochemistry experiments that target specific antigens (e.g., lipopolysaccharide) on the bacterial surface, this method requires no prior knowledge of antigens expressed by the tissue-associated bacteria10,17. Use of the universal 16S rRNA probe allows the unbiased detection of all bacterial species within the sample but does not allow determination of their identity. To determine the identify of detected bacteria, species or genus-specific 16S or 23S rRNA probes must be used. This protocol will also not detect fungal pathogens, such as Candida albicans, associated with host tissue. For detection of fungal pathogens, 28S or 18S rRNA probes must be used18.
Protocol
The study protocol was approved by and followed the guidelines of the Institutional Biosafety and Chemical Safety Committees of UT Dallas and UT Southwestern Medical Center. The use of biopsies from human subjects in this protocol was approved by and followed the guidelines of the Institutional Review Boards of the UT Dallas and the UT Southwestern Medical Center. All individuals involved with biopsy collection and processing have current human subject protection (HSP) and HIPPA training.
1. Tissue Preparation for Fixation and Paraffin Embedding
NOTE: Biopsies were taken from consenting women undergoing cystoscopy with electro-fulguration of trigonitis for the advanced management of recurrent urinary tract infection (rUTI). rUTI is defined as 3 UTIs in a 12-month period. Biopsy collection was performed in the operating room while the patient was under anesthesia after obtaining informed patient consent per UTSW IRB protocol STU 082010-016. All samples were coded and de-identified before experimentation.
- Obtain a cold-cup (~1 mm3) biopsy from the bladder trigone with urologic forceps via flexible cystoscope and place immediately into a sterile 2 mL cryovial containing 1.5 mL of 4% v/v paraformaldehyde (PFA) prepared in 1x sterile phosphate buffered saline (PBS).
NOTE: 4% PFA in 1x PBS can be made in advance and stored at -20 °C until needed. - Fix biopsy for 6 h at room temperature or for 16-24 h at 4 °C.
NOTE: Fixation duration should be calculated based on the size of the tissue sample and over-fixation should be avoided. It is not advised to use glutaraldehyde as a fixative as it introduces autofluorescence. - In a sterilized hood or biosafety cabinet, remove fixative by pipetting and replace with 1.5 mL of sterile 1x PBS. Keep the samples at 4 °C overnight or immediately perform tissue processing and paraffin embedding19.
- Use a sterilized microtome to section tissue blocks at 5 μm thickness and adhere paraffin tissue sections to charged glass microscope slides. A minimum of two slides per biopsy will be required for the 16S rRNA FISH protocol.
NOTE: The biopsy tissue should be cross sectionally arranged relative to the cutting plane in order to ensure visualization of urothelial layers in all sections. It should also be noted that section thickness should be optimized for bacterial community detection. Thinner sections or sampling of multiple serial sections may be required in the case of infections where tissue-resident bacteria are extremely scarce (e.g., <1 tissue-resident bacterium per 1,000 mammalian cells).
2. Fluorescence In Situ Hybridization with Universal 16S rRNA Probes
NOTE: Two slides per biopsy are required. One slide is needed for the universal 16S rRNA probe and one slide for a control probe with a scrambled sequence. This is important for distinguishing true signal from background signal during microscopy since the bladder epithelium is auto-fluorescent in multiple channels. In addition to the scramble probe, blocking with 0.1% Sudan Black B prior to mounting may reduce background autofluorescence inherent in the tissue20.
- Preparation of reagents and fume hood
- Clean an empty fume hood (or appropriately fitted biosafety cabinet) with 70% ethanol.
- Prepare the hybridization buffer comprised of 0.9 M sodium chloride (NaCl), 20 mM Tris-HCl (pH 7.2), 0.1% sodium dodecyl sulfate (SDS) in 10 mL of sterile-filtered water.
NOTE: Sterile-filtered water may be prepared by passing autoclaved distilled water through a 0.22 μM filter. The hybridization buffer can be stored at room temperature, but the SDS may precipitate from solution. If SDS precipitates, warm the solution in a 50 °C water bath prior to use. - Prepare at least 100 mL each of 95% and 90% ethanol in sterile-filtered water in washed, autoclaved bottles.
- Dissolve fluorescently labeled lyophilized probes in 10 mM Tris-HCl (pH 8.0) and 1 mM ethylenediaminetetraacetic acid (EDTA) buffer (TE) prepared in filter-sterilized nuclease free water to a final concentration of 100 μM. Prepare a dilution of 1 μM in TE buffer for use in this protocol. Store both 100 μM concentrated stock and 1 μM stock reconstituted probes at -20 °C protected from light.
NOTE: Do not dissolve fluorescently labeled probes in water. Buffering is required to prevent hydrolysis of the NHS ester bond conjugating the fluorophore to the nucleotide probe. - Clean five Coplin jars with 70% ethanol, allow to dry, label as follows: xylenes I, xylenes II, 95% EtOH, 90% EtOH, ddH2O, and fill with 100 mL of the appropriate solution. This will help avoid confusion in later steps.
- Tissue de-paraffinization and rehydration
- In the hood, place two slides per biopsy into a vertical slide rack.
- Place a vertical slide rack into the xylenes I Coplin jar (containing 100 mL of xylenes) for 10 min.
- Remove the slide rack from xylenes I and blot the bottom on a paper towel to remove excess xylenes. Place into the xylenes II Coplin jar for 10 min.
NOTE: Never work with xylenes outside of a certified fume hood. - Rehydrate deparaffinized tissue sections in successive ethanol washes (95% and 90%) for 10 min each in the respectively labeled Coplin jars.
NOTE: During this stage, warm Hybridization Buffer to 50-56 °C in a water bath - Remove the slide rack from the 90% ethanol wash, blot the bottom on paper towels to remove excess ethanol, and place in the ddH2O Coplin jar containing 100 mL of filter-sterilized ddH2O for 10 min.
- While waiting for the ddH2O wash, dilute the probes to 10 nM in hybridization buffer to create the staining solution. Prepare 150 μL of staining solution per slide.
NOTE: Protect staining solution from light by wrapping the tubes in aluminum foil and storing in a drawer. When working with fluorescently labeled probes on the benchtop, consider turning off overhead lights when able. Probes diluted in hybridization buffer should not be re-used.
- Hybridization and counter-staining
- Prepare a humidifying chamber for each probe by placing soaked, crumpled Kimwipe and sterile water to the reservoir of a P1000 tip box. Place the tip holder cartridge on-top - this is where the slides will sit.
NOTE: It is important to use a humidifying chamber to prevent the biopsy sections from drying out during hybridization. It should be noted that commercially available humidifier chambers are designed to maintain a stable, humid atmosphere. However, the technique detailed here sufficiently controls humidity at substantially less cost. - Remove the slides from the slide rack and place onto a fresh paper towel (tissue-side up). Use a Kimwipe to dry the slide. Be careful to only gently dab near (not on) the biopsy section to wick away water. Using a hydrophobic pen, draw a border around the biopsy section and place the slide tissue-side up in the humidifying chamber.
NOTE: Work quickly so that the tissues do not dry out before hybridization. - Place the humidifying chamber into an incubator set to 50 °C. Pipette 50-150 μL of the staining solution directly on top of the tissue so that the rectangle made by the hydrophobic border around the tissue is filled. Be careful to not add too much solution as to overflow the hydrophobic border. Close the box gently.
- Incubate overnight (~16 h) at 50 °C in the dark. If the incubator has a window, cover it with aluminum foil to create a dark environment.
NOTE: A hybridization temperature below the melting temperature of the FISH probe is required for reliable signal. 50 °C is the optimal temperature for the universal 16S rRNA probe but may not be optimal for other probes. - The following morning, prepare at least 500 mL of Wash buffer comprised of 0.9 M NaCl, 20 mM Tris-HCl (pH 7.2) in ddH2O and filter-sterilize into a sterile bottle with a vacuum bottle-top filter. Warm to 50-56 °C in a water bath.
- Remove the slides from the humidifying chambers and carefully wick away any remaining hybridization solution with a Kimwipe. Place the slides in a vertical staining rack.
- Place the staining rack into an opaque Coplin jar containing 100 mL of pre-warmed Wash buffer for 10 min. If the Coplin jars are not opaque (e.g., glass), place them in the dark during incubation steps — perhaps under a box or in a drawer.
- Repeat the wash step twice with fresh Wash buffer in new Coplin jars.
- During the wash steps, prepare the counter-stain by diluting a 100 μg/mL stock solution of Hoechst 33342 1:1,000 in wash buffer. To the same tube, add Alexa-555 wheat germ agglutinin (WGA) to a final concentration of 5μg/mL and Alexa-555 Phalloidin to a final concentration of 33 nM. Store in the dark until ready for use.
NOTE: Hoechst, Alexa-555 WGA, and Alexa-555 Phalloidin label DNA, mucin/uroplakins, and actin, respectively and may be stored long-term in the dark per the manufacturer's instructions. Fluorescent labels used for probes and counterstains can be customized for the filter sets available for the microscope to be used. - Remove the slides from the last wash and gently wick away excess wash buffer with a Kimwipe. Place the slides tissue-side up on a paper towel and add 50-150 μL of counter-stain directly on top of the tissue so that the hydrophobic border is filled, but not overflowing. Cover up to four slides with a cryobox-top and incubate for 10 min at room temperature.
- Place the sides back into the staining rack and wash twice more in Coplin jars with fresh Wash buffer, for 10 min each.
- Thoroughly dry the slides after the last wash and place tissue-side up on a paper towel under a cryobox-top. Squeeze one drop of mounting media directly on top of the tissue. Gently place an appropriately sized coverslip (will depend on biopsy size) on top. Gently press out any bubbles as they will interfere with imaging and allow the cover-slipped slides to cure overnight in the dark.
- The next day, seal the edges of the coverslip to the slide with a light coat of clear nail polish. Allow to dry for 10 min in the dark and then store in the dark at 4 °C for confocal microscopy.
- Prepare a humidifying chamber for each probe by placing soaked, crumpled Kimwipe and sterile water to the reservoir of a P1000 tip box. Place the tip holder cartridge on-top - this is where the slides will sit.
3. Visualization of 16S rRNA FISH by Confocal Microscopy
NOTE: For this protocol, best results are achieved with a laser-scanning confocal microscope with 63x and 100x objectives. Proper filter sets for visualization of Hoechst, Alexa-488, and Alexa-555 fluorescence are required. However, standard fluorescent microscopy can be used if a confocal microscope is unavailable. This protocol is for a laser scanning confocal microscope.
- Switch on the confocal microscope and the computer software associated with the microscope per manufacturer instructions.
- Load the slide and visualize with the 10x objective in the blue (DAPI/Hoechst) channel. Focus carefully until nuclei are visible.
- Once focused, change the objective to high magnification (63x or 100x). Add oil on top of the cover slip. Refocus with the new objective, making sure that the objective lens comes in contact with oil while focusing.
NOTE: Use only fine focusing at high magnification (63x or 100x). Oil should only be used for an oil objective. - Quickly assess each slide through the eye-piece in the green (eGFP/Alexa-488) channel to determine which slides are FISH positive and which are FISH negative.
NOTE: It is best if this initial assessment/scoring is done blinded by a separate individual to reduce experimental bias. - To image the stained biopsies, start with a FISH positive slide and switch to the computer visualization mode. Select the 405 (Hoechst), 488 (Alexa-488), 555 (Alexa-555) channels. Set the pinhole using the longest wavelength channel, in this case 555. Find the correct focal plane for visualization of labeled bacteria in the 488 channel. Without changing the focal plane, set the gain, laser power, and offset for each channel such that the signal is not saturated, and the background is not over-corrected. Acquire the image in all three channels.
NOTE: Use the same settings for the 488 channel to image every slide in an experiment. The laser power may require adjustment in the 405 and 555 channels if the optimal focal plane for bacterial visualization changes between fields. - Repeat on additional fields until acquiring images of the entire epithelial surface.
NOTE: You may have to change the focal plane slightly to visualize labeled bacteria in different fields, but never change the gain, laser power, or offset for the 488 channel between fields. If working on a confocal microscope, it may be informative to capture a Z-stack so that the three-dimensional localization of the bacteria within the tissue may be analyzed. - Process images and quantify labeled bacteria within or associated with the tissue using ImageJ or similar software. Minimal processing of the images is recommended (e.g., splitting/merging channels and converting into image files), although background correction may be performed if necessary. All corrections or other alterations should remain consistent between images.
Representative Results
The protocol has been optimized for the unbiased detection of bacteria associated with the bladder mucosa in paraffin-embedded bladder biopsy sections. Figure 1 depicts representative confocal micrographs from an experiment using this protocol on sections of bladder biopsies obtained from women with recurrent urinary tract infection. Two serial sections were hybridized with either the universal 16S rRNA (upper panels) or scramble (lower panels) probes. Images from the same region of the tissue were taken and bacteria (green) are clearly visible in the tissue hybridized with the 16S rRNA probes and not with the scramble probe. Figure 2 represents a false positive result. Signal corresponding to autofluorescent collagen or elastin is detected in the 405 and 488 channels in both the 16S rRNA and scramble probe-hybridized biopsy sections highlighting the importance of always using a scramble probe control.
| Probe | Sequence | Tm | Fluorophore | Linkage |
| Universal 16S rRNA | 5'-GCTGCCTCCC GTAGGAGT-3' | 54.9 | Alexa-488 (5' and 3') | NHS Ester |
| Scramble | 5'-ACTCCTACGG GAGGCAGC-3' | NA | Alexa-488 (5' and 3') | NHS Ester |
Table 1: FISH probe sequences and characteristics. Tm indicates melting temperature and NHS is an abbreviation for N-hydroxysuccinimide.
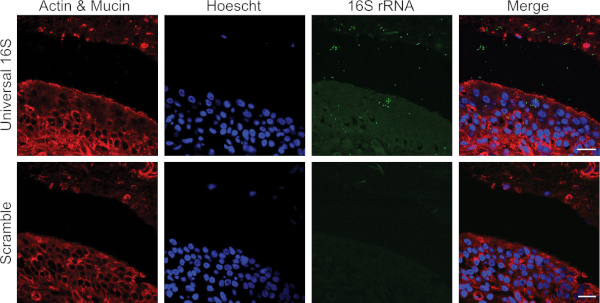
Figure 1: Representative confocal micrographs of FISH of universal 16S rRNA and scramble probe in a human bladder biopsy. Actin and Mucin are labeled in red, cellular nuclei are labeled in blue, and bacteria in green. Tissue-associated bacteria are only detected with the 16SrRNA probe and not the scramble. Images taken at 63x magnification. Scale bar = 20 μm. Please click here to view a larger version of this figure.
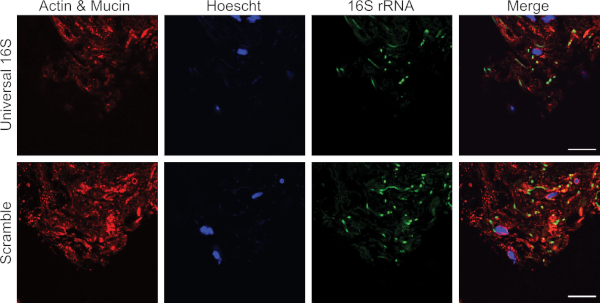
Figure 2: Representative confocal micrographs of false-positive green autofluorescence in a human bladder biopsy. Actin and mucin are labeled in red, cellular nuclei are labeled in blue, and bacteria and autofluorescent components of the extracellular matrix (e.g., collagen and elastin) are green. Green fluorescence is observed with both the 16S rRNA and scramble probes indicating a false-positive result. Images are taken at 63x magnification. Scale bar = 20 μm. Please click here to view a larger version of this figure.
Discussion
Here, we describe a protocol for the detection of tissue-associated bacteria in human bladder biopsies by 16S rRNA FlSH. This protocol can be easily adapted for biopsies taken from other tissues, such as the gastrointestinal tract or skin, and can be extended to tissues harvested from a variety of mammalian model organisms. The protocol described here can also be adapted to for the use of multiple fixation (e.g., formalin, ethanol, methacarn) and tissue preparation techniques (e.g., paraffin or resin embedded, and cryopreserved tissues). The double-labeled universal 16S rRNA probe allows for the unbiased detection of all bacterial species present within tissue and can provide valuable insight into how pathogens and the microbiota spatially interact with mucosal surfaces in disease and healthy states. Using resources such as probeBase, PhylOPDb or the PROBE_DESIGN tool of the ARB software package for the selection or design of species or genera-specific 16S or 23S rRNA probes, this protocol can be adapted for the detection of specific bacterial species or genera within tissue15,21,22. An important future direction for this method is multiplexing using species- or genera-specific probes labeled with different, discrete fluorophores for evaluation of microbial diversity within the bladder mucosa.
The primary limitation of this method for use on human specimens is the availability of biopsied tissue. Institutional Review Board approval and informed patient consent are required to obtain biopsies and direct collaboration with the clinician performing the procedure is necessary for optimal sample collection and access to patient metadata. The CEFT procedure itself destroys the bladder epithelium so we were able to justify biopsy of these areas before the procedure. A fume hood or appropriately fitted biosafety cabinet is required for this protocol due to the use of toxic xylenes in the deparaffinization step and the need to maintain a sterile environment throughout the procedure. A fluorescent microscope, preferably confocal, with a 63x or a 100x objective and appropriate filter sets for visualization of Hoechst, Alexa-555, and Alexa-488 is required for this protocol. The representative results depicted in Figure 1 were imaged using a laser scanning confocal microscope. Similar laser scanning microscopes should produce comparable images. This protocol is limited by its ability to only detect tissue-associated bacteria and not, for example, fungi. Probes specific the fungal 18S or 28S rRNA must be used to identify fungal pathogens within tissue18.
Critical steps to this protocol include maintaining a sterile environment throughout the procedure and ensuring that the tissue does not dry out between hybridization and staining steps. If the tissue dries out during the procedure, the signal may be dampened or the tissue may fall off of the slide during a wash step. It is also critical to always use two serial sections for this protocol — one for the 16S rRNA probe and one for the scramble probe. Without this control, it may be very difficult to distinguish false positives and the data obtained may not be useful or informative. If this protocol is being adapted for use with a probe other than the universal 16S rRNA probe, care must be taken to select an appropriate hybridization temperature, approximately 5 °C lower than the predicted melting temperature of the probe. To maintain signal intensity, the tissue must not be exposed to light for long periods of time after the probe has been added and must not be overexposed during microscopy. Lastly, during microscopy the same settings for the channel corresponding to the fluorophore conjugated to the FISH probe must be kept consistent between the experimental (16S rRNA probe) and control (scramble probe) slides. Visualizing the spatial relationship of bacteria within mucosal surfaces of patient-derived tissues is critical to understanding and building clinically relevant hypotheses about the host-pathogen interactions underlying infectious disease.
Acknowledgements
We would like to thank Kim Orth and Marcela de Souza Santos for protocol advice and Amanda Arute for technical support. This work was partially supported by the Cecil H. and Ida Green Chair in Systems Biology Science held by K.P.
Materials
| Name | Company | Catalog Number | Comments |
| Alexa-555 Phalloidin | Invitrogen | A34055 | Staining Actin |
| Alexa-555 Wheat Germ Agglutinin (WGA) | Invitrogen | W32464 | Staining Mucin |
| Bottle top filters | Fisher Scientific | 09-741-07 | Sterilization |
| Coplin Jar | Simport | M900-12W | Deparaffinization/washing |
| Coverslips | Fisher Scientific | 12-548-5M | Microscopy |
| Ethanol | Fisher Scientific | 04-355-224 | Rehydration |
| Ethylenediaminetetraacetic Acid, Disodium Salt Dihydrate | Fisher Scientific | S311-500 | TE |
| Frosted Slides | Thermo Fisher Scientific | 12-550-343 | |
| Hoechst 33342, Trihydrochloride, trihydrate | Invitrogen | H21492 | Staining Nucleus |
| Hydrophobic marker | Vector Laboratories | H-4000 | Hydrophobic barrier |
| Kimwipes | Fisher Scientific | 06-666-11 | |
| Oil Immersol 518 F | Fisher Scientific | 12-624-66A | Microscopy |
| Paraformaldehyde (16%) | Thermo Scientific | TJ274997 | Fixation |
| ProLong Gold antifade reagent | Invitrogen | P36934 | Mounting medium |
| Sodium Chloride | Fisher Scientific | BP358-10 | Hybridization buffer/PBS |
| Sodium Dodecyl Sulphate | Fisher Scientific | BP166-500 | Hybridization buffer |
| Sodium Phosphate Dibasic Hepahydrate | Fisher Scientific | S373-500 | PBS |
| Sodium Phosphate Monobasic Monohydrate | Fisher Scientific | S369-500 | PBS |
| Syringe | VWR | 75486-756 | Sterilization |
| Tris-HCl | Fisher Scientific | BP152-5 | TE/Hybridization buffer |
| Xylene | Fisher Chemical | X3P-1GAL | Deparaffinization |
| 0.22 micron syringe filter | Fisher Scientific | 09-754-29 | Sterilization |
References
- Abraham, S. N., Miao, Y. The nature of immune responses to urinary tract infections. Nature Reviews Immunology. 15 (10), 655-663 (2015).
- Wolfe, A. J., et al. Evidence of uncultivated bacteria in the adult female bladder. Journal of Clinical Microbiology. 50 (4), 1376-1383 (2012).
- Grist, M., Chakraborty, J. Identification of a mucin layer in the urinary bladder. Urology. 44 (1), 26-33 (1994).
- Wu, X. R., Kong, X. P., Pellicer, A., Kreibich, G., Sun, T. T. Uroplakins in urothelial biology, function, and disease. Kidney International. 75 (11), 1153-1165 (2009).
- Ingersoll, M. A., Albert, M. L. From infection to immunotherapy: host immune responses to bacteria at the bladder mucosa. Mucosal Immunology. 6 (6), 1041-1053 (2013).
- Flores-Mireles, A. L., Walker, J. N., Caparon, M., Hultgren, S. J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nature Reviews Microbiology. 13 (5), 269-284 (2015).
- Lewis, A. J., Richards, A. C., Mulvey, M. A. Invasion of Host Cells and Tissues by Uropathogenic Bacteria. Microbiology Spectrum. 4 (6), (2016).
- Horsley, H., et al. Enterococcus faecalis subverts and invades the host urothelium in patients with chronic urinary tract infection. PLoS One. 8 (12), (2013).
- Rosen, D. A., et al. Utilization of an intracellular bacterial community pathway in Klebsiella pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression. Infection and Immunity. 76 (7), 3337-3345 (2008).
- Rosen, D. A., Hooton, T. M., Stamm, W. E., Humphrey, P. A., Hultgren, S. J. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 4 (12), e329 (2007).
- Hussain, S. A., Alhalabi, F., Zimmern, P. E. Long-term efficacy of fulguration of trigonitis for recurrent urinary tract infections in women. Urological Science. 26 (3), 197-201 (2015).
- De Nisco, N. J., et al. Direct Detection of Tissue-Resident Bacteria and Chronic Inflammation in the Bladder Wall of Postmenopausal Women with Recurrent Urinary Tract Infection. Journal of Molecular Biology. , (2019).
- Amann, R. I., et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Applied and Environmental Microbiology. 56 (6), 1919-1925 (1990).
- Vaishnava, S., et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 334 (6053), 255-258 (2011).
- Greuter, D., Loy, A., Horn, M., Rattei, T. probeBase--an online resource for rRNA-targeted oligonucleotide probes and primers: new features 2016. Nucleic Acids Research. 44 (D1), D586-D589 (2016).
- Monici, M. Cell and tissue autofluorescence research and diagnostic applications. Biotechnology Annual Review. 11, 227-256 (2005).
- Mysorekar, I. U., Hultgren, S. J. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proceedings of the National Academy of Sciences of the United States of America. 103 (38), 14170-14175 (2006).
- Frickmann, H., Lakner, A., Essig, A., Poppert, S. Rapid identification of yeast by fluorescence in situ hybridisation from broth and blood cultures. Mycoses. 55 (6), 521-531 (2012).
- Fischer, A. H., Jacobson, K. A., Rose, J., Zeller, R. Paraffin embedding tissue samples for sectioning. Cold Spring Harbor Protocols. 2008, (2008).
- Sun, Y., et al. Sudan black B reduces autofluorescence in murine renal tissue. Archives of Pathology & Laboratory Medicine. 135 (10), 1335-1342 (2011).
- Ludwig, W., et al. ARB: a software environment for sequence data. Nucleic Acids Research. 32 (4), 1363-1371 (2004).
- Jaziri, F., et al. PhylOPDb: a 16S rRNA oligonucleotide probe database for prokaryotic identification. Database (Oxford). , (2014).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved