Establishing Stable Binary Cultures of Symbiotic Saccharibacteria from the Oral Cavity
* These authors contributed equally
In This Article
Summary
We demonstrate a method for isolating difficult-to-grow members of the novel bacterial phylum, Saccharibacteria, by filtering dental plaque and co-culturing with host bacteria.
Abstract
Many bacterial species cannot be cultured in the laboratory using standard methods, posing a significant barrier to studying the majority of microbial diversity on earth. Novel approaches are required to culture these uncultured bacteria so that investigators can effectively study their physiology and lifestyle using the powerful tools available in the laboratory. The Candidate Phyla Radiation (CPR) is one of the largest groups of uncultivated bacteria, comprising ~15% of the living diversity on earth. The first isolate of this group was a member of the Saccharibacteria phylum, 'Nanosynbacter lyticus' strain TM7x. TM7x is an unusually small bacterium that lives as a symbiont in direct contact with a bacterial host, Schaalia odontolytica, strain XH001. Taking advantage of the unusually small cell size and its lifestyle as a symbiotic organism, we developed a protocol to rapidly culture Saccharibacteria from dental plaque. This protocol will show how to filter a suspension of dental plaque through a 0.2 µm filter, then concentrate the collected Saccharibacteria cells and infect a culture of host organisms. The resulting coculture can be passaged as any normal bacterial culture and infection is confirmed by PCR. The resulting binary culture can be maintained in the laboratory and used for future experiments. While contamination is a possibility, the binary culture can be purified by either further filtering and reinfection of host, or by plating the binary culture and screening for infected colonies. We hope this protocol can be expanded to other sample types and environments, leading to the cultivation of many more species in the CPR.
Introduction
Culturing novel species of bacteria and bringing them into the laboratory allows for powerful experiments to better understand their physiology and broader interactions within their microbial community. While there are culture-free methods of interrogating these questions (e.g., "meta-omics"), the complex interactions of diverse microbial populations make it difficult to tease apart single variables and reach meaningful conclusions. While culturing bacteria has many benefits, there are many potential barriers to isolating a bacterium and growing it in pure culture. Potential specific growth requirements include pH, oxygen tension, vitamins, growth factors, signaling molecules or even direct cell contact to elicit growth1. However, it is believed that specific auxotrophies are the primary deterrent to culturing new species of bacteria. Standard media formulations lack many nutrients required by uncultivated bacteria, such as specific vitamins or carbons sources. These missing molecules can be key to the physiology of the uncultured bacteria and are usually provided by either another organism in the microbial community or a host organism. For example, complex carbohydrates such as mucins can be provided by animal hosts. Adding these to media has allowed cultivation of several bacteria from animal guts, including Akkermansia muciniphila and Mucinivorans hirudinis2,3,4. Many pathogenic bacteria have evolved the ability to use iron bound to hemin in animal cells, including the oral pathogen Porphyromonas gingivalis5. In the laboratory, the growth of Porphyromonas and other organisms, can be stimulated by the addition of hemin6.
Recently, many breakthroughs in culturing novel isolates of bacteria have come through co-culturing, using a "feeder" organism to provide specific factors to uncultured bacteria necessary for their growth. An elegant study by Vartoukian and colleagues showed that siderophores, iron binding molecules produced by bacteria, stimulated the growth of several novel oral isolates. Pyoverdines, a type of siderophore produced by pseudomonad species, were shown to significantly facilitate the growth of a novel Prevotella species7. In the same study, the first oral isolate for the phylum Chloroflexi was cultivated, also using F. nucleatum as a helper for providing some as yet unknown compound7. More recently, a bacterium from the genus Ruminococcaceae was isolated using Bacteroides fragilis as helper organism8. It was later shown that gamma aminobutyric acid (GABA), an inhibitory neurotransmitter, was required for growth on laboratory media. Using feeder organisms has proved to be a key strategy to mimicking specific microenvironments where uncultured bacteria grow, being more efficient than continuously reformulating growth media with different additives in varying concentrations.
One of the largest groups of uncultured bacteria are in the "Candidate Phyla Radiation" (CPR), a monophyletic group of several candidate bacterial phyla9,10. As of this writing, only members of the Saccharibacteria phylum within the CPR have been successfully cultured in the laboratory. The first isolate, 'Nanosynbacter lyticus' strain TM7x, was isolated using the antibiotic streptomycin, which had been predicted to enrich for the uncultured TM711,12. A key discovery of this work was that the new isolate grew as a parasite growing in direct contact with a bacterial host, Schaalia odontolytica, and microscopy showed these parasites were ultrasmall bacteria.
Using these clues, we devised a method to quickly establish binary cocultures of Saccharibacteria with their partners by filtering dental plaque and other oral samples through a 0.2 µm filter, collecting cells in the filtrate by centrifugation and using them to infect cultures of candidate host bacteria. This method has the advantage of avoiding enrichment cultures, which can be overwhelmed with fast-growing organisms. It also avoids the use of antibiotics, which could stop the growth of either the targeted Saccharibacteria species or their hosts. Using the method demonstrated here, we have successfully cultured 32 isolates from the Saccharibacteria phylum.
Protocol
When developing this protocol, IRB approval was sought and approved (#14-10) and informed consent was obtained from all subjects.
1. Preparation
- When working with human subjects, obtain necessary IRB approval and informed consent.
- Start cultures of host bacteria with enough time to grow to early stationary phase. For example, inoculate 2-5 mL of tryptic soy broth with 0.1% yeast extract added (TSBY) with Arachnia propionica and incubate at 37 °C for 24 h.
- Assemble filter holders with track-etched 0.2 µm filter membrane. Wrap the assembly in foil and sterilize by autoclaving.
- Sterilize centrifuge tubes and cap assemblies.
2. Obtain sample of oral bacteria
NOTE: While many oral samples contain Saccharibacteria (e.g., saliva, swabs of tonsils, tongue scrapings) dental plaque routinely is the most successful.
- Take a plaque scraping using a sterile paper point, Gracey curette, or, if self-sampling, use a sterile toothpick or pipette tip. Transfer plaque to a suitable buffer, such as maximal recovery diluent (MRD, 0.85% NaCl, 0.1% peptone) or PBS. If not processed immediately, keep the sample on ice until ready to proceed.
- Vigorously resuspend the dental plaque in MRD buffer using a combination of vortexing and pipetting with a small pipet tip.
- Add resuspended plaque sample to an additional 9 mL of MRD buffer.
3. Prepare filtrate from oral sample
- Using aseptic technique, unwrap a sterile filter assembly. Untwist a quarter turn and retighten the filter holder to be sure threads are properly engaged and the apparatus is closed properly. Using a syringe, wash the membrane by passing 10 mL of MRD buffer through the apparatus. Improper assembly is revealed at this step by fluid leaking out of the filter holder. If leakage occurs, obtain another sterile filter assembly and repeat the wash step.
- Apply the sample to the washed filter. Remove the plunger from a syringe and attach it to the filter apparatus. Pour the dispersed dental sample, now in 10 mL of MRD buffer, into a syringe and load it onto the filter.
- Place a sterile centrifuge tube beneath the filter apparatus to catch the filtrate, then apply light pressure to the plunger to push the sample through the filter.
- Repeat the procedure with another 10 mL of MRD buffer to wash the membrane, collecting the flow-through in the same tube as the filtered sample. Aseptically cap the tube.
4. Concentrate Saccharibacteria cells by centrifugation
- Make an orientation mark on the tube and cap and place the tube in a high-speed centrifuge with the mark on the upper side. The pellet formed from centrifugation is usually invisible. The marking will help determine where the pellet of Saccharibacteria cells is located when centrifugation is done and the tube removed from the centrifuge.
- Centrifuge the samples at 60,000 x g for 1 h at 4 °C.
NOTE: This force and time is sufficient to pellet all Saccharibacteria cells. However, Saccharibacteria cells can be at least partially pelleted by centrifuging for as little as 20 min at 20,000 x g. - Carefully remove the tubes from centrifuge. Pour out the liquid from the tube, keeping the pellet on the upper side of the tube.
- Resuspend the usually invisible pellet in 1-2 mL of MRD buffer by vigorous vortexing.
5. Infect host cultures with Saccharibacteria-enriched filtrate
- Prepare culture tubes by aliquoting 2 mL of appropriate growth media (e.g., TSBY, BHI, etc.) into tubes.
- Add 200 µL of overnight culture of host organisms to each tube. Add 100-200 µL of resuspended, filtered sample to each tube.
- Incubate combined samples as appropriate for the host organism (e.g., 37 °C, in an aerobic atmosphere for A. propionica).
- Passage the cells every two to three days by transferring 200 µL of binary culture to 2 mL of fresh growth medium in a new tube. If it appears that the passaged cultures show no growth (i.e., the turbidity/optical density of the culture doesn't increase after passage into fresh media) the Saccharibacteria could be overwhelming or killing all the host organism. To remedy this, add 200 µL of uninfected host culture when passaging the cells. Repeat for at least 5 passages.
6. Confirm infection by PCR
- After 5 passages, confirm infection with PCR. Five passages will ensure than any non-growing Saccharibacteria cells have been depleted beyond the limit of detection using 25 cycles of PCR.
- Prepare a PCR mastermix. A suggested recipe using is as follows, for each tube needed, prepare 12.5 µL 2x PCR buffer, 0.75 µL 10 µM Forward primer (580F12 - AYT GGG CGT AAA GAG TTG C), 0.75 µL 10 µM Reverse primer (1177R13- GAC CTG ACA TCA TCC CCT CCT TCC), 1 µL 25 mM MgCl2 and 9 µL water. Mix by vortexing.
- Aliquot 24 µL of mastermix into a 0.2 mL PCR tube. Add 1 µL of Saccharibacteria-infected culture to the PCR reaction. Place the tube in a thermocycler and perform the following protocol: Initial denaturation 95 °C for 5 min, 25 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, final elongation at 72 °C for 2 min, then final hold at 4 °C.
- Load and run the PCR products in a 1% agarose gel. A band of ~600 bases will indicate the presence of Saccharibacteria.
7. Check for purity and remove contaminating organisms
- Confirm purity of positive cultures by plating the coculture of Saccharibacteria on nutrient agar sufficient for the growth of the host organism. Perform a 10-fold serial dilution of the culture in sterile buffer (e.g., MRD or PBS) and spread 100 µL on an agar plate. Perform dilutions such that there will be approximately 20-200 colonies growing on the agar plate.
- Incubate the culture in appropriate growth conditions and observe for contaminating organisms.
NOTE: Axenic Saccharibacteria colonies have never been observed, so only the host organism is observed growing on these plates. More than one colony type usually indicates contamination. Should contamination occur, the culture should be re-filtered to remove the unwanted contaminants and re-inoculated onto fresh host. - Occasionally a double infection occurs, where two Saccharibacteria species infect the same host. To separate the Saccharibacteria species, resuspend individual colonies in 20 µL sterile PBS and use 1 µL of suspension in a PCR reaction to detect colonies with Saccharibacteria infections.
- Transfer the suspensions that gave a positive result to growth medium to start a binary culture. The success rate will depend on the titer of each Saccharibacteria species in the culture. It may be necessary to screen more than 50 colonies to find an infected one for propagation.
8. Storage of cultures
- Grow a binary culture of sufficient volume (10-50 mL) overnight.
- Pellet cells by centrifugation (4,000 x g for 10 min).
NOTE: High-speed centrifugation is not necessary here as Saccharibacteria cells will be attached to the larger, heavier cells of the host and will pellet with them. - Resuspend cells in cryoprotectant. Growth media supplemented with either 5% DMSO or 20% glycerol is usually sufficient. Ensure that the host organism is compatible with cryoprotectant media (e.g., Growth of A. propionica is inhibited by glycerol and frozen stocks may not revive). Test the viability of an uninfected host culture with cryoprotectant to ensure stocks can survive freezing.
- Resuspend cell pellet in the same volume of culture (10-50 mL). Aliquot 0.5 mL into labeled cryovials. Freeze cell cultures at -80 °C.
Representative Results
PCR for detection of Saccharibacteria may appear negative (i.e., no product seen) in initial infection cultures due to a low number of Saccharibacteria symbionts. However, after a few passages a strong PCR product should appear showing that a stable infection has occurred (Figure 1A). Conversely, some infections will initially appear positive by PCR, but diminish to undetectable after 1-4 passages (not shown). This indicates that a large amount of Saccharibacteria cells were present in the initial filtrate but were diluted out by passage and none were able to enter into a stable symbiosis with the host culture. Testing several host species with the same Saccharibacteria filtrate from the oral cavity will usually have a low success rate (Figure 1B) as the symbiont-host interaction is very specific. A researcher may also test the same host with filtrates from several different subjects. If a good host organism (such as Arachnia propionica or Schaalia odontolytica) is used, a success rate of 50% can be expected.
Testing by PCR is crucial. Experienced researchers have attempted to confirm infection by microscopy, only to report false positives. The Saccharibacteria cells are small and difficult to distinguish between vesicles or irregular cell envelope bulges. A PCR signal stable through several passages is key to confirm a successful infection.
As the infected cultures grow, normal turbid growth should appear. As the symbiosis establishes itself, infected cultures will appear less turbid than cultures of uninfected host organisms (Figure 2A). In some cases, infected cultures may appear to stop growing completely and not become turbid at all. This may be due to an infection where the Saccharibacteria cells are overwhelming the host organism. Adding "fresh" (uninfected) host to these cultures should provide a sufficient population of host to support the continued growth of the Saccharibacteria parasites.
Overgrowth, or excessively turbid cultures, may indicate contamination by either a laboratory contaminant or another small oral bacterium that was able to pass through the 0.2 µm filter. Campylobacter and Capnocytophaga spp. are typical oral contaminants of these experiments. This can be confirmed by plating the culture and looking for colonies that are atypical of the host organism followed by 16S rRNA sequencing. If contamination is seen, filtering these cultures through a 0.2 µm filter is usually sufficient to remove the contaminants. Saccharibacteria cells in the filtrate can be concentrated by centrifugation and used to re-infect a pure host culture.
Another way to purify a contaminated culture is by picking infected colonies from plating an infected culture. Colonies of infected hosts can sometimes be identified by irregular colony shape compared to uninfected colonies (Figure 2B-D). These irregular colonies are dependent on the titer of Saccharibacteria in the binary culture and a smaller proportion of colonies will appear irregular if the titer is low. This can make it easy to identify infected colonies, which can be picked and used to start a pure binary culture. If no irregular colonies are seen on plating from a Saccharibacteria infected culture, it is possible that the symbiont is at a low titer or does not cause irregularly shaped colonies. In such a case, PCR screening colonies with a normal appearance can show infection by Saccharibacteria, but at a low rate (2-10% of all colonies).
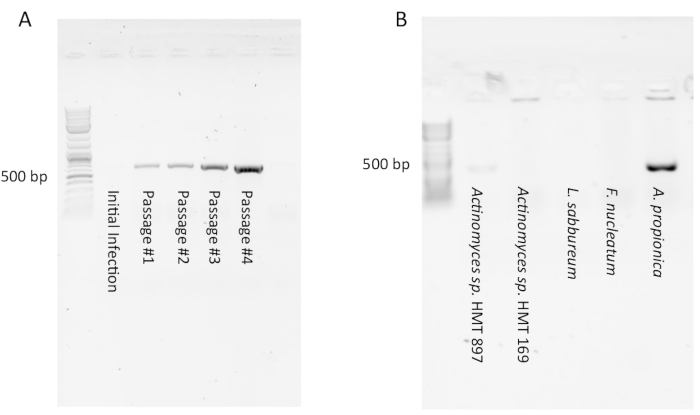
Figure 1: Typical PCR results of Saccharibacteria infections of host cultures. (A) A PCR product indicating presence of Saccharibacteria may not appear with the initial infection but can appear and become stronger in subsequent passages as the coculture establishes itself. (B) Most hosts infected with Saccharibacteria-enriched filtrate will not support their growth due to the specificity of the symbiosis. In this example, only A. propionica was successfully infected. Please click here to view a larger version of this figure.
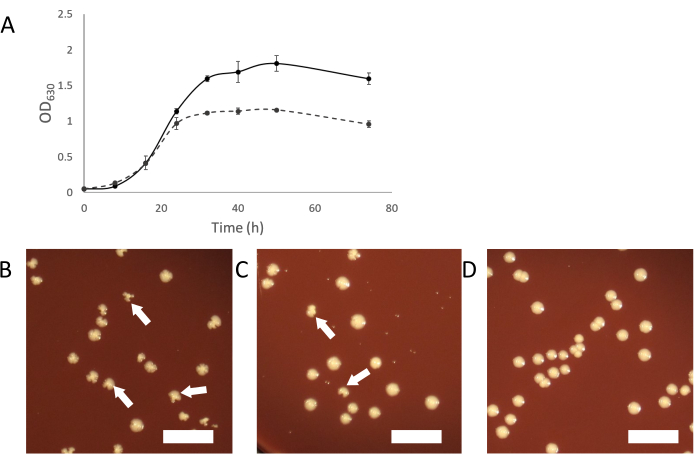
Figure 2: Growth characteristics of Saccharibacteria infected cultures. (A) Growth curves of infected and uninfected cultures of A. propionica showing infected culture will not grow to the same density as the uninfected control and appear less turbid. (B-D) Plating of cocultures can produce irregular colonies, caused by Saccharibateria infections. Irregular colonies will decrease in proportion relative to the titer of Saccharibacteria in the coculture. (B) Host with a high level of Saccharibacteria. (C) 10-fold diluted Saccharibacteria (D) Uninfected host culture. White arrows indicate examples of irregular colonies. Scale bar= 1 cm. Please click here to view a larger version of this figure.
Discussion
Our method of filtering plaque and applying it to pure cultures of host organisms is largely based on earlier observations on the first cultured Saccharibacteria, 'Nanosynbacter lyticus' strain TM7x11,14,15. Given the small cell size, we deduced that they could be separated from dental plaque using a filter and concentrated with centrifugation. Second, as these organisms live as parasites, providing these cells pure cultures of hosts would allow them to enter into a symbiosis and grow as binary cultures.
One advantage of this method is that it does not require an enrichment culture or selective pressure. 'Nanosynbacter lyticus' strain TM7x was cultured from an enrichment culture using streptomycin as a selective agent, which sequencing had suggested would be effective at enriching for Saccharibacteria. Fortuitously, the host for 'Nanosynbacter lyticus' Schaalia odontolytica, is known to be resistant to streptomycin16. Using antibiotics as a selective agent could also prevent the host organism from growing, which in turn would preclude the growth of Saccharibacteria.
A larger issue of using enrichment cultures is that fast-growing organisms will quickly displace organisms of interest. In the oral cavity, for example, Streptococcus species can grow quickly and, if sugar is present in the growth medium, produce enough acid to acidify the medium, further selecting against organisms of interest. By avoiding an enrichment culture and selective antibiotics, our method provides a general approach that could be applied to a wider range of Saccharibacteria and potential hosts without the complications of these other methods.
There are some obstacles to the method presented here. First, this method assumes that Saccharibacteria live in a binary culture. We have not tested combinations of trinary or ternary cultures to gauge their effectiveness, but there are likely Saccharibacteria that require growth factors that a single host organism cannot supply. Testing the vast combinations of oral bacteria that could support the growth of Saccharibacteria would be a daunting task. Second, the method assumes all Saccharibacteria are small enough to pass through a 0.2 µm filter. It could be that other Saccharibacteria are larger than believed and the filter is selecting against these organisms. A filter with a larger pore size could be used, but this runs the risk of allowing more unwanted oral bacteria into the infected co-culture. Lastly, it is very difficult to find host species outside of those that have already been published. Thus far, the only successful hosts are species from the genera Actinomyces, Schaalia, Arachnia and Cellulosimicrobium, all members of the phylum Actinobacteria15,17,18. However, these hosts only support the growth of specific Saccharibacteria. To culture more species of Saccharibacteria, many more hosts must be explored.
We hope the method presented here will aid future research of the Saccharibacteria and other CPR organisms. Metagenomic sequencing suggests that these organisms also have small genomes and are suspected to be symbionts or rely on the local microbial community to supply metabolites and other factors critical to their survival19. A similar filtering strategy could be used to isolate these organisms, provided they are small enough and that their host organisms(s) can be cultured. The methods described here are a first step to bringing the powerful tools of laboratory culture to this large and diverse group of bacteria.
Acknowledgements
The authors wish to thank Anne Tanner, Bruce Paster, Heike Boisvert, Xuesong He and Batbileg Bor for helpful discussions and for providing bacterial strains. We thank Susan Yost and Jessica Woods for microbial technical assistance. Research reported in this publication was supported by The National Institute of Dental and Craniofacial Research of the National Institutes of Health under award numbers R37 DE016937 (FED), R01 DE024468 (FED) and T32 DE007327 (AJC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Materials
| Name | Company | Catalog Number | Comments |
| Agarose | Fisher Scientific | BP160-100 | |
| Alphaimager | Cell Biosciences | FluorChem HD2 | Or equivalent UV gel imaging system |
| Aluminum foil | Fisher Scientific | 01-213-101 | |
| Brain Heart Infusion Broth (dehydrated powder) | Becton-Dickinson | 211059 | Or other growth media suitable for target organisms |
| Centrifuge Rotor 70-Ti | Beckman Coulter | 337922 | |
| Cryovials | Fisher Scientific | 12-567-500 | |
| DMSO | Fisher Scientific | BP231-100 | |
| Electrophoresis Power Supply | Bio-Rad | 1645052 | |
| Electrophoresis Rig | Bio-Rad | 1704467 | |
| Filter Forceps | Millipore Sigma | XX6200006P | Not essential, helps ensure filters are not punctured during handling |
| Glycerol | Fisher Scientific | G33-500 | |
| GoTaq Green Mastermix | Promega | M7122 | |
| Mastercycler Pro Thermocycler | Eppendorf | 950040025 | Or equivalent thermocycler for PCR |
| MgCl2 solution 25mM | Promega | A3513 | |
| Molecular Biology grade water | Fisher Scientific | BP2819100 | |
| O2 Control InVitro Glove Box | Coy Laoratories | 031615 | If needed for microaerobic organisms |
| Optima L-100 XP High Speed Centrifuge | Beckman Coulter | 8043-30-1124 | |
| P-10 micro pipette | Gilson | F144802 | |
| P-1000 micro pipette | Gilson | F123601G | |
| P-2 micro pipette | Gilson | F144801 | |
| P-20 micro pipette | Gilson | F123600 | |
| P-200 micro pipette | Gilson | F123602G | |
| PBS | Fisher Scientific | BP399500 | |
| PCR tubes 0.2 mL | Fisher Scientific | 14-230-205 | |
| Peptone | Fisher Scientific | BP1420-500 | |
| Pipette tips - 10 μL | Fisher Scientific | 02-717-157 | |
| Pipette tips - 1000 μL | Fisher Scientific | 02-717-166 | |
| Pipette tips - 20 μL | Fisher Scientific | 02-717-161 | |
| Pipette tips - 200 μL | Fisher Scientific | 02-717-165 | |
| Polycarbonate filters - 47mm, 0.2 μm pore size | Millipore | GTTP04700 | |
| Screw-cap conical centrifuge tubes 15 mL | Falcon | 352096 | Or other tube suitable for bacterial culture |
| Sodium chloride | Fisher Scientific | BP358-1 | |
| Swin-Lok Filter - 47mm | Whatman | 4200400 | |
| SYBR Safe DNA Gel stain | ThermoFisher Scientific | S33102 | |
| Syringes - 20 mL | Fisher Scientific | 14-955-460 | |
| TAE Buffer (50x) concentrate | Fisher Scientific | P1332500 | |
| Thickwall Polycarbonate 25 x 89 mm (26.3mL capacity) centrifuge tubes with caps | Beckman Coulter | 355618 | |
| Tryptic Soy Blood Agar Plates | Northeast Laboratory Services | P1100 | Or other agar plate sufficient for growth of host organisms |
| Tryptic Soy Broth (dehydrated powder) | Becton-Dickinson | 211825 | Or other growth media suitable for target organisms |
| Vinyl Anaerobic Chamber | Coy Laboratories | 032714 | If needed for anaerobic organisms |
| Vortex mixer | Scientific Industries | SI-0236 | |
| Yeast Extract | Fisher Scientific | BP1422-500 |
References
- Stewart, E. J. Growing unculturable bacteria. Journal of Bacteriology. 194 (16), 4151-4160 (2012).
- Bomar, L., Maltz, M., Colston, S., Graf, J. Directed culturing of microorganisms using metatranscriptomics. mBio. 2 (2), 00012 (2011).
- Derrien, M., Vaughan, E. E., Plugge, C. M., de Vos, W. M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. International Journal of Systematic and Evolutionary Microbiology. 54, 1469-1476 (2004).
- Nelson, M. C., Bomar, L., Maltz, M., Graf, J. Mucinivorans hirudinis gen. nov., sp. nov., an anaerobic, mucin-degrading bacterium isolated from the digestive tract of the medicinal leech Hirudo verbana. International Journal of Systematic and Evolutionary Microbiology. 65, 990-995 (2015).
- Scott, J. C., Klein, B. A., Duran-Pinedo, A., Hu, L., Duncan, M. J. A two-component system regulates hemin acquisition in porphyromonas gingivalis. PLoS One. 8 (9), 0073351 (2013).
- Martin, B., et al. New growth media for oral bacteria. Journal of Microbiological Methods. 153 (22), 10-13 (2018).
- Vartoukian, S. R., et al. In vitro cultivation of 'unculturable' oral bacteria, facilitated by community culture and media supplementation with siderophores. Plos One. 11 (1), 0146926 (2016).
- Strandwitz, P., et al. GABA-modulating bacteria of the human gut microbiota. Nature Microbiology. 4 (3), 396-403 (2019).
- Hug, L. A., et al. A new view of the tree of life. Nature Microbiology. 1, 16048 (2016).
- Castelle, C. J., Banfield, J. F. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell. 172 (6), 1181-1197 (2018).
- He, X., et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proceedings of the National Academy of Sciences of the United States of America. 112 (1), 244-249 (2015).
- Hugenholtz, P., Tyson, G., Webb, R. I., Wagner, A. M., Blackall, L. L. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Applied and Environmental Microbiology. 67 (1), 411-419 (2001).
- Brinig, M., Lepp, P. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Applied and Environmental Microbiology. 69 (3), 1687-1694 (2003).
- Bor, B., et al. Insights obtained by culturing saccharibacteria with their bacterial hosts. Journal of Dental Research. , 689-694 (2020).
- Bor, B., et al. Phenotypic and physiological characterization of the epibiotic interaction between TM7x and its basibiont actinomyces. Microbial Ecology. 71 (1), 243-255 (2016).
- Skopek, R. J., Liljemark, W. F., Bloomquist, C. G., Rudney, J. D. Dental plaque development on defined streptococcal surfaces. Oral Microbiology and Immunology. 8 (1), 16-23 (1993).
- Murugkar, P. P., Collins, A. J., Chen, T., Dewhirst, F. E. Isolation and cultivation of candidate phyla radiation Saccharibacteria (TM7) bacteria in coculture with bacterial hosts. Journal of Oral Microbiology. 12 (1), 1814666 (2020).
- Cross, K. L., et al. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nature Biotechnology. 37 (11), 1314-1321 (2019).
- Kantor, R., et al. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio. 4 (5), 00708-00713 (2013).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved