Two-Dimensional Visualization and Quantification of Labile, Inorganic Plant Nutrients and Contaminants in Soil
In This Article
Summary
This protocol presents a workflow for sub-mm 2D visualization of multiple labile inorganic nutrient and contaminant solute species using diffusive gradients in thin films (DGT) combined with mass spectrometry imaging. Solute sampling and high-resolution chemical analysis are described in detail for quantitative mapping of solutes in the rhizosphere of terrestrial plants.
Abstract
We describe a method for two-dimensional (2D) visualization and quantification of the distribution of labile (i.e., reversibly adsorbed) inorganic nutrient (e.g., P, Fe, Mn) and contaminant (e.g., As, Cd, Pb) solute species in the soil adjacent to plant roots (the ‘rhizosphere’) at sub-millimeter (~100 µm) spatial resolution. The method combines sink-based solute sampling by the diffusive gradients in thin films (DGT) technique with spatially resolved chemical analysis by laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS). The DGT technique is based on thin hydrogels with homogeneously distributed analyte-selective binding phases. The variety of available binding phases allows for the preparation of different DGT gel types following simple gel fabrication procedures. For DGT gel deployment in the rhizosphere, plants are grown in flat, transparent growth containers (rhizotrons), which enable minimal invasive access to a soil-grown root system. After a pre-growth period, DGT gels are applied to selected regions of interest for in situ solute sampling in the rhizosphere. Afterwards, DGT gels are retrieved and prepared for subsequent chemical analysis of the bound solutes using LA-ICP-MS line-scan imaging. Application of internal normalization using 13C and external calibration using matrix-matched gel standards further allows for the quantification of the 2D solute fluxes. This method is unique in its capability to generate quantitative, sub-mm scale 2D images of multi-element solute fluxes in soil-plant environments, exceeding the achievable spatial resolution of other methods for measuring solute gradients in the rhizosphere substantially. We present the application and evaluation of the method for imaging multiple cationic and anionic solute species in the rhizosphere of terrestrial plants and highlight the possibility of combining this method with complementary solute imaging techniques.
Introduction
Nutrient acquisition by crop plants is a key factor in determining crop productivity. The processes governing efficient uptake of nutrients by crops have been studied intensely, especially the mechanisms controlling nutrient availability to and nutrient internalization by plant roots at the soil-root interface, the rhizosphere, are recognized for their role in crop nutrient acquisition. Important processes for plant nutrient uptake include: nutrient transport towards the root; dynamic sorption equilibria between species dissolved in the soil porewater and species bound to solid soil surfaces; microbial competition for nutrients; microbial mineralization of nutrients that are contained in soil organic matter; and nutrient internalization into the root symplasm. The uptake of inorganic trace metal(oid) contaminants is largely controlled by the same mechanisms.
Depending on nutrient and contaminant availability, plant demand and diffusivity in soil, differential nutrient patterns in the rhizosphere can be observed. For strongly sorbing elements with comparatively high internalization rates (e.g., P, Fe, Mn, Zn, As, Cd, Pb), depletion of the labile (i.e., reversibly adsorbed) element fraction compared to the bulk soil is found, with depletion zone widths often being ≤1 mm, while for more mobile nutrients such as NO3-, depletion zones can extend up to several centimeters1. Moreover, accumulation of elements such as Al and Cd has been observed when availability exceeds plant uptake rates2,3.
Given the importance of rhizosphere processes in nutrient and contaminant cycling, several techniques for measuring the plant-available element fraction at high spatial resolution have been developed4,5. However, measuring small-scale labile solute distributions has proven to be challenging for several reasons. A major difficulty is to sample very small (low µL range) volumes of soil and/or porewater at defined positions adjacent to living plant roots to resolve the steep nutrient gradients in the rhizosphere. One approach to address this problem is to use micro-suction cups for the extraction of porewater samples6. With this method, A. Göttlein, A. Heim and E. Matzner7 measured soil porewater nutrient concentrations in the vicinity of Quercus robur L. roots at a spatial resolution of ~1 cm. A difficulty of analyzing µL volumes of soil or soil solution is, that these small sample volumes, in combination with the low concentrations of all but the major nutrient species, require highly sensitive chemical analysis techniques.
An alternative system, capable of resolving nutrient gradients at a resolution down to ~0.5 mm, is to grow a root mat on the surface of a soil block, with a thin hydrophilic membrane layer separating soil from the roots8,9. In this configuration, solutes can pass through the membrane and roots can take up nutrients and contaminants from the soil while root exudates can diffuse into the soil. After the establishment of a dense root layer, the soil block can be sampled and sliced to obtained soil samples for subsequent extraction of element fractions. In this way, one-dimensional nutrient, and contaminant gradients, averaged across a relatively large area (~100 cm2) can be analyzed.
A further challenge is to obtain samples of the labile, plant-available element fraction, since most chemical soil extraction techniques operate very differently compared to the mechanisms by which plants take up nutrients and contaminants. In many soil-extraction protocols, soil is mixed with an extractant solution with the aim to establish a (pseudo-)equilibrium between dissolved and sorbed element fraction. However, plants continuously internalize nutrients and, therefore, often progressively deplete the rhizosphere soil. Although equilibrium extraction protocols have been widely adopted as soil tests as they are easy to implement, the extracted nutrient fraction often does not represent the plant-available nutrient fraction well10,11,12,13. Sink methods which continuously deplete the sampled soil for nutrients have been proposed as advantageous methods and may better resemble the underlying nutrient uptake mechanism by mimicking the root uptake processes10,11,14,15.
In addition to the methods described above, genuine imaging applications, capable of measuring continuous parameter maps with resolutions ≤100 µm across fields of view of several cm2 have been developed for specific elements and soil (bio)chemical parameters5. Autoradiography can be used to image the element distribution in the rhizosphere provided that suitable radioisotopes are available16. Planar optodes enable visualization of important soil chemical parameters such as pH and pO217,18,19, and enzyme activity or total protein distributions can be mapped using fluorescent indicator imaging techniques such as soil zymography20,21,22,23 and/or root blotting methods24. While zymography and autoradiography are limited to the measurement of a single parameter at a time, pH and pO2 imaging using planar optodes can be done concurrently. The more traditional root mat techniques provide 1D information only, while micro suction cups provide point measurements or low resolution 2D information, however both approaches allow for multi-element analysis. More recently, P. D. Ilhardt, et al.25 presented a novel approach using laser induced breakdown spectroscopy (LIBS) to map 2D total multi-element distributions at a resolution of ~100 µm in soil-root core samples where the natural element distribution was preserved by careful sample preparation.
The only technique capable of targeted 2D sampling of multiple nutrient and contaminant solutes at high spatial resolution is the diffusive gradients in thin films (DGT) technique, a sink-based sampling method that immobilizes labile trace metal(loid) species in situ on a binding material embedded in a hydrogel layer26,27. DGT was introduced as a chemical speciation technique for measuring labile solutes in sediments and waters, and was soon adopted for its use in soils28. It enables sub-mm scale multi-element solute imaging, which was initially demonstrated in a river sediment29, and has been developed further for its application in plant rhizospheres30,31,32,33.
For DGT sampling, a gel sheet of a size of approximately 3 cm x 5 cm is applied onto a single plant root that is growing in the surface layer of a soil block, with a hydrophilic membrane separating the gel from the soil. During the contact time, labile nutrients and/or contaminants diffuse towards the gel and are bound immediately by the binding material incorporated in the gel. In this way, a concentration gradient, and thus a continuous net flux towards the gel is established and prevailed during the sampling time. After sampling, the hydrogel can be removed and analyzed using an analytical chemical technique allowing for spatially resolved analysis. A highly specialized and frequently used technique for this purpose is laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS). In some early studies, micro particle induced X-ray emission (PIXE) was also used29. DGT sampling combined with LA-ICP-MS analysis allows for multi-element chemical imaging at a spatial resolution of ~100 µm. If highly sensitive ICP-MS techniques (e.g., sector field ICP-MS) are employed, exceptionally low limits of detection can be achieved. In a study on the effect of liming on Zn and Cd uptake by maize15, we were able to map labile Cd in the maize rhizosphere in uncontaminated soil with a limit of detection of 38 pg cm-2 of Cd per gel area. DGT, planar optodes, and zymography rely on diffusion of the target element from soil into a gel layer, which can be exploited for combined application of these methods in order to simultaneously, or consecutively, image a large number of parameters relevant for plant nutrient and contaminant uptake. Detailed information on analytical chemical aspects of DGT imaging, on the potential of combining DGT and other imaging methods, and on its applications is comprehensively reviewed in ref.34,35.
In this article we describe how to carry out a solute imaging experiment using the DGT technique on roots of terrestrial plants in a unsaturated soil environment, including plant cultivation, gel fabrication, gel application, gel analysis and image generation. All steps are elaborated in detail, including notes on critical steps and experimental alternatives.
Protocol
1. Fabrication of DGT gels
NOTE: Several DGT gel types are available for 2D imaging of labile solute species at high (sub-mm) spatial resolution35. Here, the fabrication of three well-characterized high resolution (HR)-DGT binding gels used in solute imaging applications is briefly summarized. Laboratory procedures for trace element analysis, as well as detailed fabrication procedures of all presented HR-DGT gels are described in the Supporting Information (SI) sections S1 and S2.
- Polyurethane-based mixed anion and cation binding gel (HR-MBG; SI S2.1)31
- Prepare a polyurethane gel suspension with homogeneously dispersed zirconium (IV) hydroxide and iminodiacetate (IDA) phases.
- Coat the gel suspension in a thin film onto a glass plate and initiate gel formation by solvent evaporation to obtain a 0.1 mm-thin, tear-proof mixed anion and cation binding gel (HR-MBG).
- Polyacrylamide-zirconia anion binding gel (HR-ABG; SI S2.2)36
- Fabricate a 0.4 mm-thin agarose cross-linked polyacrylamide gel (APA) following established gel casting procedures37 (see SI S2.4 for a detailed protocol of the APA fabrication).
- Precipitate zirconium (IV) hydroxide phases into the precast APA gel to obtain a 0.4 mm-thin anion binding gel (HR-ABG).
- Polyacrylamide-iminodiacetate cation binding gel (HR-CBG; SI S2.3)38
- Prepare a polyacrylamide gel suspension with homogeneously dispersed IDA phases.
- Cast the gel between two glass plates and initiate the polymerization reaction to obtain a 0.4 mm-thin cation binding gel (HR-CBG) where the IDA phases are settled to one side of the gel.
2. Plant cultivation
NOTE: The experimental system uses rhizotrons4 (Figure 1) to grow plants in unsaturated soil for solute imaging. First, rhizotron soil filling and watering are described, then details on the experimental plant growth are given. Details on the rhizotron design and soil substrate preparation before filling into the rhizotron are presented in the SI section S3.
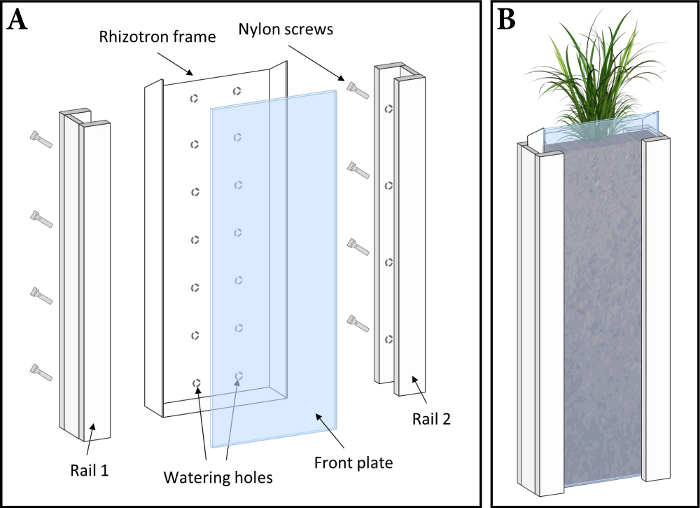
Figure 1: Rhizotron design (not to scale). (A) Exploded view of a rhizotron growth container. (B) Assembled rhizotron during plant growth. Please click here to view a larger version of this figure.
- Rhizotron soil filling
- Before filling the rhizotron with pre-moistened soil (gravimetric water content,
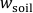 , is known; see SI S3.2), close the watering holes at the back of the rhizotron using adhesive tape and remove the front plate and its fixation rails and screws.
, is known; see SI S3.2), close the watering holes at the back of the rhizotron using adhesive tape and remove the front plate and its fixation rails and screws. - Weigh the empty rhizotron (excluding the front plate, rails, and screws), eight 5 cm x 11 cm acrylic plates and 16 clamps (Table of Materials) and record the sum of weights.
- Attach one small acrylic plate at the bottom of the rhizotron using two clamps on each side, with the pressure of the clamps directed onto the rhizotron frame, so the plate does not bend inwards and the volume is constant.
- Incline the rhizotron slightly towards the small plastic plate and fill with pre-moistened soil up to a height of ~4 cm (Figure 2A). Distribute the soil inside the rhizotron by agitating the rhizotron slightly and gently compress the soil by a few mm (depending on the specific soil characteristics) with a compaction tool (Figure 2B).
- Repeat 2.1.3 - 2.1.4 until the rhizotron is filled with soil (Figure 2C). Leave a gap of ~3 cm at the top for the subsequent planting of seedlings into the rhizotron.
- Weigh the soil-filled rhizotron (including 8 small plates and 16 clamps) and record the weight. From this, subtract the empty rhizotron weight obtained in 2.1.2 and record the weight difference, i.e., the mass of moist soil,
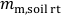 (g), in the rhizotron.
(g), in the rhizotron. - Calculate the mass of dry soil in the rhizotron,
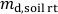 (g), according to Eq. 1, and then calculate the dry soil bulk density,
(g), according to Eq. 1, and then calculate the dry soil bulk density, 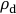 (g cm-3), in the rhizotron according to Eq. 2.
(g cm-3), in the rhizotron according to Eq. 2.
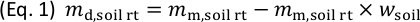
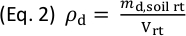
Here,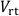 (cm3) is the total inner volume of the rhizotron.
(cm3) is the total inner volume of the rhizotron.
NOTE: Typical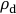 values in the rhizotron are between 1.0-1.4 g cm-3. Do not exceed 1.5 g cm-3 as root growth might be impeded above this value.
values in the rhizotron are between 1.0-1.4 g cm-3. Do not exceed 1.5 g cm-3 as root growth might be impeded above this value. - Place the soil-filled rhizotron on a support box and remove all clamps and small plates from the rhizotron (Figure 2D). Carefully clean the rhizotron frame (i.e., edges) using tissue paper, as remaining soil particles on the frame can cause leaks.
NOTE: The exposed soil surface must be homogeneous and leveled with the rhizotron frame without any cracks or gaps. If not, empty the rhizotron and repeat 2.1.2 - 2.1.8. - Cut two pieces of polytetrafluoroethylene (PTFE) (Table of Materials) foil to 22 cm x 13 cm each. In addition, cut a piece of plastic foil to 46 cm x 15 cm. Record the sum of PTFE and plastic foil weights. Place the first piece of PTFE foil on the top half of the exposed soil surface in the rhizotron, extending ~1 cm over the soil level at the top of the rhizotron.
- Carefully fix the PTFE foil to the rhizotron frame using adhesive tape (Figure 2E). Start by fixing one corner at the top of the rhizotron first, followed by its opposite corner and finally the two corners further down the rhizotron. Apply tension when fixing the corners 2-4 to ensure a flat foil surface. If folds emerge, open and re-fix the tape at individual corners (not all at once) until all folds are removed and the PTFE foil is flat and contiguous with the soil surface.
- Place the second piece of PTFE foil on the lower end of the rhizotron, overlapping the upper PTFE foil piece by ~1 cm. Repeat 2.1.10 for fixing the second PTFE foil to the rhizotron.
- Place the plastic foil (46 cm x 15 cm) onto the PTFE foils. Fix the plastic foil using the fixation procedure as detailed in 2.1.10.
- Place a front plate onto the soil-filled and foil-covered rhizotron. Place one rail around each side of the rhizotron and tighten the screws by hand to fix the rails and, thereby, the front plate to the rhizotron. The screws are positioned towards the closed side of the rhizotron, i.e., the side with the watering holes (Figure 1A).
- Before filling the rhizotron with pre-moistened soil (gravimetric water content,
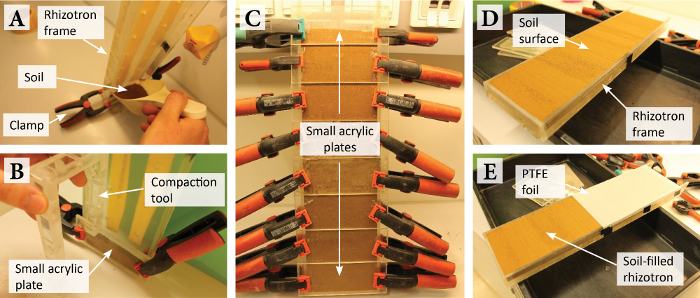
Figure 2: Rhizotron assembly and filling to grow plants in soil for solute imaging in the rhizosphere. (A) Soil filling into the rhizotron. (B) Compaction of the filled soil using a compaction tool. (C) Soil-filled rhizotron with small acrylic plates and clamps. (D) Soil-filled rhizotron with exposed soil surface. (E) Soil-filled rhizotron partly covered with a protective PTFE foil. Please click here to view a larger version of this figure.
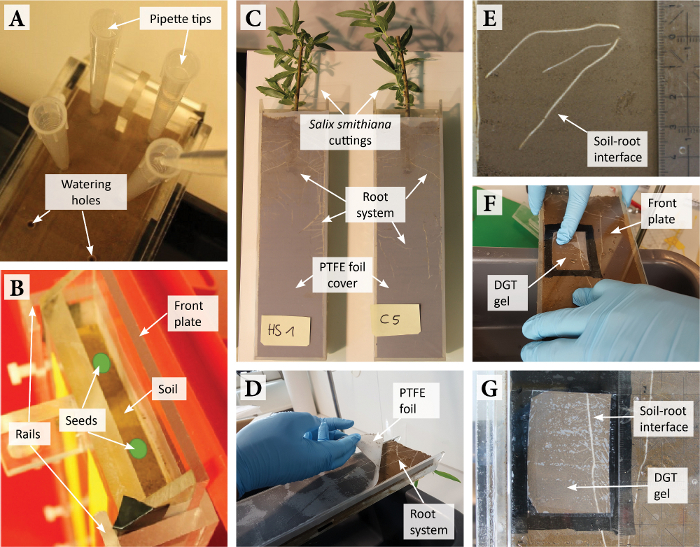
Figure 3: Rhizotron handling and DGT gel application. (A) Soil watering using 10 mL pipette tips in the watering holes in the back of the rhizotron. (B) Planting of seedlings (indicated as green spots) into the soil-filled and closed rhizotron. (C) Rhizotron planted with Salix smithiana cuttings and removed front plate and plastic foil cover. (D) Carefully peeling off the PTFE foil cover before DGT gel application. (E) High-resolution photo of the soil-root interface ROI. (F) Application of the front plate equipped with the DGT gel onto the rhizotron. (G) Photo of the ROI with the DGT gel applied during solute sampling. Please click here to view a larger version of this figure.
- Watering the soil
- Determine the water holding capacity (WHC) of the experimental soil in the rhizotron. To this end, manufacture two rhizotrons with an open bottom and fill them with soil as specified in section 2.1. Fully saturate these open, soil-filled rhizotrons by immersion in a water container for 16 h and drain the rhizotrons for 8 h.
- Take a composite soil sample of ~50 g from random locations in the open-bottom rhizotrons and dry the sample at 105 °C to determine
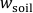 according to Eq. S1. The determined
according to Eq. S1. The determined 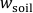 corresponds to the WHC of the soil and is, therefore, expressed as
corresponds to the WHC of the soil and is, therefore, expressed as 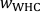 (g g-1).
(g g-1). - Define the target
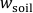 in the experimental rhizotron. During the growth phase, set a
in the experimental rhizotron. During the growth phase, set a 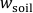 of 60 % of the WHC (i.e., the WHC factor,
of 60 % of the WHC (i.e., the WHC factor, 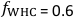 ) to supply plants with an adequate amount of water while avoiding anoxic conditions in the rhizotron.
) to supply plants with an adequate amount of water while avoiding anoxic conditions in the rhizotron. - Calculate the total mass of water to be added,
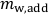 (g), to irrigate the soil in the rhizotron at the target
(g), to irrigate the soil in the rhizotron at the target 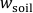 according to Eq. 3. Account for the mass of water present in the soil in the rhizotron,
according to Eq. 3. Account for the mass of water present in the soil in the rhizotron, 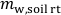 (g).
(g).
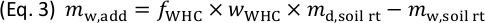
- Divide
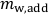 by the number of rhizotron watering holes (here 14) to obtain the mass of water to be added in each watering hole. Add water by pushing 10 mL pipette tips into the watering holes and letting the water flow into the soil by gravity (Figure 3A).
by the number of rhizotron watering holes (here 14) to obtain the mass of water to be added in each watering hole. Add water by pushing 10 mL pipette tips into the watering holes and letting the water flow into the soil by gravity (Figure 3A).
- Plant growth
- Pre-germinate seeds of the experimental plant (e.g., on wet filter paper) according to the specific seed germination requirements until the radicle emerges (up to 1 cm long).
- Plant up to two seedlings into the rhizotron as indicated in Figure 3B. At planting, add ~5 mL of water directly to the seedlings to support their growth. Cover the top opening of the rhizotron for the first ~two days after planting with a transparent, moisture-retaining film (Table of Materials). Wrap the rhizotron in aluminum foil to prevent microphytic growth.
NOTE: Apart from seedlings, plant cuttings can also be cultivated in rhizotrons. - Transfer the planted rhizotron into a growth room, with environmental conditions (i.e., temperature, humidity, light intensity) set to the specific plant requirements. Incline the rhizotron at 25°-35° to ensure root development alongside the front plate via gravitropism.
- During plant growth, gravimetrically maintain the target water content in the rhizotron by periodical watering every 2-4 days using the rhizotron watering holes as detailed in 2.2.5. Keep the soil surface at the top opening moist by regular additions of ~5 mL water.
NOTE: If plants are grown for extended periods and the plant biomass is expected to substantially decrease the amount of water added to the rhizotron by the proposed method, account for the weight of the plant biomass by growing plants in separate rhizotron replicates and harvesting and weighing the plant tissues at defined intervals. - Once roots reach a suitable location along the front plate, preferentially in the center of the rhizotron, apply DGT gels for sampling the rhizospheric solute distribution.
3. Sampling the solute distribution
- Gel application
- Increase
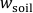 in the rhizotron from 60 %
in the rhizotron from 60 % 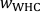 to 80 %
to 80 % 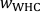 24 h before gel application as detailed in 2.2.4.-2.2.5. This ensures good soil-gel contact and allows solute diffusion into the gel while avoiding anoxic soil conditions during solute sampling.
24 h before gel application as detailed in 2.2.4.-2.2.5. This ensures good soil-gel contact and allows solute diffusion into the gel while avoiding anoxic soil conditions during solute sampling. - Take a new, acid-cleaned front plate, align it on the rhizotron used for sampling, and mark the regions of interest (ROIs) on the plate. Transfer the plate into a laminar flow bench or any other dust- and metal-free environment, unmarked side facing up.
- Cut the DGT binding gel on an acrylic support to the required rectangular size corresponding to the ROI, usually around 3 cm × 5 cm, using PTFE-coated razor blades. Cut the gel by pressing rather than sliding the razor blade to ensure a clear cut. Place the rectangular gel piece on the unmarked side of the plate at the marked location of the ROI.
NOTE: If HR-MBG is used, a 100 µm-thin spacer foil may be added underneath the gel to ensure that the gel is in good contact with the soil-root system. - Cut a 10 µm-thin polycarbonate membrane (0.2 µm pore size; Table of Materials) to a size which extends the gel size by ≥1 cm at each side and place the membrane onto the gel. Apply some water to remove air bubbles from the stack.
- Fix the membrane along all four edges using vinyl electrical tape (Table of Materials). In the process, carefully remove air bubbles trapped in between the gel and the membrane using plastic tweezers. The tape must only get in contact with the membrane and not with the gel.
NOTE: If the tape comes in contact with the gel, air bubbles are trapped between the gel and the membrane, or the final membrane surface shows folds, the gel/membrane stack needs to be reassembled as the diffusive flux of solutes into the gel can be impaired35 (repeat 3.1.3 - 3.1.5). - Place the rhizotron on a stand, remove the rails and carefully lift off the front plate (Figure 3C). Remove the plastic foil, cut off the PTFE foil at the edges of the rhizotron and slowly peel off the PTFE foil to avoid disturbance of the soil-root system (Figure 3D).
- Take an orthogonal photo of the ROI using a digital single-lens reflex (DSLR) camera (Table of Materials) to facilitate the interpretation and presentation of the solute distribution based on the soil structure and root morphology (Figure 3E). Use a camera stand and, if available, a macro lens. Align the camera so that the center of the photo corresponds to the center of the ROI, and the camera’s focus plane is parallel to the soil surface. Include a scale bar (e.g., a ruler) in the photo.
- Attach the plate equipped with the gel/membrane stack to the open rhizotron (Figure 3F). Therefore, align one edge of the plate with an edge of the rhizotron and gently ‘bend’ the plate towards the soil. This mode of application helps to avoid air bubbles between the gel/membrane stack and the soil-root system. Fix the front plate using the rails and screws.
NOTE: This step is critical and needs to be carried out carefully. The plate cannot be moved after establishing contact between the gel/membrane stack and the soil-root system without displacing soil and roots. - Record the exact starting time of gel deployment and take a photo of the deployed gel at the ROI as given in 3.1.7 (Figure 3G). Wrap the rhizotron in aluminum foil and transfer into the growth room until the end of the solute sampling period (often 24 h).
- Increase
- Gel retrieval
- Place the rhizotron on a support box, carefully lift off the front plate and rinse the gel/membrane stack on the plate with water to wash off adhering particles (Figure 4A). Record the exact end time of gel deployment. Sample soil from the ROI to determine the actual wsoil in the rhizotron according to Eq. S1.
- Transfer the front plate into the laminar flow bench, gel/membrane stack facing up. Retrieve the gel from the front plate by carefully removing first the tape along all four edges and then the polycarbonate membrane covering the gel (Figure 4B). Apply water to help the gel to float freely on a thin film of water on the plate with the soil-contact side facing up.
NOTE: It is critical to keep track of the gel orientation. The gel side exposed to the soil and roots must always face up (toward the user).
- Gel drying
- Cut a rectangular piece of gel blotting paper (Table of Materials) and place a slightly smaller piece of a polyethersulfone membrane (0.45 µm pore size; Table of Materials) on top.
- Transfer the gel from the plate onto the gel blotting paper/membrane stack using plastic tweezers, soil-contact side facing up. The gel must be relaxed (i.e., not stretched) and completely flat, without any air bubbles between gel and membrane. Apply some water onto the gel blotting paper/membrane stack to facilitate the gel transfer and positioning.
- Cover the gel blotting paper/membrane/gel stack completely with a piece of plastic foil and label the gel sample and its orientation on the plastic foil (Figure 4C). Place the gel blotting paper/membrane/gel/plastic foil stack in a vacuum gel dryer (Table of Materials) and dry until the stack is fully desiccated (typically 48-72 h). For HR-MBG, set the temperature to 50-55 °C, for HR-ABG and HR-CBG best dry at room temperature.
- Remove the gel blotting paper from the dried stack and transfer the gel, which is now inseparably merged with the polyethersulfone membrane, into a zip bag. The plastic foil cover stays on the gel until shortly before the LA-ICP-MS analysis.
- Include a gel piece from the original gel sheet as a method blank, which does not get exposed to soil. Process the method blank gel identical to the sample gel following 3.3.2 - 3.3.4.
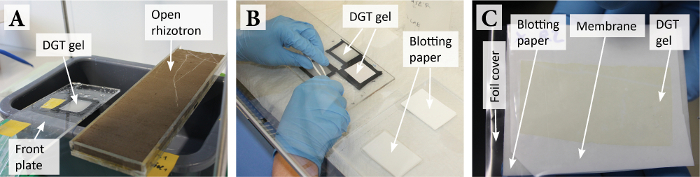
Figure 4: DGT gel retrieval and preparation for drying upon solute sampling. (A) Plate with the DGT gel and rhizotron directly after solute sampling. (B) Retrieval of the DGT gel from the plate in a laminar flow bench. (C) Stack of gel blotting paper/polyethersulfone membrane/DGT gel/plastic foil cover for gel drying. Note that the gel is slightly colored after its deployment on the rhizosphere soil. Please click here to view a larger version of this figure.
4. Chemical analysis of the DGT binding gel
NOTE: In this protocol, analysis of the solute distribution on the DGT binding gel is accomplished by LA-ICP-MS using a nanosecond 193 nm ArF excimer LA system equipped with a two-volume ablation cell coupled to a quadrupole ICP-MS (Figure 5). All instruments are listed in the Table of Materials. Alternatively, nanosecond 213 nm or 266 nm solid-state LA systems can be applied36,39,40,41,42,43. If enhanced sensitivity or mass resolution is required, sector field ICP-MS is an alternative to quadrupole ICP-MS15,44. Details on the preparation of DGT gel standards for external calibration and coupling of the LA system to the quadrupole ICP-MS are presented in the SI sections S4 and S5.

Figure 5: LA-ICP-MS setup for DGT gel analysis. (A) Nanosecond 193 nm ArF excimer LA system and quadrupole ICP-MS. (B) Dried gels mounted onto glass plates and fixed on the LA sample stage ready for introduction into the ablation cell. (C) Nebulizer gas (Ar) from ICP-MS and aerosol carrier gas (He or Ar) from ablation cell connected to the ICP via a two-way Y-splitter and torch adapter fitting. Please click here to view a larger version of this figure.
- Sample preparation for LA-ICP-MS
- Transfer the dried gel samples, standards, and method blanks (which are merged with the polyethersulfone membrane support) onto individual pieces of double-sided adhesive tape (Table of Materials), gel side facing up. Crop excess tape parts to save space in the ablation cell.
- Mount the dried gels onto glass plates. Use individual glass plates for each gel sample, standard series, or method blank to allow for flexible arrangement on the LA sample stage (typical size of 10 cm × 10 cm). Use a glasscutter to adjust the glass plate size as needed. Fix the glass plates with the gels on the LA sample stage (Figure 5B) using plasticine (Table of Materials). Level the gel surface by adjusting the stage floor and lock the sample stage into the ablation cell.
- LA-ICP-MS line-scan analysis
- Couple the LA system to the ICP-MS as specified in the SI section S5. In the LA software (Table of Materials), set the camera light settings (i.e., ‘Ring’, ‘Coax’ and ‘Transmitted’) to illuminate the gel surface in the ablation cell and focus onto the gel surface by adjusting the z-axis distance. Move to random locations across the cell to ensure that all gel surfaces are in focus.
- Set the LA parameters in the ‘Laser Setup’ window of the LA software. Typical settings used in LA-ICP-MS line-scan analysis of DGT gels are: mode continuous; energy output 20-30%; repetition rate 10-20 Hz; spot diameter 100-200 µm; scan speed 150-250 µm s-1.
NOTE: The parameters need to be optimized for the type of gel used and may vary. Parameters can also be enhanced for increasing signal to noise ratio, spatial resolution, or reducing acquisition times depending on the experimental and instrumental setup. Use a relatively low energy output (≤40 %) and repetition rate (≤25 Hz) to avoid penetrating through the gels into the backing materials39. Make sure that the laser scan speed is ≤ the spot diameter divided by the total ICP-MS scan cycle duration (see 4.2.6) to avoid compression of the datapoints and thus a loss of resolution in scan direction45. For example, when setting a spot diameter of 200 µm and a total ICP-MS scan cycle duration of 0.25 s, the scan speed should be ≤800 µm s-1. - Select the line tool and draw a single, ~1 mm-long line pattern across a gel standard. Right click the line pattern in the ‘Scan Patterns’ window and verify that the LA parameters set in 4.2.3. have been adopted. Use the ‘Duplicate Scans’ tool to duplicate this line four times, with a interline distance (distance between the line center) larger than the spot diameter (Figure 6). This approach offers a total of five parallel lines per gel standard (n = 5). Repeat this step for each gel standard, calibration blank and method blank.
- Move to the gel sample and draw a single line along the top edge of the rectangular area to be analyzed. Duplicate the line to create parallel lines for the entire sample area as specified in 4.2.3. Use an interline distance of 300-400 µm. Make sure the focus (z-axis distance) is set correctly for each start and end point of each line.
NOTE: The spatial resolution of the analysis is higher along the scan direction compared to the interline distance. Therefore, start and end points of the lines may best follow the direction of the analyte gradients. For rhizosphere gradients, this is usually perpendicular to the root axis (Figure 6). - In the ICP-MS software (Table of Materials), set up a time-resolved ‘Data Only’ method in the ‘Method’ screen, select one or more suitable isotope(s) for every analyte and include 13C as internal normalization standard31,36,40,41,42. Verify that detection of the isotopes is not impaired by interferences46.
- Set the total scan cycle duration of the ICP-MS method (‘Est. Reading Time’) to ≤0.5 s with appropriate dwell times per isotope (typically between 10-50 ms). Set the ICP-MS ‘Readings’ to 1 and change the ‘Readings/Replicate’ value to set the total measurement time per sample (‘Est. Sampling Time’), i.e., per individual ablation line. This depends on the specific LA parameters and line distances set in 4.2.2 - 4.2.4.
- For data reporting use an intensity versus time data collection mode and set the ‘File Write Option’ to ‘New Per Sample’ to create an individual data file (here .xl) for each ablation line.
- Set up a ‘Batch’ sample sequence in the ICP-MS ‘Sample’ screen, with each sample entry corresponding to an individual ablation line set in 4.2.3 - 4.2.4.
- Click ‘Analyze Batch’ to initiate the sample sequence on the ICP-MS, which will wait with data acquisition until it is triggered by the first laser pulse (see SI S5 for details on the trigger configuration).
- In the LA software, select all lines to be analyzed and verify that the ICP-MS method (‘Est. Sampling Time’, see 4.2.6.) matches the duration of the individual line ablations, which is typically different for gel samples, standards and method blanks. Repeat 4.2.6. to adjust the ICP-MS method if necessary.
- Click ‘Emission’ in the ‘Laser Energy’ window to recharge the laser head, and then click ‘Run’ to open the ‘Run Experiment’ window. Here, select ‘Selected Patterns Only’, set the ‘Washout Delay’ to 20-30 s, tick the ‘Enable Laser During Scans’ box, and set the ‘Laser Warmup Time’ to 10 s.
- Click ‘Run’ in the ‘Run experiment’ window to start the line-scan analysis and monitor the raw signal intensity in counts per second (cps) for each isotope on the ICP-MS in real time. Each line should start (‘Laser Warmup Time’, 10 s) and end (‘Washout Delay’, 20-30 s) with a gas blank.
- Monitor the laser fluence (J cm-2) during analysis to assess the laser stability. If the fluence varies largely, abort the analysis, and verify that the laser source and/or its mirror system are fully functional.
- After the analysis, stop the ICP-MS plasma and set the carrier gas flow rate on the LA system to 0 mL min-1. Remove the gels from the ablation cell and store in zip bags for further use.
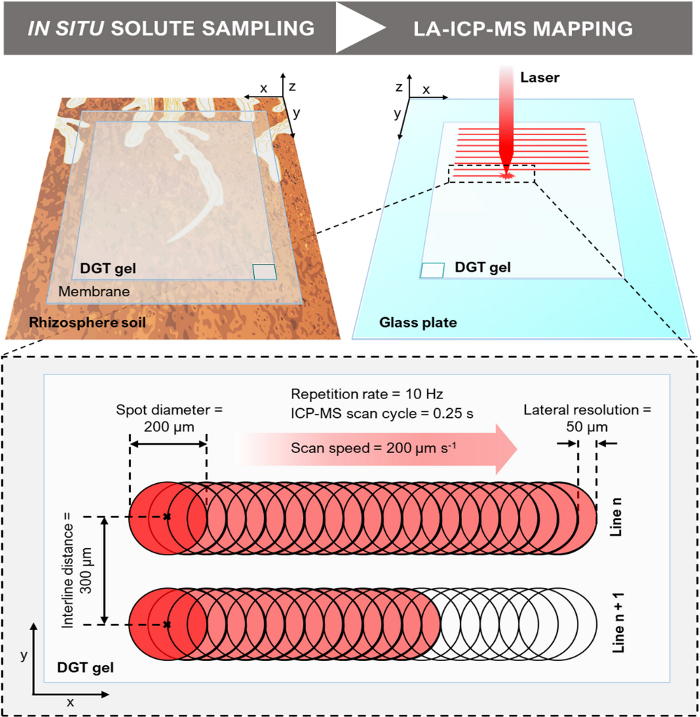
Figure 6: Schematic of the DGT LA-ICP-MS experimental design (not to scale). The illustration depicts the DGT-based in situ solute sampling in the rhizosphere and the LA-ICP-MS mapping of the solute distribution on the gel surface, including a close-up showing exemplary line-scan dimensions and parameters. Note that the DGT gel is horizontally flipped when transferred from the rhizosphere soil onto the glass plate, as indicated by the position of the rectangle at the bottom corner of the DGT gel. Please click here to view a larger version of this figure.
- Data processing and calibration
- Import the raw data file (.xl) for each ablated line in spreadsheet software (Table of Materials). The raw data table shows the ICP-MS readings (datapoints) for each isotope in cps and the corresponding time points in s. List all lines next to each other in different columns.
- Evaluate the line-scan data for signal stability of the internal standard (13C) and adequate washout times.
- Calculate an average gas blank for each isotope from all gas blank values recorded before the line ablations (i.e., during the laser warm-up time) and subtract the average gas blank from the corresponding raw intensities for each isotope to correct for the background signal.
- Apply internal normalization by dividing the signal intensity of each isotope (cps) by the signal intensity of the internal standard (cps) for each datapoint to correct for variations in the amount of material ablated and instrumental drift.
- Crop data before the start and after the end of each ablated line to remove the background signal. Transpose the data table to obtain a grid matrix where each row corresponds to an ablated line and each column to a normalized isotope intensity value. Separate the matrices for each isotope into individual worksheets.
- Calculate the average normalized signal intensity ratio per isotope for the calibration standards and calibration blank and compute the calibration function (y = ax + b) using a linear regression model with the gel standard analyte loadings (µg cm-2; see SI S4) as x-values. Evaluate the calibration function of each isotope for linearity47.
- Apply the calibration function to the sample data matrix. Convert the normalized signal intensity ratios,
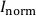 , to gel analyte loadings,
, to gel analyte loadings, 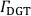 (µg cm-2), and subsequently to time-averaged solute fluxes,
(µg cm-2), and subsequently to time-averaged solute fluxes, 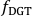 (pg cm-2 s-1), for each isotope and datapoint according to Eq. 4 and Eq. 5:
(pg cm-2 s-1), for each isotope and datapoint according to Eq. 4 and Eq. 5:
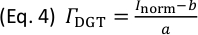
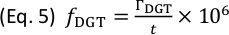
Here, a is the slope of the calibration line, b is the intercept of the calibration line, and t (s) is the gel deployment time during solute sampling. - Save the calibrated sample data matrix for each analyte as txt-file.
- Image generation
NOTE: Make sure to avoid any pixel interpolation operation during all steps of image generation, as this may lead to artificially smoothed solute flux gradients in the resulting images.- Import the calibrated sample data matrix (.txt) as text image in the image analysis software (Table of Materials).
- Calculate the distance per pixel in x-direction (i.e., lateral resolution) by multiplying the laser scan speed with the total ICP-MS scan cycle duration (e.g., assuming a scan speed of 200 µm s-1 and a total scan cycle duration of 0.25 s, the lateral resolution equates to 50 µm). The distance per pixel in y-direction equates to the interline distance (Figure 6).
- Calculate the aspect ratio correction factor of the image. Therefore, divide the distance per pixel in y-direction (e.g., 300 µm) by the distance per pixel in x-direction (e.g., 50 µm). Apply the obtained y/x correction factor (in this example 6) under ‘Scale’. Apply the distance per pixel (in µm or mm) in x-direction as scaling under ‘Set scale’.
- Apply a ‘Look Up Table’ i.e., pseudo-color scale for better visualization of chemical gradients in the solute image and adjust the image ‘color balance’ to control the lower/upper limits of the display range. Add a ‘Calibration bar’ and save the solute image as tiff-file.
- Copy the solute image using the ‘Copy to System’ command in the image analysis software and paste into desktop publishing software (Table of Materials). Scale-match, align and compose the solute image with the photo of the ROI obtained in 3.1.7.
NOTE: Before copying, resize the solute image by e.g., factor 10 via applying a ‘X Scale’ of 10 and a ‘Y Scale’ of 10 under ‘Set scale’ to ensure sufficient pixels for high-resolution publishing.
| DGT gel fabrication | Plant cultivation | In situ solute sampling | LA-ICP-MS solute flux mapping |
| HR-MBG 1 week | Soil preparation 1 week | Gel application 1 hour per gel | Sample preparation 1 hour per gel |
| HR-ABG 3 days | Rhizotron assembly 2 hours per replicate | Solute sampling period variable, typically 24 hours | LA-ICP-MS analysis 1 day per gel |
| HR-CBG 3 days | Plant growth dependent on study | Gel retrieval 1 hour per gel | Data processing 4 hours per gel |
| Gel drying 2-3 days | Image generation 10 min per image |
Table 1: Approximate times for general steps of the DGT LA-ICP-MS technique.
Representative Results
To demonstrate the capability and data detail of the DGT imaging method, we compiled the sub-mm, 2D flux distribution of multiple labile nutrient and contaminant solute species in soil adjacent to roots of Fagopyrum esculentum and Salix smithiana (Figure 7). Approximate times for general procedural steps of the protocol are presented in Table 1.
Solute images in Figure 7 were generated in three different studies using either HR-MBG or HR-CBG binding gels. The chemical images show the 2D solute flux distribution at a spatial resolution of 82-120 µm along the x-axis and 300-400 µm along the y-axis, depending on the LA-ICP-MS parameters used. Because no interpolation was applied during image calibration and resizing, single pixels represent measured datapoints. Alignment of the solute images with a photographic image of the ROI reveals that the sub-mm, 2D solute flux distribution of different elements is highly variable according to soil structure and root morphology. This can be attributed to the differential biogeochemical behavior of the elements in the soil-rhizosphere-plant system, and their interaction with the soil matrix and the plant roots.
In Figure 7A labile inorganic Mg, Al, P, Mn and Fe solute fluxes were visualized around a young F. esculentum root grown in carbonate-free soil fertilized with NH4NO3. The sub-mm solute distribution showed zones of decreased Al, P and Fe fluxes alongside older root sections due to root uptake, and highly increased Mg, Al, P, Mn and Fe fluxes at the root apex due to localized P mobilization processes of the F. esculentum root. Note that the root tip is located somewhat behind the soil surface and therefore hardly visible in the photographic image. Figure 7B shows the distribution of labile trace metals, including Mn, Fe, Zn, Cd and Pb around a root of metal-tolerant S. smithiana grown in a soil moderately contaminated with Zn, Cd and Pb. The solute images visualized distinct depletion particularly of Zn, Cd and Pb at the immediate root position, showing that S. smithiana roots act as a localized sink for labile trace metals in contaminated soil. Besides, localized Zn, Cd and Pb flux increases can be observed, indicating accumulation of these trace metals at the immediate soil-root interface.
In addition to multi-elemental solute imaging, the presented method can also be combined with complementary diffusion-based imaging techniques such as planar optodes34. This is demonstrated in Figure 7C, where the distribution of labile trace metals in the rhizosphere of S. smithiana was co-localized with the distribution of pH using a combined, single-layer planar optode-DGT cation binding gel33. The soil substrate was fertilized with (NH4)2SO4, leading to a pH decrease along the root axes by ~1 unit as compared to bulk soil. The pH decreases were co-localized with increased solute fluxes of Mn, Fe, Co, Ni, Cu and Pb, suggesting pH-induced metal solubilization.
Moreover, these example results show some of the potential imaging artefacts that may be obtained. For example, structural soil inhomogeneities, e.g., observed as a horizontal line in the lower third of the ROI image of Figure 7A, can cause soil-gel contact discontinuities resulting in limited diffusion at this location into the binding gel. Conversely, excessive soil compaction in the rhizotron can lead to poor porosity resulting in an artificial shift of the soil redox status towards anoxia. This is illustrated in Figure 7B, where extensive areas of highly elevated Mn and Fe fluxes in the solute images visually matched with a dense layer of soil in the ROI image. This suggests a decreased soil redox potential due to high soil compaction, resulting in reductive dissolution and solubilization of the highly redox-sensitive elements. Careful rhizotron filling and visual inspection of the soil surface directly after filling is therefore recommended.
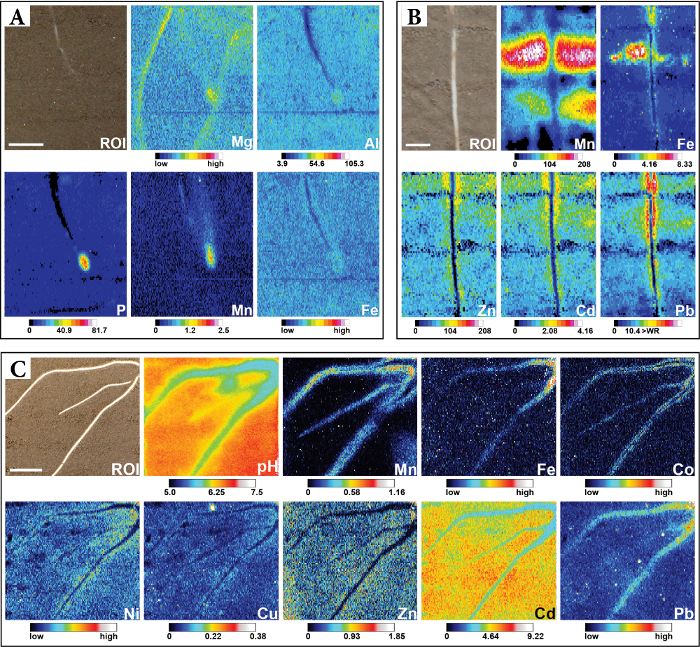
Figure 7: Sub-mm 2D distribution of labile nutrient and contaminant solute species across different soil-root interfaces. (A) Distribution of anionic P and cationic Mg, Al, Mn, and Fe solutes around a young F. esculentum root. Co-localized sampling of anionic and cationic solutes was achieved using HR-MBG for 24 h at a soil water saturation of ~75% WHC. The Al, P and Mn images are displayed as calibrated fDGT values (pg cm-2 s-1), whereas Mg and Fe images show 13C-normalized intensities. Scale bar represents 1 cm. This figure is adapted from ref.48. (B) Distribution of Mn, Fe, Zn, Cd and Pb around a S. smithiana root grown in soil moderately contaminated with Zn, Cd and Pb. Cationic trace metal solutes were sampled using HR-CBG for 20 h at a soil water saturation of ~80 % WHC. All images are displayed as calibrated fDGT values (pg cm-2 s-1). Scale bar represents 0.5 cm. This figure is adapted from ref.3. (C) Distribution of pH and multiple cationic solutes around S. smithiana roots grown in soil spiked with Cd. Co-localization of pH and solute dynamics was achieved using a modification of the HR-MBG protocol, allowing for simultaneous solute sampling and planar optode imaging33. The Mn, Cu, Zn and Cd images are displayed as calibrated fDGT values (pg cm-2 s-1), whereas Fe, Co, Ni and Pb images show 13C-normalized intensities. Scale bar represents 1 cm. This figure is adapted from ref.33. The presented figures are reproduced from the cited articles3,33,48 licensed under CC BY. Please click here to view a larger version of this figure.
Supplemental File 1. Please click here to download this file.
Discussion
The solute imaging protocol presented here is a versatile method to visualize and quantify 2D nutrient and contaminant fluxes in soil-plant environments. It is unique in its capability to generate sub-mm scale multi-element images of labile solute species at the soil-root interface, exceeding the achievable spatial resolution of alternative methods for measuring solute gradients in the rhizosphere substantially4. The targeted in situ sampling approach of DGT, in combination with a highly sensitive chemical analysis method such as LA-ICP-MS, facilitates the detailed investigation of solute flux dynamics around individual plant roots grown in soil or similar substrates. Due to the sink-based sampling process, the obtained images reflect the lability of the visualized solutes, and therefore are an estimation of their plant-availability10. Although the method-inherent measurement of solute fluxes bears considerable advantages like the interpretability as plant-available nutrient fractions, flux measurements are much less straight-forward to understand than porewater concentration measurements. The standard DGT sampling geometry in bulk soil applications (specifically the 0.8 mm-thick diffusion gels used in that setup) allows for comparing the actual porewater concentration, csoln, and a time-averaged porewater concentration estimate by a bulk DGT measurement, cDGT, and for the interpretation of these parameters regarding the resupply dynamics of a solute species. However, such comparison cannot be done based on imaging DGT application with very thin diffusion layers, as the derived cDGT values are unrealistically small34. DGT imaging results are therefore not always simple and quick to interpret and are often not directly comparable to more conventional porewater concentration measurements.
When applying the method, a few critical steps need to be carefully considered, mainly related to filling and watering the rhizotron growth containers. During filling the soil into the rhizotron, it is very important to avoid compacting the soil too much, as the plant roots cannot penetrate strongly compacted soil and root growth will be inhibited. We have observed roots avoiding strongly compacted soil and growing along the inner edges of the rhizotron growth container, where the soil is usually less compacted. In this case, individual roots located in the center of the rhizotrons, where DGT gels can be applied conveniently, may not develop at all, effectively inhibiting successful gel application. In our laboratory, experience showed that dry soil bulk densities of 1.0-1.4 g cm-3 allow unimpeded root development. Moreover, excessive soil compaction is also a potential source of artefacts regarding the solubility of redox-sensitive elements and biogeochemically associated species. As the total pore volume is reduced and the pore diameter distribution is shifted towards lower diameters in highly compacted soil, less air-filled larger-diameter pore volume is available, which may lead to reductive conditions locally. Consequently, MnIII/IV- and FeIII-oxides may be reduced, leading to increased Mn2+ and Fe2+ fluxes. The dissolution of Fe-oxides, which are important sorption sites e.g., for phosphate and micronutrients, may liberate sorbed and/or co-precipitated species and thereby cause artificially elevated fluxes of the biogeochemically associated species. A similar issue may arise if the growth containers are watered too much. Evaporation via the small soil surface area at the top of the growth container is low and the soil may remain water-saturated for up to several weeks after planting, which may also cause redox artefacts.
Another important consideration is the chemical functionality of the fabricated HR-DGT binding gel. By following the protocol, thin gels with a homogeneous distribution of binding phases are obtained. If the gels have areas of inhomogeneous material distribution (e.g., holes in the gel or aggregates of binding phases) these areas need to be removed or, if too extensive, the gel fabrication protocol needs to be repeated. If prepared correctly, the gel must be able to bind the target solute species that diffuse into the gel immediately and quantitatively27, which is determined by the analyte-specific gel binding capacity. While exceeding the gel capacity is less problematic in uncontaminated soils, it should be considered in metal-contaminated soils and saline soil environments. Saturation of the gel binding phases will not only impair quantitative solute sampling, but also result in lateral diffusion of solutes between binding phases in the gel, leading to an indefinite localization of small-scale solute flux features. Thus, if very high quantities of labile nutrient/contaminant species are expected in the target soil environment, preliminary tests should be performed. For estimating expected DGT loadings, bulk soil DGT piston sampling followed by gel elution and wet-chemical analysis can be applied15,49. If necessary, DGT deployment times may be adjusted to reduce the gel contact time and thus avoid gel saturation above capacity thresholds. Conversely, preliminary tests can also be helpful to identify required gel contact times and/or LA-ICP-MS sensitivities if very low solute loadings are expected, which may be important for mapping trace element solutes at natural soil background levels15. Besides, correct DGT gel functioning should be verified before its experimental application via controlled loading of gels in the preparation of DGT LA-ICP-MS calibration standards. The gel standard provides a matrix-matched reference gel analyte loading that can be used to assess if the sample gel loading determined by LA-ICP-MS is within the expected range. If unable to obtain a signal which is different from the gas and method blank background noise, the operator must ensure that laboratory procedures for trace element analysis were implemented and all protocol steps were performed correctly. Sometimes, the DGT gel is accidentally flipped after solute sampling with the soil-exposed, loaded side facing towards the glass plate rather than the laser beam, resulting in low signal intensities and erroneously flipped features in the final solute flux images.
During the LA-ICP-MS analysis, a large quantity of data is generated, which takes considerable time to evaluate. In our lab, we use in-house data evaluation scripts tailored for our target data output format using standard spreadsheet software. After semi-automated sorting and calibration, image plotting is conducted using open source, open access image analysis tools (ImageJ, Fiji50). This approach allows full control over data sorting, evaluation, and presentation, which is essential because the collected data correspond to rectangular, and not quadratic pixels, which needs to be properly displayed in the generated solute maps. Moreover, during data processing, any pixel interpolation should be carefully avoided. Pixel interpolation leads to smoothed gradients in the chemical images, resulting in softened, often circular element distribution features and is therefore an undesirable alteration of the original data. Pixel interpolation is a standard procedure in re-scaling and re-formatting operations in many image processing software products but can be deselected usually.
In conclusion, the described method is a significant advancement for understanding nutrient and contaminant dynamics in natural soil-rhizosphere-plant systems. In addition to DGT-only applications, the method can be combined with other, diffusion-based imaging techniques like planar optodes3,33,42,43,48,51 and zymography20,21,22,23,24, and may be developed further for including additional elements and soil parameters.
Acknowledgements
This study was co-funded by the Austrian Science Fund (FWF): P30085-N28 (Thomas Prohaska) and the Austrian Science Fund (FWF) and the Federal State of Lower Austria: P27571-BBL (Jakob Santner).
Materials
| Name | Company | Catalog Number | Comments |
| (NH4)2S2O8 (ammonium persulfate; APS) | VWR | 21300.260 | ≥98.0%, analytical reagent |
| 2-(N-morpholino)-ethanesulfonic acid (MES) | Sigma-Aldrich | M8250-100G | ≥99.5% |
| Acrylamide solution | Sigma-Aldrich | A4058-100ML | 40%, for electrophoresis |
| Analyte salts | n/a | n/a | Use water soluble analyte salts of analytical grade or higher |
| Buechner funnel | VWR | 511-0065 | 13 cm plate diameter |
| Chemical equilibrium modelling software | KTH Sweden | n/a | Visual MINTEQ |
| Clamp | Local warehouse | n/a | |
| Desktop publishing software | Adobe Inc. | n/a | InDesign CS6 |
| DGT cross-linker | DGT Research Ltd | n/a | 2%, agarose derivative |
| DGT piston sampler | DGT Research Ltd | n/a | 2 cm diameter exposure window |
| Digital single-lens reflex (DSLR) camera | Canon Inc. | n/a | Canon EOS 1000D |
| Dispersion device | IKA | 3737000 | Ultra-Turrax T10 Basic |
| Double-sided adhesive tape | Tesa | 56171 | |
| Ethanol | Sigma-Aldrich | 34923 | Puriss. p.a., absolute, ≥99.8% |
| Gel blotting paper | Whatman | 10426981 | Blotting Papers, Grade GB005, 20 × 20 cm, 1.5 mm thickness |
| Gel drier | UniEquip | n/a | UNIGELDRYER 3545 |
| High-pressure microwave system | Anton Paar | n/a | Multiwave 3000 |
| HNO3 | VWR | 1.00456.2500P | 65%, ISO for analysis |
| Horizontal shaker | GFL | 305 | |
| HydroMed D4 | AdvanSource Biomaterials Corp. | n/a | Ether-based hydrophilic urethane |
| ICP-MS software | Perkin Elmer | n/a | Syngistix |
| Image analysis software | National Institutes of Health (NIH) | n/a | ImageJ Fiji, freely available at https://fiji.sc/ |
| Knife-coating device | BYK | 5561 | Single Bar 6″, 0.5 mils |
| LA software | Elemental Scientific Lasers | n/a | ActiveView |
| LA system | Elemental Scientific Lasers | n/a | NWR193 |
| Laminar flow bench | Telstar Laboratory Equipment B.V. | n/a | Class II biological safety cabinet |
| Magnetic stirrer | IKA | 0003582400 | C-MAG MS 7 |
| Moisture-retaining film | Bemis Company, Inc. | PM999 | Parafilm M, 4" x 250' |
| N,N,N’,N’-tetramethylethylenediamine (TEMED) | Sigma-Aldrich | T9281-50ML | BioReagent, suitable for electrophoresis, ~99% |
| NaNO3 | Sigma-Aldrich | 229938-10G | 99.995% trace metals basis |
| NaOH | Sigma-Aldrich | 1064980500 | Pellets for analysis |
| Overhead shaker | GFL | 3040 | |
| Perfluoroalkoxy alkane (PFA) vials | Savillex | 200-015-20 | 15 mL Standard Vial, Rounded Interior |
| pH meter | Thermo Scientific | 13-644-928 | Orion 3-Star Benchtop pH Meter |
| pH probe | Thermo Scientific | 8157BNUMD | Orion ROSS Ultra pH/ATC Triode |
| Plastic cutter | DGT Research Ltd | n/a | Use empty cross-linker vials from DGT research Ltd |
| Plastic tweezers | Semadeni | 602 | |
| Plasticine | Local stationary shop | n/a | non-drying plastic modelling mass based on paraffin wax and bulking agents |
| Polycarbonate membrane discs | Whatman | 110606 | Nuclepore Hydrophilic Membrane, 25 mm diameter, 0.2 µm pore size, 10 µm thickness |
| Polycarbonate membrane sheet | Whatman | 113506 | Nuclepore Hydrophilic Membrane, 8 × 10 in, 0.2 µm pore size, 10 µm thickness |
| Polyethersulfone membrane discs | Pall Corporation | 60172 | Supor 450 Membrane Disc Filters, 25 mm diameter, 0.45 µm pore size, 0.14 mm thickness |
| Polyethersulfone membrane sheet | Pall Corporation | 60179 | Supor 450 Membrane Disc Filters, 293 mm diameter, 0.45 µm pore size, 0.14 mm thickness |
| PTFE foil | Haberkorn | n/a | 50 µm thickness |
| PTFE spacer | Haberkorn | n/a | Variable thicknesses available |
| PTFE-coated razor blades | Personna GEM | 62-0178 | Stainless steel single edge blades (coated) |
| PTFE-coated Tygon tubing | S-prep GmbH | SP8180 | 0.32 cm inner diameter |
| Quadrupole ICP-MS | Perkin Elmer | N8150044 | NexION 2000B |
| Quantitative filter paper, 454 | VWR | 516-0854 | Particle retention 12-15 µm |
| Spreadsheet software | Microsoft Corporation | n/a | Microsoft Excel 2016 (v16.0) |
| Stainless-steel cutter | Local locksmithery | n/a | 2.5 cm diameter |
| Suspended particulate reagent-iminodiacetate (SPR-IDA) | Teledyne CETAC Technologies | n/a | 10 µm diameter polystyrene beads, 10 % (w/v) bead suspension |
| Transistor-transistor logic (TTL) cable | n/a | n/a | Consult ICP-MS technician to identify a suitable TTL cable for a specific instrument |
| Two-volume cell | Elemental Scientific Lasers | n/a | Two-volume cell 1 |
| Vinyl electrical tape | 3M | n/a | Scotch Super 33+ |
| Water purification system | Termo Electron LED GmbH | n/a | TKA-GenPure |
| ZrOCl2 × 8H2O | Alfa Aesar | 86108.30 | 99.9 %, metals basis |
References
- Hinsinger, P., Gobran, G. R., Gregory, P. J., Wenzel, W. W. Rhizosphere geometry and heterogeneity arising from root-mediated physical and chemical processes. New Phytologist. 168 (2), 293-303 (2005).
- Jungk, A. . Plant Roots: The Hidden Half. , 587-616 (2002).
- Hoefer, C., Santner, J., Puschenreiter, M., Wenzel, W. W. Localized metal solubilization in the rhizosphere of Salix smithiana upon sulfur application. Environmental Science & Technology. 49 (7), 4522-4529 (2015).
- Luster, J., Göttlein, A., Nowack, B., Sarret, G. Sampling, defining, characterising and modeling the rhizosphere-the soil science tool box. Plant and Soil. 321 (1), 457-482 (2009).
- Oburger, E., Schmidt, H. New Methods To Unravel Rhizosphere Processes. Trends in Plant Science. 21 (3), 243-255 (2016).
- Göttlein, A., Hell, U., Blasek, R. A system for microscale tensiometry and lysimetry. Geoderma. 69 (1), 147-156 (1996).
- Göttlein, A., Heim, A., Matzner, E. Mobilization of aluminium in the rhizosphere soil solution of growing tree roots in an acidic soil. Plant and Soil. 211 (1), 41-49 (1999).
- Hinsinger, P., Gilkes, R. J. Dissolution of phosphate rock in the rhizosphere of five plant species grown in an acid, P-fixing mineral substrate. Geoderma. 75 (3), 231-249 (1997).
- Wenzel, W. W., Wieshammer, G., Fitz, W. J., Puschenreiter, M. Novel rhizobox design to assess rhizosphere characteristics at high spatial resolution. Plant and Soil. 237 (1), 37-45 (2001).
- Degryse, F., Smolders, E., Zhang, H., Davison, W. Predicting availability of mineral elements to plants with the DGT technique: a review of experimental data and interpretation by modelling. Environmental Chemistry. 6 (3), 198-218 (2009).
- Mason, S., McNeill, A., McLaughlin, M. J., Zhang, H. Prediction of wheat response to an application of phosphorus under field conditions using diffusive gradients in thin-films (DGT) and extraction methods. Plant and Soil. 337 (1), 243-258 (2010).
- Six, L., Smolders, E., Merckx, R. The performance of DGT versus conventional soil phosphorus tests in tropical soils-maize and rice responses to P application. Plant and Soil. 366 (1), 49-66 (2013).
- Mason, S. D., McLaughlin, M. J., Johnston, C., McNeill, A. Soil test measures of available P (Colwell, resin and DGT) compared with plant P uptake using isotope dilution. Plant and Soil. 373 (1), 711-722 (2013).
- Freese, D., Lookman, R., Merckx, R., van Riemsdijk, W. H. New Method for Assessment of Long-Term Phosphate Desorption from Soils. Soil Science Society of America Journal. 59 (5), 1295-1300 (1995).
- Smolders, E., Wagner, S., Prohaska, T., Irrgeher, J., Santner, J. Sub-millimeter distribution of labile trace element fluxes in the rhizosphere explains differential effects of soil liming on cadmium and zinc uptake in maize. Science of The Total Environment. , 738 (2020).
- Bhat, K. K. S., Nye, P. H. Diffusion of phosphate to plant roots in soil. Plant and Soil. 38 (1), 161-175 (1973).
- Glud, R. N., Ramsing, N. B., Gundersen, J. K., Klimant, I. Planar optrodes: a new tool for fine scale measurements of two-dimensional O2 distribution in benthic communities. Marine Ecology Progress Series. 140, 217-226 (1996).
- Blossfeld, S., Gansert, D. A novel non-invasive optical method for quantitative visualization of pH dynamics in the rhizosphere of plants. Plant, Cell & Environment. 30 (2), 176-186 (2007).
- Larsen, M., Borisov, S. M., Grunwald, B., Klimant, I., Glud, R. N. A simple and inexpensive high resolution color ratiometric planar optode imaging approach: application to oxygen and pH sensing. Limnology and Oceanography: Methods. 9 (9), 348-360 (2011).
- Spohn, M., Carminati, A., Kuzyakov, Y. Soil zymography - A novel in situ method for mapping distribution of enzyme activity in soil. Soil Biology and Biochemistry. 58, 275-280 (2013).
- Spohn, M., Kuzyakov, Y. Spatial and temporal dynamics of hotspots of enzyme activity in soil as affected by living and dead roots-a soil zymography analysis. Plant and Soil. 379 (1), 67-77 (2014).
- Heitkötter, J., Marschner, B. Soil zymography as a powerful tool for exploring hotspots and substrate limitation in undisturbed subsoil. Soil Biology and Biochemistry. 124, 210-217 (2018).
- Guber, A. K., Kravchenko, A. N., Razavi, B. S., Blagodatskaya, E., Kuzyakov, Y. Calibration of 2-D soil zymography for correct analysis of enzyme distribution. European Journal of Soil Science. 70 (4), 715-726 (2019).
- Lin, V. S., et al. Non-destructive spatial analysis of phosphatase activity and total protein distribution in the rhizosphere using a root blotting method. Soil Biology and Biochemistry. 146, 107820 (2020).
- Ilhardt, P. D., et al. High-resolution elemental mapping of the root-rhizosphere-soil continuum using laser-induced breakdown spectroscopy (LIBS). Soil Biology and Biochemistry. 131, 119-132 (2019).
- Davison, W., Zhang, H. In situ speciation measurements of trace components in natural waters using thin-film gels. Nature. 367 (6463), 546-548 (1994).
- Davison, W. . Diffusive Gradients in Thin-Films for Environmental Measurements. , (2016).
- Zhang, H., Davison, W., Knight, B., McGrath, S. In Situ Measurements of Solution Concentrations and Fluxes of Trace Metals in Soils Using DGT. Environmental Science & Technology. 32 (5), 704-710 (1998).
- Davison, W., Fones, G. R., Grime, G. W. Dissolved metals in surface sediment and a microbial mat at 100-μm resolution. Nature. 387 (6636), 885-888 (1997).
- Santner, J., et al. High-resolution chemical imaging of labile phosphorus in the rhizosphere of Brassica napus L. cultivars. Environmental and Experimental Botany. 77, 219-226 (2012).
- Kreuzeder, A., Santner, J., Prohaska, T., Wenzel, W. W. Gel for simultaneous chemical imaging of anionic and cationic solutes using diffusive gradients in thin films. Analytical Chemistry. 85 (24), 12028-12036 (2013).
- Kreuzeder, A., Santner, J., Zhang, H., Prohaska, T., Wenzel, W. W. Uncertainty evaluation of the diffusive gradients in thin films technique. Environmental Science and Technology. 49 (3), 1594-1602 (2015).
- Hoefer, C., Santner, J., Borisov, S. M., Wenzel, W. W., Puschenreiter, M. Integrating chemical imaging of cationic trace metal solutes and pH into a single hydrogel layer. Analytica Chimica Acta. 950, 88-97 (2017).
- Santner, J., Larsen, M., Kreuzeder, A., Glud, R. N. Two decades of chemical imaging of solutes in sediments and soils--a review. Analytica Chimica Acta. 878, 9-42 (2015).
- Santner, J., Williams, P. N., Davison, W. . Diffusive Gradients In Thin-Films For Environmental Measurements. , 174-215 (2016).
- Guan, D. X., et al. Novel Precipitated Zirconia-Based DGT Technique for High-Resolution Imaging of Oxyanions in Waters and Sediments. Environmental Science & Technology. 49 (6), 3653-3661 (2015).
- Zhang, H., Davison, W. Performance Characteristics of Diffusion Gradients in Thin Films for the in Situ Measurement of Trace Metals in Aqueous Solution. Analytical Chemistry. 67 (19), 3391-3400 (1995).
- Warnken, K. W., Zhang, H., Davison, W. Performance characteristics of suspended particulate reagent-iminodiacetate as a binding agent for diffusive gradients in thin films. Analytica Chimica Acta. 508 (1), 41-51 (2004).
- Warnken, K. W., Zhang, H., Davison, W. Analysis of Polyacrylamide Gels for Trace Metals Using Diffusive Gradients in Thin Films and Laser Ablation Inductively Coupled Plasma Mass Spectrometry. Analytical Chemistry. 76 (20), 6077-6084 (2004).
- Gao, Y., Lehto, N. A simple laser ablation ICPMS method for the determination of trace metals in a resin gel. Talanta. 92, 78-83 (2012).
- Lehto, N. J., Davison, W., Zhang, H. The use of ultra-thin diffusive gradients in thin-films (DGT) devices for the analysis of trace metal dynamics in soils and sediments: a measurement and modelling approach. Environmental Chemistry. 9 (4), 415-423 (2012).
- Williams, P. N., et al. Localized flux maxima of arsenic, lead, and iron around root apices in flooded lowland rice. Environmental Science & Technology. 48 (15), 8498-8506 (2014).
- Lehto, N. J., Larsen, M., Zhang, H., Glud, R. N., Davison, W. A mesocosm study of oxygen and trace metal dynamics in sediment microniches of reactive organic material. Scientific Reports. 7 (1), 11369 (2017).
- Zitek, A., Aléon, J., Prohaska, T. . Sector Field Mass Spectrometry for Elemental and Isotopic Analysis. , 152-182 (2015).
- Lear, J., Hare, D., Adlard, P., Finkelstein, D., Doble, P. Improving acquisition times of elemental bio-imaging for quadrupole-based LA-ICP-MS. Journal of Analytical Atomic Spectrometry. 27 (1), 159-164 (2012).
- May, T. W., Wiedmeyer, R. H. A table of polyatomic interferences in ICP-MS. Atomic Spectroscopy. 19 (5), 150-155 (1998).
- Raposo, F. Evaluation of analytical calibration based on least-squares linear regression for instrumental techniques: A tutorial review. TrAC Trends in Analytical Chemistry. 77, 167-185 (2016).
- Kreuzeder, A., et al. In situ observation of localized, sub-mm scale changes of phosphorus biogeochemistry in the rhizosphere. Plant and Soil. 424 (1), 573-589 (2018).
- Hooda, P. S., Zhang, H., Davison, W., Edwards, A. C. Measuring bioavailable trace metals by diffusive gradients in thin films (DGT): soil moisture effects on its performance in soils. European Journal of Soil Science. 50 (2), 285-294 (1999).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Wagner, S., et al. Arsenic redox transformations and cycling in the rhizosphere of Pteris vittata and Pteris quadriaurita. Environmental and Experimental Botany. 177, 104122 (2020).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved