A subscription to JoVE is required to view this content. Sign in or start your free trial.
Quantification and Whole Genome Characterization of SARS-CoV-2 RNA in Wastewater and Air Samples
In This Article
Summary
This protocol aims to quantify SARS-CoV-2 RNA in wastewater and air samples to be used for wastewater-based epidemiology studies and to assess the exposure risk to SARS-CoV-2 in indoor and outdoor aerosols. This protocol also describes a tiled amplicon long-template sequencing approach for SARS-CoV-2 whole genome characterization.
Abstract
Wastewater-based epidemiology has emerged as a promising and efficacious surveillance system for SARS-CoV-2 and other infectious diseases in many nations. The process typically involves wastewater concentration, nucleic acid extraction, amplification of selected genomic segments, and detection and quantification of the amplified genomic segment. This methodology can similarly be leveraged to detect and quantify infectious agents, such as SARS-CoV-2, in air samples. Initially, SARS-CoV-2 was presumed to spread primarily through close personal contact with droplets generated by an infected individual while speaking, sneezing, coughing, singing, or breathing. However, a growing number of studies have reported the presence of SARS-CoV-2 RNA in the air of healthcare facilities, establishing airborne transmission as a viable route for the virus. This study presents a composite of established protocols to facilitate environmental detection, quantification, and sequencing of viruses from both wastewater and air samples.
Introduction
In December 2019, a novel disease called COVID-19 emerged, caused by a previously unknown coronavirus, SARS-CoV-21. The resulting global pandemic has presented a significant challenge to clinical and public health laboratories worldwide, as a large number of individuals require testing to accurately assess virus transmission and prevalence in the community. However, in many regions, achieving the necessary level of testing in a timely and spatially comprehensive manner is economically unfeasible2,3. Current surveillance systems based on individual clinical diagnostics rely heavily on symptom severity and individual reporting, as well as the extent to which these symptoms overlap with existing diseases circulating in the population4,5,6,7,8,9,10. Consequently, a high number of asymptomatic cases contributes to a significant underestimation of disease burden7,11.
Due to these challenges, wastewater-based epidemiology (WBE) for COVID-19 surveillance was proposed as a complementary surveillance strategy. WBE was first described in 200112, and was initially used to trace cocaine and other illegal drugs13. This approach relies on the assumption that it is possible to calculate the initial concentration of any substance that is stable in wastewater and excreted by humans8,12. WBE has been successfully implemented in many countries as a complementary and efficient surveillance system for SARS-CoV-23,8,14,15,16. The majority of methods to detect human viruses in aquatic environments follow these steps: concentration, nucleic acid extraction, amplification of the genomic segment (or segments) chosen, and detection/quantification of the amplified genomic segment3.
Another important environment for the detection and quantification of SARS-CoV-2 is in air samples. Initially, SARS-CoV-2 was thought to be transmitted mainly through close personal contact with respiratory droplets from aerosols generated by an infected person while speaking, sneezing, coughing, singing, or breathing17. However, several studies began to report the presence of SARS-CoV-2 RNA in the air, especially in healthcare facilities and other enclosed spaces18,19,20,21. Evidence of SARS-CoV-2 viability in air samples taken indoors in hospitals and other enclosed spaces has been found when the virus concentration was sufficiently high22,23,24. Outdoor studies have generally found no evidence of SARS-CoV-2, except in crowded outdoor spaces21,25,26,27,28,29. As of now, airborne transmission of SARS-CoV-2 has been recognized as a mode of transmission30,31. A recent review study shows the differences between outdoors, where risks of airborne transmission are minimal outside of crowded areas, and indoors, where larger risks could be present in poorly ventilated environments in which strong sources (i.e., number of infected people) could be present. A recent comprehensive review study has highlighted the substantial differences between the risks of airborne transmission in outdoor versus indoor environments, particularly in crowded areas with poor ventilation. The study indicates that the risk of airborne transmission is minimal in outdoor environments, where there is a larger volume of air available for the dilution and dispersion of virus particles32. These findings have important implications for public health policies and guidelines related to COVID-19. By recognizing the significant differences in transmission risks between indoor and outdoor environments, policymakers can develop more effective strategies to mitigate the spread of the virus and protect public health.
There are a variety of methods and protocols for the detection, quantification, and sequencing of SARS-CoV-2 from different environmental samples. This method article aims to present a combination of well-established protocols that allow laboratories with different capacity levels to perform environmental detection, quantification, and sequencing of viruses from wastewater and air samples.
Protocol
All methods described here have been published elsewhere and contain small modifications from the original methods.
1. Wastewater collection and sample pre-processing
NOTE: Due to the low concentrations of SARS-CoV-2 RNA in environmental samples, the implementation of a concentration step is crucial for a successful detection33,34,35. Described here is the first reported method for the detection of SARS-CoV-2 in wastewater36.
- Collection and concentration of wastewater sample(s)
- Collect 1 L of 24 h composite wastewater sample. Store the sample at 4 °C and proceed with the protocol within 24 h.
- Homogenize the sample by gently shaking the bottle. Collect 70 mL into a 100 mL centrifugal tube or divide the sample into two centrifugal tubes of 50 mL.
- Spike 20 µL of mengovirus (3.2 x 103 copies/µL) to 70 mL of each sample as an internal control of the concentration process. Alternatively, spike 10 µL of mengovirus (3.2 x 103 copies/µL) into each 50 mL centrifuge tube.
- Homogenize the sample and centrifuge at 700 x g for 10 min to remove large particles and organisms (pellet).
- Use the resulting supernatant for concentration with centrifugal ultrafiltration devices with a cutoff of 10 KDa by centrifuging at 4,000 x g for 40 min at 4 °C. Elute the concentrate by centrifuging at 700 x g for 40 min and inverting the position of the ultrafiltration device.
- Measure the volume of the resulting concentrate. The volume will change depending on the quantity of solids in the sample that clog the centrifugal filters. In this study, the concentrate volumes obtained were between 200-1,200 µL.
- Use the resulting concentrate for RNA extraction using a commercial kit or an in-house method. For the samples used in this study, a specific commercial kit for viral RNA extraction with an elution volume of 50 µL is used.
- Measure the concentration of the extracted RNA with a fluorometric RNA quantification kit. Divide the extracted RNA into aliquots and freeze at -80 °C until further analysis.
NOTE: For each RNA isolation procedure set, a negative control of isolation (NCI) should be included, containing only buffers to detect possible contamination during the extraction.
- Collection of air samples using a Coriolis compact air sampler
- Choose a sampling strategy according to the research goal by defining the sampling frequency and sampling locations.
- Set the flow rate on the air sampler (here, 50 L/min for 30 min). Please note that a flow rate of more than 200 L/min can degrade viral RNA, so it is recommended to use a flow rate below 200 L/min.
- Place a sterile cone in the air sampler before starting the collection by pulsing the start. Once the sampling is finished, add 5-15 mL of sterile phosphate-buffered saline (PBS) into the cone. Gently vortex or shake the sample by hand for 15 s. Store the samples for up to 24 h at 4 °C or at -80 °C in cryotubes.
- An optional concentration step can be performed. Clean and decontaminate the cones after each experiment using bleach.
- Extract RNA using a commercial kit or an in-house method. For the samples in this study, a specific commercial kit for viral RNA extraction with an elution volume of 50 µL is used.
- Measure the concentration of the extracted RNA with a fluorometric RNA quantification kit. Divide the extracted RNA into aliquots and freeze at -80 °C until further analysis.
NOTE: For each RNA isolation procedure set, a negative control of isolation (NCI) should be included, containing only buffers to detect possible contamination during the extraction.
2. Quantification of SARS-CoV-2 RNA by real-time-quantitative polymerase chain reaction (RT-qPCR)
NOTE: The below protocol is according to the CDC 2019-Novel Coronavirus (2019-nCoV) RT-PCR diagnostic panel37. Divide the primer/probe mix into several aliquots to avoid freezing and thawing cycles.
- Prepare the reaction mastermix for each target (mengovirus; SARS-CoV-2 [N1 and N2 genes]) as in Table 1, in a clean hood in the reagent setup room. Mix the primers/probe mix and enzyme by inversion five times. Alternatively, light pulse-vortex the primers/probe mix and enzymefive times.
NOTE: Keep all reagents cold during preparation and use. In order to minimize freeze-thaw cycles, it is highly recommended to freeze in aliquots. - Spin down (1,000 x g for 15-30 s) the tubes to collect the contents at the bottom and place the tubes in a cold rack or on ice.
- Label 1.5 mL microcentrifuge tubes for each target. Add to each microcentrifuge tube the amount of each reagent needed (volume per reaction times the number of reactions including the needed controls). Mix by pipetting up and down and centrifuge for 5 s to collect the contents at the bottom.
- Keep the microcentrifuge tubes in a cold rack and dispense 15 µL into strip PCR tubes or a 96-well plate in a cooling rack. Cover the plate and move it to the nucleic acid handling area (keep it in a cooling rack).
- Thaw an aliquot of extracted RNA and gently vortex for 5 s. Pipette 5 µL of RNA (in addition to 5 µL of non-template control [NTC], negative control of isolation [NCI], and positive control [PC]) to each reaction well or tube containing the previously prepared mastermix. Change gloves often as needed to avoid cross-contamination.
- Cover the entire reaction plate or tubes and gently vortex. Centrifuge (1,000 x g for 15-30 s) the plate or PCR tubes.
- Start the 25 µL RT-qPCR with the following cycling conditions: reverse transcriptase at 45 °C for 10 min; polymerase activation at 95 °C for 10 min; 45 cycles of denaturation at 95 °C for 15 s; and annealing/extension at 60 °C for 45s.
- Calculate the recovery efficiency of mengovirus controls as reported by Conte et al.38:
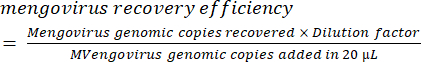 × 100
× 100
3. Sequencing variants in wastewater and data analysis
NOTE: The described protocol is a modified protocol created by Quick et al.39,40. It uses two sets of primers for SARS-CoV-2 genome amplification by PCR tiling methodology-ARTIC primers and VarSkip primers. A combination of primers is used to guarantee the best genome coverage and to minimize the possibility of novel mutations causing primers of one type to fail. In general, the protocol is divided into three parts: reverse transcription (RT) and amplicon generation, sequencing library preparation, and sequencing and data analysis.
- Reverse transcription and amplicon generation
- Ensure the input samples have a known qPCR cycling threshold (Ct) value to correctly dilute them in PCR-grade water. The Ct value is obtained during the quantification with qPCR. The dilutions are as follows: if the Ct value is 12-15, dilute the samples 1:100; if the Ct value is 15-18, dilute the samples 1:10; if the Ct value is 18-35, do not dilute the sample. Samples above a Ct value of 35 have a high chance of not working.
- In a PCR plate, pipette 16 µL of each sample to its position on the plate. Add 4 µL of reverse transcription (RT) mastermix (5x). Cover the plate and start the PCR reaction with the following conditions: 25 °C for 2 min, 55 °C for 20 min, and 95°C for 1 min.
- Prepare 10 µM dilutions of each primer set (ARTIC pool A and pool B, and VarSkip pool A and pool B). Prepare the mastermix for each set of primers (ARTIC set A and B, and VarSkip set A and B) as in Table 2. Four mastermixes will result from this.
- On a new PCR plate, pipette 20 µL of the mastermix to each corresponding well. Add 5 µL of the RT sample to each mix.
NOTE: Each sample will have four reaction mixtures-ARTIC pool A and B, and VarSkip pool A and B. - Cover the plate and start the PCR reaction as follows: polymerase activation at 98 °C for 30 s; 35 cycles of denaturation at 98 °C for 15 s; and annealing/extension at 65 °C for 4 min. Spin down (1,000 x g for 15-30 s) the plate. Combine the ARTIC reaction pools and separately combine the VarSkip reaction pools. Each sample will have two 50 µL amplicon reactions.
- Add 50 µL of bead-based reagent for PCR purification to each well and mix well by pipetting. Incubate at room temperature for 5 min.
NOTE: Before every use, mix the PCR purification reagent vigorously to resuspend the beads. - Place the plate on a magnetic separation stand and wait for the beads to form a pellet and the liquid to clear (~5 min). Pipette and discard the supernatant. Keep the PCR plate on the magnetic stand.
- Maintaining the PCR plate on the magnetic stand, add 200 µL of 80% ethanol to each sample without touching the bead pellet. Remove the ethanol.
- Repeat the previous step. Leave the plate on the magnetic stand uncovered for 30 s to allow the ethanol to evaporate.
- Remove the plate from the magnetic stand and add 15 µL of PCR-grade water to each sample. Resuspend the pellet by pipetting. Incubate at room temperature for 2 min.
- Place the plate back on the magnetic stand and allow the beads to pellet (2 min). Carefully pipette 15 µL of the supernatant from each sample to a new PCR plate.
- Measure the concentration of each sample with a fluorometric RNA quantification kit . Take approximately 50 ng of each sample in 12.5 µL to the next step. If required, dilute the sample in PCR-grade water.
- Library preparation
- Add 1.75 µL of reaction bufferfrom the Illumina DNA library preparation kit and 0.75 µL of enzyme mix to each sample. Cover the plate, vortex briefly, and spin down (1,000 x g for 15-30 s). Incubate at 21°C for 5 min and at 65 °C for 5 min.
- In a new plate, pipette 3 µL of PCR-grade water for each sample to a new PCR plate. Add 0.75 µL of the prepared samples, 1.25 µL of barcodes for sequencing library preparation, and 5 µL of T4 DNA ligase mastermix. Mix well by pipetting, briefly spin down (1,000 x g for 15-30 s) in a centrifuge and incubate at 21 °C for 20 min and at 65 °C for 10 min.
NOTE: If using less than 25 samples, double all the volumes in this step. - Pool all the samples together by adding 10 µL of each sample to the same low-binding tube. Take 480 µL to the next step.
- Add 192 µL of bead-based reagent for PCR purification to the pool and mix well by pipetting. Incubate at room temperature for 10 min.
- Place the tube on a magnetic stand and wait for the supernatant to clear and a bead pellet to form. Remove the supernatant. Remove the tube from the magnetic stand.
- Add 700 µL of the short fragment buffer (SFB) and mix by pipetting. Place back on the magnetic stand and wait for the pellet to form and the liquid to clear (~5 min). Pipette out the supernatant and discard it.
- Repeat the previous step. Leave the tube on the magnetic stand. Add 100 µL of 80% ethanol without touching the bead. Pipette out the ethanol and allow the bead to dry for 30 s.
NOTE: Do not allow the bead to over-dry. - Remove the tube from the magnetic stand and add 35 µL of PCR-grade water. Mix by pipetting and incubate at room temperature for 2 min. Place the tube on the magnetic stand and allow the bead to pellet and the liquid to clear. Pipette 35 µL of the supernatant to a new tube.
- Measure the RNA concentration of the pooled library. Pipette the volume needed to reach an amount of 30-50 ng of RNA and fill it with PCR-grade water to reach a final volume of 30 µL.
- Prepare the adapter ligation reaction mix: 30 µL of the pooled library, 5 µL of Adapter Mix II, 10 µL of the ligation reaction buffer (5x), and 5 µL of T4 DNA ligase. Mix by pipetting and spin-down (1,000 x g for 15-30 s). Incubate at room temperature for 10 min.
- Add 20 µL of the bead-based reagent for PCR purification and mix by pipetting. Incubate for 10 min at room temperature.
- Place the tube on the magnetic stand and wait for the bead pellet to form and the supernatant to become colorless. Carefully pipette out and discard the supernatant.
- Remove from the magnetic stand and add 125 µL of SFB. Mix by pipetting and place back on the magnetic stand to separate the beads. When the liquid is clear, pipette and discard the supernatant.
- Repeat the previous step. Leave the tube open on the magnetic stand for 30 s for some of the leftover liquid to evaporate.
- Remove the tube from the magnetic stand and add 15 µL of the elution buffer. Mix well by pipetting and briefly spin-down (1,000 x g for 15-30 s). Incubate at room temperature for 5 min. Place on the magnetic stand for 2 min.
- Pipette 15 µL of the supernatant into a new low-binding tube. This is the final library. Measure the concentration with a fluorometric DNA quantification kit.
NOTE: In the next step, 12 µL is needed. If the concentration is very high and less than 12 µL of the library is needed, fill up to 12 µL with the elution buffer reagent.
- Flow cell loading and sequencing
- Insert the flow cell (R9.4.1) into the real-time DNA and RNA sequencing device.
- Prepare the priming mix by adding 30 µL of flush tether (FLT) reagent to a tube of flush buffer (FB) reagent. Vortex to mix and spin down (1,000 x g for 15-30 s).
- Open the priming port cover. Using a 1,000 µL pipette, set to 200 µL and insert the tip into the priming port. Turn the volume setting wheel and increase the volume until the liquid in the tip is seen. Discard the tip.
NOTE: Only turn the wheel until a few µL of liquid in the tip is seen. Pulling larger amounts could damage the flow cell. - Slowly add 800 µL of the priming mix to the priming port, taking care not to introduce bubbles. Wait for 5 min.
- In the meantime, prepare the libraries for loading. Mix 37.5 µL of the sequencing buffer, 25.5 µL of the loading buffer, and 12 µL of the library.
NOTE: Mix the loading buffer before pipetting. The beads in this reagent settle very quickly. - Open the port cover. Pipette 200 µL of the priming mix into the priming port.
NOTE: While pipetting into the priming port with the cover port open, one will notice small drops coming from the sample port. Pipette slow enough so the sample port can pull the drops in without spilling over. - Before loading the prepared library, mix well by pipetting. Using a 100 µL pipette set to 75 µL, take the library and pipette drop by drop into the sample port. Do not touch the port and wait for each drop to be absorbed into the port before loading the next drop to avoid overflowing of the liquid.
- Close the cover port and priming port. Open the software and start the sequencing experiment. Select basecalling On. For a library with 96 multiplexed samples, 10-12 h of sequencing is usually enough to generate sufficient data.
NOTE: Using the software, one can insert the sample sheet in .csv format. The sample sheet requires the following data: flow_cell_id, position_id, sample_id, experiment_id, flow_cell_product_code, and kit. The flow cell ID is needed (listed on the side of the flow cell) together with the kit used. For the suggested protocol, the LSK109 kit should be selected.
- Sequencing data analysis
- Insert the compressed fastq reads into the wf-artic workflow41. Run the workflow separately with two different primer schemes (ARTIC/V4.1 and NEB-VarSkip/v1a). Two ". primertrimmed.rg.sorted.bam" files are created for each sample.
- Merge the BAM files into a single file with the "samtools merge" command42. Insert the merged BAM file into Freyja tool, leaving default settings, to recover relative lineage abundances43.
- To create a FASTA file, insert the merged BAM file into the "samtools mpileup" and "ivar" tools with default settings44,45. For mutation calling, load the FASTA file into the Nextclade tool46,47.
Results
The results summarized in Table 3 show examples of the detection and quantification of SARS-CoV-2 RNA in wastewater and air samples using the method described in this article. Wastewater samples were collected from wastewater treatment plants in Spain and Slovenia and were considered positive if the Ct was less than 40 in at least two of the three replicates, with quantification considered valid if the Ct had a variation of less than 5%. In Spain and Portugal, indoor and outdoor air samples were collecte...
Discussion
Microbial and viral detection and quantification using (RT-)qPCR methods have garnered widespread acceptance due to their remarkable sensitivity. However, these techniques face numerous challenges when analyzing environmental samples. Wastewater samples contain an abundance of inhibitory substances that can skew measurements and generate misleading results. To tackle these limitations and enhance precision, a complex protocol was conceived, designed, and implemented. This protocol was tailored by combining protocols from...
Disclosures
The authors have no competing economic interests or other conflicts of interest.
Acknowledgements
This work was performed with financial support from the Regional Government of Castilla y Leon and the FEDER program (projects CLU 2017-09, UIC315 and VA266P20).
Materials
| Name | Company | Catalog Number | Comments |
| Adapter+A25+A2:D19+A2:D20+A2+A2:D19 | Oxford Nanopore | EXP-AMII001 | Sequencing |
| AllPrep PowerViral DNA/RNA Kit | Qiagen | 28000-50 | RNA extraction kit |
| AMPure XP | Beckman Coulter | A63880 | PCR Purification, NGS Clean-up, PCR clean-up |
| ARTIC SARS-CoV-2 Amplicon Panel | IDT | 10011442 | SARS-CoV-2 genome amplification |
| Blunt/TA Ligase Master Mix | NEB | M0367S | Library preparation |
| CENTRICON PLUS70 10KDA. | Fisher Scientific | 10296062 | Concentration filters |
| CORIOLIS COMPACT AIR SAMPLER | Bertin Technologies | 083-DU001 | Air sampler |
| Duran laboratory bottles | Merck | Z305200-10EA | Sampling Bottles |
| Flow Cell (R9.4.1) | Oxford Nanopore | FLO-MIN106D | Sequencing |
| General labarotory consumables (tubes, qPCR plates, etc) | |||
| Ligation Sequencing Kit | Oxford Nanopore | SQK-LSK109 | Sequencing |
| LunaScript RT SuperMix Kit | NEB | E3010 | cDNA synthesis |
| Mengovirus extraction control Kit | Biomérieux | KMG | Concentration control |
| Nalgene General Long-Term Storage Cryogenic Tubes | Thermofisher | 5011-0012 | Sample storage |
| Native Barcoding Expansion 1-12 (PCR-free | Oxford Nanopore | EXP-NBD104 | Barcoding |
| NEBNext Ultra II End Repair/dA-Tailing Module | NEB | E7595 | DNA repair |
| NEBNext VarSkip Short SARS-CoV-2 Primer Mixes | NEB | E7658 | SARS-CoV-2 genome amplification |
| NEBNext Quick Ligation Reaction Buffer | NEB | B6058S | Sequencing |
| Phosphate buffered saline | Merck | P4474 | Collection buffer |
| Phosphate-buffered saline (PBS, 1X), sterile-filtered | Thermofisher | J61196.AP | Elution of air samples |
| Q5 Hot Start High-Fidelity 2X Master Mix | NEB | M0494S | hot start DNA polymerase |
| Qubit RNA HS Assay Kit | Thermofisher | Q32852 | RNA quantitation |
| SARS-CoV-2 RUO qPCR Primer & Probe Kit | IDT | 10006713 | Primer-Probe mix and qPCR positive control |
| TaqPath 1-Step RT-qPCR Master Mix | Thermofisher | A15299 | RT-qPCR kit |
References
- Naming the Coronavirus Disease (COVID-19) and the Virus that Causes it. World Health Organization Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease (2020)
- Lab Workplace Safety. Centers for Disease Control and Prevention Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/lab-safety-practices.html (2020)
- Gonçalves, J., et al. Centralized and decentralized wastewater-based epidemiology to infer COVID-19 transmission - A brief review. One Health. 15, 100405 (2022).
- Dawood, F. S., et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. The Lancet Infectious Diseases. 12 (9), 687-695 (2012).
- Gonçalves, J., Koritnik, T., Paragi, M. Assessment of weather and atmospheric pollution as a co-factor in the spread of SARS-CoV-2. Acta Bio-Medica: Atenei Parmensis. 92 (3), e2021094 (2021).
- Gonçalves, J., et al. Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. The Science of The Total Environment. 755, 143226 (2021).
- Mizumoto, K., Kagaya, K., Zarebski, A., Chowell, G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 25 (10), 2000180 (2020).
- Polo, D., et al. Making waves: Wastewater-based epidemiology for COVID-19 - approaches and challenges for surveillance and prediction. Water Research. 186, 116404 (2020).
- Shmueli, G., Burkom, H. Statistical challenges facing early outbreak detection in biosurveillance. Technometrics. 52 (1), 39-51 (2010).
- Simonsen, L., et al. Global mortality estimates for the 2009 influenza pandemic from the GLaMOR project: A modeling study. PLoS Medicine. 10 (11), e1001558 (2013).
- Oran, D. P., Topol, E. J. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative reivew. Annals of Internal Medicine. 173, 362-367 (2020).
- Daughton, C., Jones-Lepp, T. Pharmaceuticals and Personal Care Products in the Environment: Scientific and Regulatory Issues. ACS Symposium Series. , (2001).
- Zuccato, E., et al. Cocaine in surface waters: a new evidence-based tool to monitor community drug abuse. Environmental Health. 4, 14 (2005).
- Aguiar-Oliveira, M. d. e. L., et al. Wastewater-based epidemiology (WBE) and viral detection in polluted surface water: A valuable tool for COVID-19 surveillance-a brief review. International Journal of Environmental Research and Public Health. 17, 9251 (2020).
- García-Encina, P. A. Wastewater-based epidemiology (WBE). Water and Environment Journal. 35 (4), 1162-1163 (2021).
- Mao, K., Zhang, H., Pan, Y., Yang, Z. Biosensors for wastewater-based epidemiology for monitoring public health. Water Research. 191, 116787 (2021).
- Shereen, M. A., Khan, S., Kazmi, A., Bashir, N., Siddique, R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research. 24, 91-98 (2020).
- Chia, P. Y., et al. Detection of air and surface contamination by SARS-CoV-2 in hospital rooms of infected patients. Nature Communications. 11 (1), 2800 (2020).
- Lei, H., et al. SARS-CoV-2 environmental contamination associated with persistently infected COVID-19 patients. Influenza and Other Respiratory Viruses. 14 (6), 688-699 (2020).
- Razzini, K., et al. SARS-CoV-2 RNA detection in the air and on surfaces in the COVID-19 ward of a hospital in Milan, Italy. The Science of The Total Environment. 742, 140540 (2020).
- da Silva, P. G., Gonçalves, J., Nascimento, M. S. J., Sousa, S. I. V., Mesquita, J. R. Detection of SARS-CoV-2 in the indoor and outdoor areas of urban public transport systems of three major cities of Portugal in 2021. International Journal of Environmental Research and Public Health. 19 (10), 5955 (2022).
- Barbieri, P., et al. Molecular detection of SARS-CoV-2 from indoor air samples in environmental monitoring needs adequate temporal coverage and infectivity assessment. Environmental Research. 198, 111200 (2021).
- Lednicky, J., et al. Earliest detection to date of SARS-CoV-2 in Florida: Identification together with influenza virus on the main entry door of a university building, February 2020. PLoS One. 16 (1), 0245352 (2021).
- Santarpia, J. L., et al. Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Scientific Reports. 10 (1), 12732 (2020).
- Chirizzi, D., et al. SARS-CoV-2 concentrations and virus-laden aerosol size distributions in outdoor air in north and south of Italy. Environment International. 146, 106255 (2021).
- Hadei, M., et al. Presence of SARS-CoV-2 in the air of public places and transportation. Atmospheric Pollution Research. 12 (3), 302-306 (2021).
- Moreno, T., et al. Tracing surface and airborne SARS-CoV-2 RNA inside public buses and subway trains. Environment International. 147, 106326 (2021).
- Mouchtouri, V. A., et al. Environmental contamination of SARS-CoV-2 on surfaces, air-conditioner and ventilation systems. International Journal of Hygiene and Environmental Health. 230, 113599 (2020).
- Setti, L., et al. Airborne transmission route of COVID-19: why 2 meters/6 feet of inter-personal distance could not be enough. International Journal of Environmental Research and Public Health. 17, 2932 (2020).
- SARS-CoV-2 Transmission. Centers for Disease Control and Prevention (CDC) Available from: https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/sars-cov-2-transmission.html (2021)
- Coronavirus Disease (COVID-19): How is it Transmitted. World Health Organization Available from: https://www.who.int/news-room/q-a-detail/coronavirus-disease-covid-19-how-is-it-transmitted (2021)
- Dinoi, A., et al. A review on measurements of SARS-CoV-2 genetic material in air in outdoor and indoor environments: Implication for airborne transmission. The Science of the Total Environment. 809, 151137 (2022).
- Bosch, A., et al. Analytical methods for virus detection in water and food. Food Analytical Methods. 4, 4-12 (2011).
- Gonçalves, J., et al. Surveillance of human enteric viruses in coastal waters using concentration with methacrylate monolithic supports prior to detection by RT-qPCR. Marine Pollution Bulletin. 128, 307-317 (2018).
- La Rosa, G., Muscillo, M. Molecular detection of viruses in water and sewage. Viruses in Food and Water. , 97-125 (2013).
- Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., Brouwer, A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environmental Science & Technology Letters. 7 (7), 511-516 (2020).
- CDC - 2019-nCoV Real-Time RT-PCR Diagnostic Panel Fact Sheet for Healthcare Providers. Centers for Disease Control and Prevention Available from: https://stacks.cdc.gov/view/cdc/85028 (2020)
- Conte, M. Airborne concentrations of SARS-CoV-2 in indoor community environments in Italy. Environmental Science and Pollution Research International. 29 (10), 13905-13916 (2022).
- nCoV-2019 sequencing protocol v3 (LoCost). protocols.io Available from: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye (2020)
- Tyson, J. R. . Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. , (2020).
- . ARTIC SARS-CoV-2 Workflow Available from: https://github.com/epi2me-labs/wf-artic (2022)
- Li, H., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 25 (16), 2078-2079 (2009).
- . Freyja Available from: https://github.com/andersen-lab/Freyja (2022)
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 27 (21), 2987-2993 (2011).
- Grubaugh, N. D., et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biology. 20 (1), 8 (2019).
- Hadfield, J., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 34 (23), 4121-4123 (2018).
- Aksamentov, I., Roemer, C., Hodcroft, E. B., Neher, R. A. Nextclade: clade assignment, mutation calling and quality control for viral genomes. Journal of Open Source Software. 6 (67), 3773 (2021).
- Markt, R., et al. Detection and stability of SARS-CoV-2 fragments in wastewater: impact of storage temperature. Pathogens. 10 (9), 1215 (2021).
- Kocamemi, B. A., et al. First Data-Set on SARS-CoV-2 Detection for Istanbul Wastewaters in Turkey. MedRxiv. , (2020).
- Randazzo, W., et al. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Research. 181, 115942 (2020).
- Hoorfar, J., et al. Practical considerations in design of internal amplification controls for diagnostic PCR assays. Journal of Clinical Microbiology. 42 (5), 1863-1868 (2004).
- Parshionikar, S. U., Cashdollar, J., Shay Fout, G. Development of homologous viral internal controls for use in RT-PCR assays of waterborne enteric viruses. Journal of Virological Methods. 121, 39-48 (2004).
- Nalla, A. K. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. Journal of Clinical Microbiology. 58 (6), 00557 (2020).
- Hirotsu, Y., Mochizuki, H., Omata, M. Double-quencher probes improve detection sensitivity toward Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in a reverse-transcription polymerase chain reaction (RT-PCR) assay. Journal of Virological Methods. 284, 113926 (2020).
- Ahmed, W. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. The Science of The Total Environment. 728, 138764 (2020).
- Bar-Or, I., et al. Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. The Science of the Total Environment. 789, 148002 (2021).
- La Rosa, G., Bonadonna, L., Lucentini, L., Kenmoe, S., Suffredini, E. Coronavirus in water environments: Occurrence, persistence and concentration methods - A scoping review. Water Research. 179, 115899 (2020).
- Wu, F., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 5, 00614 (2020).
- Wurtzer, S., et al. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. European Communicable Disease Bulletin. 25 (50), 2000776 (2020).
- . VATar COVID-19 | Caso de Exito - Ministerio para la Transición Ecologica y el Reto Demografico Available from: https://esri.es/es-es/descubre-los-gis/casos-de-exito/administracion-/vatar-covod19-miteco-cs (2022)
- Nemudryi, A., et al. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Reports. Medicine. 1 (6), 100098 (2020).
- Rios, G., et al. Monitoring SARS-CoV-2 variants alterations in Nice neighborhoods by wastewater nanopore sequencing. The Lancet Regional Health. Europe. 10, 100202 (2021).
- Gomes da Silva, P. Environmental dissemination of SARS-CoV-2 in a University Hospital during the COVID-19 5th wave Delta variant peak in Castile-León, Spain. International Journal of Environmental Research and Public Health. 20, 1574 (2023).
- Gonçalves, J., et al. . Exposure assessment of SARS-CoV-2 and Nov GII/GII in aerosols generated by a municipal wastewater treatment plant. , (2022).
- Lednicky, J. A., et al. Isolation of SARS-CoV-2 from the air in a car driven by a COVID patient with mild illness. International Journal of Infectious Diseases. 108, 212-216 (2021).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved