Improved Methods for Preparing Transverse Sections and Unrolled Whole Mounts of Maize Leaf Primordia for Fluorescence and Confocal Imaging
In This Article
Summary
Maize leaf primordia are deeply ensheathed and rolled, making them difficult to study. Here, we present methods for preparing transverse sections and unrolled whole mounts of maize leaf primordia for fluorescence and confocal imaging.
Abstract
In maize (Zea mays) and other grasses (Poaceae), the leaf primordia are deeply ensheathed and rolled within the leaf whorl, making it difficult to study early leaf development. Here, we describe methods for preparing transverse sections and unrolled whole mounts of maize leaf primordia for fluorescence and confocal imaging. The first method uses a wire stripper to remove the upper portions of older leaves, exposing the tip of the leaf primordium and allowing its measurement for more accurate transverse section sampling. The second method uses clear, double-sided nano tape to unroll and mount whole-leaf primordia for imaging. We show the utility of the two methods in visualizing and analyzing fluorescent protein reporters in maize. These methods provide a solution to the challenges presented by the distinctive morphology of maize leaf primordia and will be useful for visualizing and quantifying leaf anatomical and developmental traits in maize and other grass species.
Introduction
Grass crops are a major source of food and biofuel for the global population1, and improving leaf anatomy has the potential to increase their productivity2,3. However, our current understanding of how leaf anatomy is regulated in grasses is limited4 and requires the analysis of leaf primordia, as many anatomical and physiological traits of the leaf are predetermined early in development5,6,7. Cellular imaging techniques, such as fluorescence and confocal imaging, are indispensable for studying grass leaf anatomy and cellular traits, but these techniques are difficult to apply to grass leaf primordia because they are deeply ensheathed and rolled within the leaf whorl. We addressed this issue by developing methods for preparing transverse sections and unrolled whole-leaf mounts for fluorescence and confocal analysis of maize leaf primordia, a model system for studying grass leaf anatomy and development2,8.
The maize leaf, like all grass leaves, consists of a strap-like blade with a sheath that wraps around the stem and developing shoot9,10,11,12,13. The leaves develop from the shoot apical meristem (SAM) in a distichous pattern, where each new leaf initiates in the opposite position of the previous leaf, resulting in two ranks of leaves along the vertical axis (Figure 1A)14 . The developmental stage of each leaf primordium is identified by its position relative to the SAM, with the closest primordium designated as plastochron1 (P1) and the following primordia designated as P2, P3, and so on (Figure 1B,C)2. During development (Figure 1D), the leaf primordium first appears as a crescent-shaped buttress around the base of the SAM (P1), and then grows into a hood-shaped primordium that extends over the meristem (P2)9,10,11. The basal margins of the hood then expand laterally and overlap each other as the tip grows upward, forming a cone-shaped primordium (P3-P5)10. The primordium then rapidly grows in length, and the sheath-blade boundary at the base becomes more prominent with the formation of the ligule, the fringe-like projection on the adaxial side of the leaf (P6/P7). Finally, the leaf unrolls as it emerges from the whorl during steady-state growth, in which the dividing cells are restricted within the small basal region of the blade, forming a gradient with expanding and differentiating cells along the proximal-distal axis (P7/P8)15. The shoot apex of a maize seedling contains multiple primordia at different stages of development, making it an excellent model for studying leaf development8.
Accurate analysis of early leaf development requires staging or the use of standardized criteria to define distinct stages of primordium development in relation to other growth or morphological parameters. Because the leaf primordia are hidden within the grass shoot, investigators typically use parameters such as the age of the plant or the size of the emerging leaves as predictors for the stages and sizes of the leaf primordia9,16. In maize, the chronological age of the plant is determined either by the number of days after planting or germination (DAP/DAG)17,18. The vegetative stage (V stage) is determined by the uppermost leaf with a visible collar, a pale line on the abaxial side between the blade and the sheath that corresponds to the position of the ligule and auricles, a pair of wedge-shaped regions at the base of the blade (Figure 1A,B)17,19 . Between 20 and 25 DAG, the SAM transitions into an inflorescence meristem and ceases to produce new leaves20. The growth rates of maize leaf primordia can vary depending on the environment and the genotype of the plant. For this reason, plant age and the size of the emerging leaves cannot accurately predict the sizes of leaf primordia; however, using these parameters can help predict the range of primordia stages and sizes for experimental purposes.
Transverse section analysis is a popular method for examining leaf anatomy and development in maize and other grasses because it allows for the sampling of multiple plastochrons in a single section across the shoot21,22,23. This method is also convenient for cellular imaging of fresh samples, as the surrounding leaves serve as a scaffold that keeps the leaf primordia in place during sectioning and mounting24. However, a disadvantage of this method is that it can be challenging to precisely locate the target plastochron and region within the primordium when sectioning from an intact shoot. Furthermore, because leaf growth varies across plastochrons and along the proximal-distal axis2,5, imprecise sampling could result in an incorrect interpretation of the developmental stage and region of the primordium in a given section. Therefore, developing a method for precise transverse section sampling is critical for ensuring the accuracy and reproducibility of anatomical and developmental analyses of grass leaf primordia.
Whole-leaf mount analysis enables the comprehensive and integrative investigation of tissue and cellular processes that occur at the whole-organ scale, such as proliferative growth25 and vein patterning26,27,28. The method provides a paradermal overview of the leaf, allowing the discovery of distinct processes and patterns that would otherwise be difficult to detect using transverse section analysis24,27. Unlike in Arabidopsis, where there are already established methods for imaging whole-leaf mounts29,30, there is currently no standard method for imaging unrolled whole-leaf mounts in grasses. A previous protocol for unrolling isolated maize leaf primordia involved uncommon materials and was not suitable for cellular imaging31. Advanced imaging techniques, such as computed tomography (CT) and magnetic resonance imaging (MRI), can acquire 3D anatomical information without isolating and unrolling the primordia11,32,33, but they are expensive and require specialized equipment. Developing a technique to overcome the constraints imposed by the rolled and conical morphology of leaf primordia in maize and other grasses would advance investigations into their anatomical and developmental traits.
Here, we present methods for preparing transverse sections and unrolled whole mounts of maize leaf primordia for fluorescence and confocal imaging. We used these methods to quantify vein number and map the spatial-temporal hormone distribution in maize leaf primordia with fluorescent proteins (FPs)24. The first method involves removing the upper portion of older leaves from maize seedlings with a wire stripper (Figure 1E). By exposing the tip of the primordium (P5-P7), it becomes possible to determine its length without having to completely remove the older surrounding leaves, thus enabling easy and accurate sectioning. The second method involves unrolling and mounting whole-leaf primordia (P3-P7) with clear, double-sided nano tape (Figure 1F). These methods are suitable for visualizing various FPs24 but need optimization for using fluorescent dyes and clearing reagents. In addition, we outline some procedures for flattening z-stacks, stitching images, and merging channels in ImageJ/FIJI34, which apply to the images produced by the two methods. These methods are useful for routine fluorescence or confocal imaging of maize leaves, but they can also be adapted for other model grass species, such as rice, Setaria, and Brachypodium.
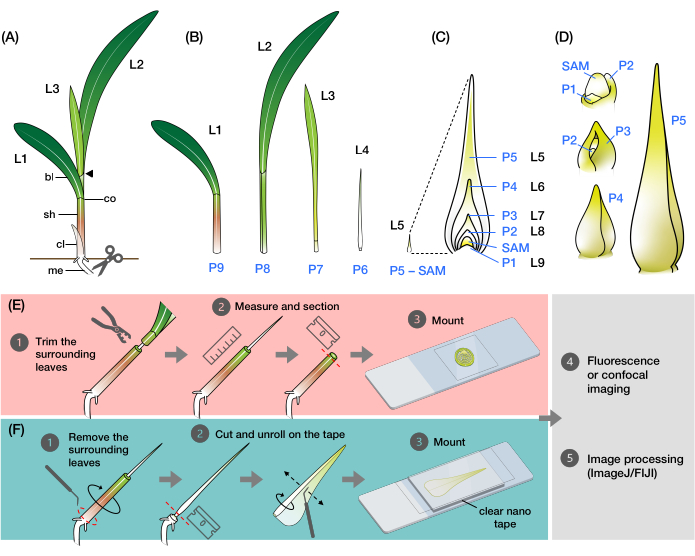
Figure 1: Organization and morphology of maize leaf primordia and overview of the methods. (A) Schematic representation of a maize seedling. Maize has a distichous phyllotaxy, with the new leaf initiating at the opposite position of the previous leaf. The leaf number indicates the chronological order in which the leaves emerged from germination (i.e., first leaf, L1; second leaf, L2; third leaf, L3; etc.). Each leaf has a distal blade and a basal sheath delineated by a collar that corresponds to the ligule and auricle. The uppermost leaf with the collar visible denotes the vegetative stage. The seedling in this example is at the V2 stage, with the L2 collar (arrowhead) visible. The scissors icon indicates the location at the mesocotyl (me) where the seedling must be cut in order to be collected. (B) Schematic representation of the dissected shoot showing isolated L1 to L4, with the leaf primordia L5 to L9 shown as an enlarged image in (C). The plastochron number indicates the position of the primordium relative to the SAM, with the youngest leaf primordium (P1) closest to the SAM and the older leaf primordia (P2, P3, P4, and so on) successively farther away2. (D) Schematic representation of the morphology of maize leaf primordia from P1 to P5. (E) Schematic overview of the method for transverse section analysis of the maize leaf primordia. (1) Trim the older leaves with a wire stripper. (2) Measure the primordium and section the shoot. (3) Mount the section on a slide for imaging and processing (4, 5). (F) Schematic overview of the method for whole-mount analysis of the maize leaf primordia. (1) Remove the surrounding leaves to extract the primordium. (2) Cut and unroll the primordium flat on the nano tape. (3) Mount the sample for imaging and processing (4, 5). Abbreviations: L = leaf; bl = blade; sh = sheath; co = collar; me = mesocotyl; V = vegetative; P = plastochron; SAM = shoot apical meristem. Please click here to view a larger version of this figure.
Protocol
1. Staging maize leaf development and designing the experiment
- Growing the plants and staging leaf development
- Determine the age and developmental stage of the plants to be used for the experiment and perform a detailed staging of leaf development.
NOTE: It is recommended that the staging be performed on plants with the same genetic background as the mutant or transgenic lines that will be used in the experiments. The SAM stops producing new leaves between 20 and 25 DAG, depending on the genetic background and growth conditions. For this reason, the methods described here are ideal for maize seedlings aged 7-14 DAG. - Grow the maize plants in a greenhouse or growth chamber following a suitable plant care protocol35.
- Collect the plants at the desired age or V stage with a small knife or pair of scissors by cutting at the mesocotyl, which is the stem beneath the soil surface (Figure 1A). Carefully insert the cutting tool into the soil beside the seedling and slide it down to the base at a 45° angle to cut the mesocotyl.
- Lift the seedling out of the soil and dust away any remaining dirt or soil particles that may be clinging to the plant. Remove the coleoptile, the protective sheath covering the emerging shoot in young seedlings, by hand.
- For each plant, record the V stage, the number of leaves, and the length of the last leaf emerging from the whorl. Take photos of the plants for future reference.
- Remove the older leaves one by one. To do this, hold the plant by the remnants of the mesocotyl or stem, and excise each leaf from the base of the sheath and gently unfurl the sheath in a circling fashion with a dental probe (Figure 1B).
NOTE: See step 2.1 for a quick method for removing the surrounding leaves. - Under a stereo microscope at around 2x magnification, carefully dissect the rest of the shoot apex on damp wipes to prevent the sample from drying out. Gently excise and unfurl each leaf primordium with a dental probe with a bent needle or a pair of fine forceps until the SAM, P1, and P2 are visible (Figure 1D). Record the appearance and measurement of each plastochron.
NOTE: See an example of leaf development staging in maize seedlings in Supplementary File 1.
- Determine the age and developmental stage of the plants to be used for the experiment and perform a detailed staging of leaf development.
- Planning the experiment
NOTE: Planning the experiment carefully can improve efficiency. For instance, in transverse section analysis, determining the approximate location to cut when imaging specific plastochrons or regions can reduce the dissection time. Figure 2 shows an example of standardized sampling. Furthermore, optimizing the imaging parameters based on the properties of the FPs or fluorescent probes can improve the efficiency of the experiments.- Utilize the staging information as a guide to focus the experiment on specific plastochrons, primordium regions, and tissues. Use the FPs or other fluorescent probes of interest to follow sections 2 and/or 3 below on a few samples.
NOTE: See available maize FP marker line36-42 seed stocks at https://www.maizegdb.org/data_center/stock. - Determine the optimal image acquisition parameters for each FP reporter or probe, including the appropriate detector or filter, excitation and emission wavelengths, pinhole size, and other settings. Save the settings to reproduce the working conditions for each of the experiments.
NOTE: See Linh and Scarpella30 for some technical guidelines for confocal laser scanning microscopy of developing leaves. - Plan the imaging time based on the half-life and other properties of the FP reporter43.
NOTE: The time between dissection and imaging must also be considered when planning an experiment, as fresh plant samples are subjected to desiccation and cellular changes during this period.
- Utilize the staging information as a guide to focus the experiment on specific plastochrons, primordium regions, and tissues. Use the FPs or other fluorescent probes of interest to follow sections 2 and/or 3 below on a few samples.
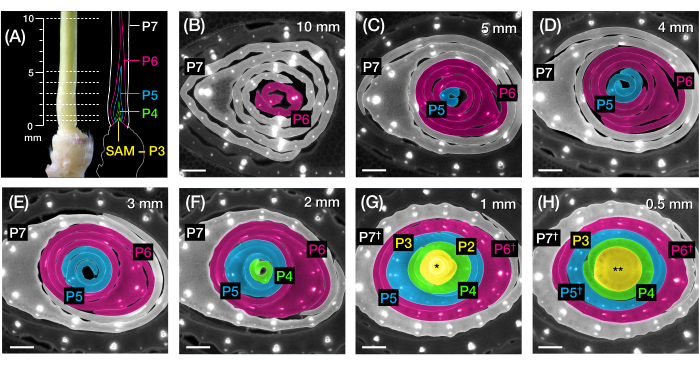
Figure 2: Sampling scheme for transverse section analysis of maize leaf primordia. (A, left) Proximal shoot of a 7 DAG maize seedling, showing an exposed fourth leaf (L4) at P7. The broken lines indicate the seven sampling points along the primordium, from 0.5 mm to 10 mm. (A, right) Schematic representation of the leaf primordia, with the projected size and position of each plastochron: P7 (white); P6 (magenta); P5 (blue); P4 (green); and P3 until the SAM (yellow). (B-H) Fluorescence images of transverse sections representing the sampling points shown in A from 10 mm (B) to 0.5 mm (H). The primordia are pseudocolored according to the plastochron color scheme in (A). The sections were imaged with an epifluorescence microscope using a longpass-emission UV filter for autofluorescence. Scale bar = 200 µm (B-H). This figure has been modified and reproduced with permission from Robil and McSteen24. Abbreviations: DAG = days after germination; P = plastochron; SAM = shoot apical meristem; † = leaf sheath or pre-ligule proper; * = shoot apical meristem; ** = stem. Please click here to view a larger version of this figure.
2. Imaging transverse sections of maize leaf primordia
- Stripping older surrounding leaves and measuring the leaf primordium
- Collect the seedling by carefully cutting it at the mesocotyl beneath the soil surface with a small knife or pair of scissors, as described in steps 1.1.3-1.1.4 (Figure 1A).
- Determine the correct wire stripper hole size to be used and where the cut will be made along the shoot. Make sure that the hole is large enough to fit the target primordium.
- Begin cutting at the more distal part of the shoot and gradually work toward the base until the tip of the primordium is exposed.
- To make the cut, hold the wire stripper with the jaws facing the shoot. Position the shoot to the selected hole and squeeze the stripper handles together to cut through the leaf whorl.
NOTE: To avoid accidentally cutting the target primordium, carefully align the center of the shoot with the hole in the wire stripper. For larger plants, it is recommended to manually remove some of the surrounding leaves by hand before using the stripper.
CAUTION: Wire strippers have sharp jaws that can cause cuts or other injuries if not used carefully. - While continuing to squeeze the handles together, slide the stripper away from the shoot to trim the upper portion of the leaves. Ensure that the target region in the primordium remains ensheathed by the surrounding leaves (Supplementary Figure S1C,D).
NOTE: After trimming the upper leaves, the shoot is left with an exposed primordium tip (P5, P6, or P7, depending on the position of the cut and the size of the wire stripper hole used; Figure 1E). This primordium will serve as the reference for estimating the size and stage of younger primordia. P6 is the most practical reference to use in maize seedlings due to its size range and pin-like morphology. - Use a ruler to measure the length from the tip of the primordium to the base of the shoot. Record the measurement.
NOTE: The length of the primordium will be slightly overestimated as there are a few millimeters of stem at the base of the primordia (Figure 2A).
- Freehand sectioning and imaging of the leaf primordium
- Under a stereo microscope at around 0.8x magnification, hold the shoot steady on a smooth surface, such as a glass cutting board or other scratch-resistant material. Use a razor blade or scalpel to cut thin sections (0.25-0.8 mm) of the shoot at the desired points along the length of the primordium.
- Make an initial cut slightly above the target region to discard the distal portion of the shoot, and then obtain thin sections around the target region. To make clean cuts, hold the blade perpendicular to the shoot and slide it inward with a smooth, even motion.
NOTE: There may be a few millimeters of sectioning leeway for larger primordia (P6 or P7). However, for smaller primordia (P5 and earlier), 1 mm can make a significant difference in sampling, as these primordia are stacked up within the first few millimeters of the base of the shoot (see Figure 2A,F-H). It is recommended to use a double-edged razor blade for finer sectioning.
CAUTION: Exercise caution when using razor blades and scalpels to avoid any accidental cuts or injuries. - Replace the razor blade regularly, since the leaf sheaths can easily dull the blade. Cover the shoot with damp wipes after each round of sectioning to prevent the sample from drying out.
- Mount the leaf section on a clean glass slide (75 mm x 25 mm), use a pipette to apply a drop of water (or 50% glycerol) directly to the section, and place a coverslip (22 mm x 22 mm) on top.
- If using a fluorescent dye, apply the dye solution to the section, place a coverslip on top, and incubate it as needed.
NOTE: Depending on the type of dye, additional steps for processing the leaf sections may be necessary before proceeding to the next step. These references44,45,46,47 provide general details and uses of fluorescent dyes in plant cell imaging. A nucleus-staining fluorescent dye, 5-ethynyl-2′-deoxyuridine (EdU), has been used effectively on transverse sections of maize leaf primordia48. By contrast, plasma membrane-staining dyes, such as Fei Mao 4-64 (FM 4-64) and propidium iodide, do not produce satisfactory results (Figure 3A-D). - Place the slide on the stage of an epifluorescence or confocal laser scanning microscope and adjust the focus and settings as needed to visualize the fluorophore30. See Table 1 for the illumination and image acquisition settings used for selected maize FP reporter lines.
NOTE: Thick sections may produce high levels of background autofluorescence. Consider some imaging and specimen preparation strategies that can help mitigate this problem30,49 (see Table 2).
CAUTION: Use caution and wear protective eyewear when working with powerful light sources and lasers to reduce the risk of eye damage. - Image through the leaf section at different magnifications and detection channels (including the bright-field illumination)30. Use 4x magnification to image the entire transverse section and 20x or 40x magnification to image specific tissues. Reduce the exposure time at higher magnifications for epifluorescence imaging to protect the samples from rapid photobleaching (see Table 1).
- Capture images of the entire section or specific regions of interest (ROIs). If necessary, acquire a z-stack, which is a series of images captured at different focal depths30.
- To acquire a z-stack, first determine the upper and lower positions in the specimen, then the number of optical sections that must be captured between these positions. Fewer optical sections result in a larger z-step size, which is the distance between each section. Depending on the properties of the FP and the magnification used, capture three to 25 optical sections with z-step sizes between 1 and 12 µm for imaging transverse sections of maize leaf primordia.
NOTE: To flatten the z-stack, use an appropriate volume rendering technique50 that can be accomplished through the microscope software or by using ImageJ/FIJI34 (see steps 4.1 and 4.2). - Save all the image files and their associated metadata, if available.
Table 1: Illumination and image acquisition settings used for fluorescence and confocal imaging of selected maize FP reporters. Abbreviations: FP = fluorescent protein; TRITC = tetramethylrhodamine; FITC = fluorescein isothiocyanate; WLL = white light laser; Ar-ion = argon ion laser; HyD = hybrid detector; AU = Airy unit; Hz = Hertz, scan line per second. Please click here to download this Table.
3. Imaging unrolled whole mounts of maize leaf primordia
- Dissecting the primordia and preparing the glass slide with nano tape
- Collect the seedling by carefully cutting it at the mesocotyl beneath the soil surface with a small knife or a pair of scissors, as described in steps 1.1.3-1.1.4 (Figure 1A).
- Prepare the glass slide (75 mm x 25 mm) with the clear, double-sided nano tape (Supplementary Figure S1H) before dissecting the plant. Cut a rectangular piece of nano tape and stick it to the center of a clean glass slide. Do not remove the protective plastic film on the top side of the tape.
NOTE: Determine the size of the tape to use according to the size of the samples to image (see examples of oversized samples in Supplementary Figure S1I,J). The nano tape is available in either acrylic or polyurethane gel material. Both types are effective for fluorescence and confocal imaging24. To prevent discoloration of the tape, protect it from prolonged exposure to light by storing it in a dark container. - Begin dissecting the shoot by removing the upper portion of the older leaves with a wire stripper, as described in step 2.1.
- Hold the shoot by the mesocotyl and carefully remove the surrounding leaves with a dental probe to extract the primordia. To do this, remove the leaves from the base of the shoot and unfurl them one at a time until the primordium of interest is exposed.
- Alternatively, use the wire stripper to directly extract the leaf primordia by making a cut at the basal region of the shoot (Supplementary Figure S1E-G).
NOTE: This method carries a higher risk of snapping the primordia.
- Mounting and imaging the primordium.
- Remove the protective film from the tape. Lay the exposed primordium on the tape. Use a razor blade to cut the primordium at the base, discarding the basal stem and the hypocotyl (Figure 1F).
- Unroll the primordium using a dental probe with a bent needle (tip diameter of 0.25-0.6 mm). Use a microprobe (tip diameter of 0.15-0.2 mm) for primordia smaller than 3 mm (P3 to early P4).
- Position the needle tip parallel to the surface and unfurl the outer basal margin, gently pressing it against the tape.
- For smoother unrolling, lubricate the inner surface (adaxial) of the leaf by dipping the tip of the needle into 100% glycerol.
- With the outer margin adhering to the tape, align the needle tip parallel to the long axis of the leaf. Gently slide the needle sideways to unroll and flatten the leaf onto the tape (Figure 1F).
NOTE: Breaking or damaging the leaf while unrolling is common, especially in larger primordia with a robust midrib (see Figure 3E-G). Unrolling the leaf vigorously can bruise its surface and produce artifacts during imaging (Figure 3H). See Table 2 and the discussion for recommended solutions to these problems. - Apply a drop of water to the unrolled primordium. Immediately place a coverslip on top of the water drop and the primordium. Gently press down the edges of the coverslip so that they adhere to the tape.
NOTE: It is recommended to use large rectangular coverslips (50 mm x 24 mm or 60 mm x 24 mm) with extra areas for adhering to the tape. Minimize the formation of air bubbles, which can distort the leaf and cause uneven light refraction (see Figure 3I-K). Make sure that the coverslip adheres completely to the tape, as the margins of the mounted primordium may roll back in under a loosely adhered coverslip (see Figure 3L). - Place the slide on the stage of an epifluorescence (see Supplementary Figure S1K) or confocal laser scanning microscope and adjust the focus and settings as needed to visualize the fluorophore30. See Table 1 for the illumination and image acquisition settings used for the selected maize FP reporter lines.
- Image the entire leaf or a specific ROI through various detection channels30. To image the entire leaf, either manually capture mosaic images of the leaf or use the tiling operation of the microscope at low magnification (4x or 10x).
NOTE: Even at the lowest magnification, maize leaf primordia may be too large to image with an epifluorescence or confocal laser scanning microscope, resulting in longer imaging times that may result in photobleaching (see Figure 3M) or increased background autofluorescence. Therefore, it is recommended to use a fluorescence stereo microscope for imaging whole-leaf samples larger than 5 mm. - For confocal imaging, obtaining z-stacks that encompass the thickness of the leaf will be necessary (see step 2.2.9).
- Save all the image files and their associated metadata, if available.
Table 2: Troubleshooting common problems in imaging transverse sections and whole mounts of maize leaf primordia. Please click here to download this Table.
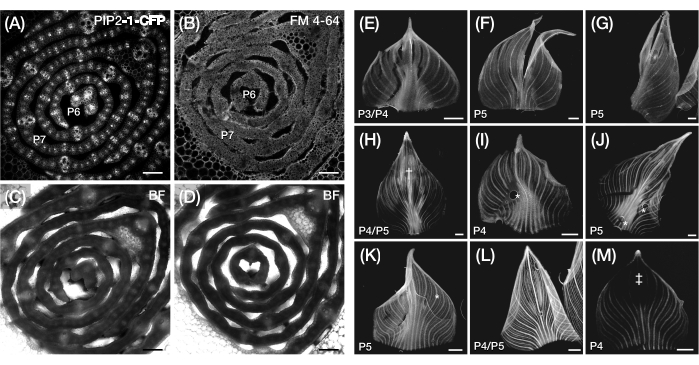
Figure 3: Suboptimal transverse section and whole-mount preparation of maize leaf primordia. (A-D) Representative confocal images of transverse sections of leaf primordia with a plasma membrane marker, PIP2-1-CFP (A), and a plasma membrane-binding fluorescent dye, FM 4-64 (B), with corresponding bright-field images (C,D). When compared to PIP2-1-CFP, FM 4-64 displays suboptimal visualization of cell outlines. (E-M) Representative fluorescence images of whole mounts of leaf primordia showing the presence of tearing (E-G), bruised surfaces† (H), air bubbles* (I-K), rolled back margins (L), and photobleached regions‡ (M). The leaf primordia express DII-Venus (E-G), GAR2-YFP (H-J), mDII-Venus (K), PIN1a-YFP (L), and DR5-RFP (M). Scale bar = 200 µm (A-D); 500 µm (E-M). Figure 3A has been modified and reproduced with permission from Robil and McSteen24, while Figure 5B-M are unpublished data from the authors. Abbreviations: P = plastochron; YFP = yellow fluorescent protein; RFP = red fluorescent protein; CFP = cyan fluorescent protein; PIP2-1-CFP = pZmPIP2-1::ZmPIP2-1:CFP; DII-Venus = pZmUbi:DII:YFP-NLS; GAR2-YFP = pZmGAR2::ZmGAR2:YFP; mDII-Venus = pZmUbi:mDII:YFP-NLS; PIN1a-YFP = pZmPIN1a::ZmPIN1a:YFP; DR5-RFP = DR5rev::mRFPer; BF = bright-field. Please click here to view a larger version of this figure.
4. Processing the images using ImageJ/FIJI
- Flattening z-stacks using maximum intensity projection (MIP)
- Launch ImageJ/FIJI and open the stack of images, or create a new stack from multiple separate images by selecting Image | Stacks | Images to Stack from the menu.
- Select the image stack. From the menu, select Image | Stacks | Z-Project.... In the Z-Project dialog box, choose Max Intensity from the Projection type drop-down menu. Click OK to create the maximum intensity z-projection.
NOTE: The flattened image is a 2D projection of the pixels with the highest intensity across the z-stacks50. This method is suitable for visualizing the fluorophore but not the anatomical features in bright-field images of thick transverse sections and whole-leaf mounts. - Save the image as TIFF (Tagged Image File Format) by selecting File | Save As | Tiff. Enter a name for the file and click Save.
NOTE: It is recommended to use TIFF for storing fluorescence and confocal images, as it retains the quality and detail when the image is saved.
- Flattening z-stacks using the Extended Depth of Field (EDF) plugin51
- Install the EDF plugin52. Download the Extended_Depth_Field.jar file (version 17.05.2021) from http://bigwww.epfl.ch/demo/edf/ and place it in the "plugins" folder of ImageJ/FIJI.
- Launch ImageJ/FIJI and open the stack of images, or create a new stack from multiple separate images by selecting Image | Stacks | Images to Stack from the menu.
- Select the image stack. From the menu, select Plugins | Extended Depth of Field | EDF Easy Mode. In the Extended Depth of Field dialog box, use the sliders under Speed/Quality to set the desired quality and processing speed, and the Height-map reg to set the level of smoothness for the topography. Click Run to create the reconstructed image.
NOTE: EDF combines the stack into a sharp, composite 2D image by projecting the best focus51. This technique is useful for overcoming the limited depth-of-focus and is suitable for reconstructing bright-field images of thick transverse sections or whole-leaf mounts. Use caution when applying this filter to visualize the fluorophore because, unlike MIP, EDF adjusts the contrast of the image, affecting pixel intensity. - Save the image as a TIFF file by selecting File | Save As | Tiff. Enter a name for the file and click Save.
- Stitching images together using the Grid/Collection Stitching plugin53
- Save the collection of images or image stacks to be stitched together in a single directory.
- Launch ImageJ/FIJI and select Plugins | Stitching | Grid/Collection Stitching from the menu. In the Grid/Collection Stitching dialog box, choose Unknown position from the Type drop-down menu, and then All files in directory from the Order. Click OK.
- In the Grid Stitching: Unknown position, All files in directory dialog box, specify the location of the images by typing or pasting the directory path in the Directory text field or by clicking Browse… to locate the directory. Click OK to begin the stitching process.
NOTE: The Unknown Position option in the Grid/Collection Stitching plugin reconstructs a composite image from a set of contiguous images by analyzing their overlapping regions. Therefore, this option is useful for stitching mosaics of images of transverse sections or whole-leaf mounts that are not obtained using a tile scanning operation. - When the stitching is finished, a new window will appear with the stitched image. Save the image as a TIFF file by selecting File | Save As | Tiff. Enter a name for the file and click Save.
NOTE: Stitching large montages of 2D and 3D images can also be accomplished using image reconstruction commands in the microscope software or through the Bio-Formats Import Options in ImageJ/FIJI.
- Combining multiple channels using the Merge Channels command
- Launch ImageJ/FIJI and open the images or image stacks to be merged.
NOTE: Image stacks can be flattened first using steps 4.1 or 4.2. MIP is generally recommended for flattening fluorophore channels, whereas EDF is better suited for flattening bright-field channels or other channels with varying depth-of-focus. - Adjust the brightness and contrast of the images with Image | Adjust | Brightness/Contrast. Use the sliders or input fields to adjust the parameters, then click Apply. Alternatively, use a linear histogram stretching method by selecting Process | Enhance Contrast…. Set the percentage of Saturated pixels, select Normalize, then click OK.
NOTE: When quantifying or comparing fluorophore signal intensity between images, it is generally best not to adjust the brightness or contrast because this can change the relative pixel intensities in the image, which can affect the measurements and comparisons. - To differentiate between channels, apply pseudocolors to specific images using a lookup table (LUT). To do this, select the image, then from the ImageJ/FIJI menu, select Image, click Lookup Tables, and choose the appropriate LUT from the drop-down list.
- From the menu, select Image | Color | Merge Channels. For each channel (C1, C2, C3…), use the drop-down list to select the image to be assigned to that channel. Click OK to merge the channels.
- Save the new image as TIFF by selecting File | Save As | Tiff. Enter a name for the file and click Save.
- Launch ImageJ/FIJI and open the images or image stacks to be merged.
Representative Results
Transverse section analysis of maize leaf primordia
We used protocol section 2 to quantify vein number and characterize hormone response patterns in transverse sections of maize leaf primordia with FPs (Figure 4)24. To assess the role of the plant hormone auxin in leaf growth and vein formation, we quantified the number of veins in the leaf primordia of an auxin-deficient maize mutant, vanishing tassel254. In early plastochrons, developing veins exhibit distinct cellularization in the median cell layer of the maize leaf primordium21,22. However, identifying and counting veins using conventional histological techniques22 can be labor-intensive and time-consuming. Thus, to quantify the veins, we utilized the maize auxin efflux protein marker pZmPIN1a::ZmPIN1a:YFP41 (PIN1a-YFP hereafter), which marks developing veins and procambial strands (PCSs; Figure 4A). Using protocol section 2, we were able to standardize the transverse section sampling by measuring the primordia prior to sectioning (Figure 4B,C). We discovered a trend in which vt2 has a wider primordium and more veins than normal (Figure 4D,E)24, which is consistent with data from fully expanded leaves55, indicating that the defect in vt2 began early in leaf development. Using protocol section 2, we were also able to systematically examine the expression patterns of hormone response FP reporters in the leaf primordia (see examples of the images in Figure 4F-I). Through a standardized transverse section sampling scheme, we mapped the distribution of auxin, cytokinin (CK), and gibberellic acid (GA) responses across different plastochrons and regions of leaf primordia, and discovered novel response patterns that we hypothesize have implications for leaf growth and vein formation24. Therefore, these representative results demonstrate the utility of protocol section 2 for transverse section analysis of maize leaf primordia.
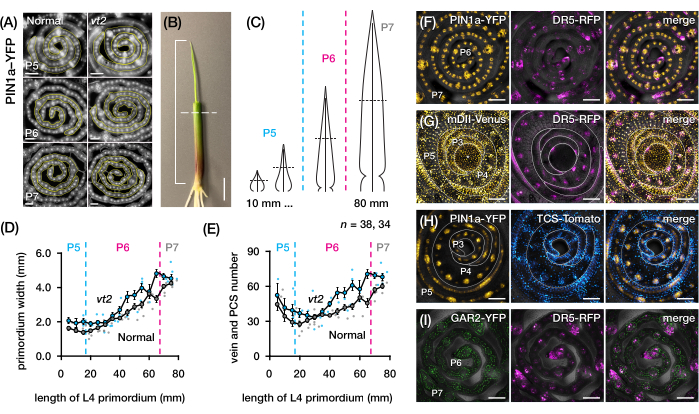
Figure 4: Representative results for transverse section analysis of maize leaf primordia. (A-E) Quantification of vein number and primordium width in leaf primordia of normal and vanishing tassel2 with the auxin efflux protein marker PIN1a-YFP. (A) Representative fluorescence images of transverse sections of P5 to P7 expressing PIN1a-YFP in developing veins and procambial strands. The transverse sections were imaged with an epifluorescence microscope using a FITC filter (495-519 nm excitation). The number of veins and the primordium width were quantified using the multi-point and freehand line tools in FIJI/ImageJ, respectively. (B) A maize seedling with the upper leaf whorls removed with a wire stripper to expose the tip of the fourth leaf (L4). The bracket spans the projected primordium length, with the dashed line indicating the mid-length. (C) Schematic diagram of varying primordium shapes from P5 to P7, illustrating how vein number and primordium width at the mid-length (horizontal dashed line) can vary depending on the developmental stage of the primordium. (D,E) Measurement of primordium width (D) and vein number (E) at the mid-length section of the L4 of normal and vt2. The trendline represents 10 mm rolling averages of measurements ± standard error of the mean (SEM). (F-I) Representative confocal images of transverse sections of leaf primordia expressing combinations of PIN1a-YFP, auxin response reporter, DR5-RFP, cytokinin response reporter, TCS-Tomato, gibberellic acid-responsive marker, GAR2-YFP, and mDII-Venus, a mutated version of the auxin signaling input reporter DII-Venus. The FP channels are superimposed on the bright-field channel in each image. Scale bar = 200 µm (A); 10 mm (B); 100 µm (F-I). This figure has been modified and reproduced with permission from Robil and McSteen24. Abbreviations: DAG = days after germination; P = plastochron; vt2 = vanishing tassel2; PCS = procambial strand; YFP = yellow fluorescent protein; FITC = fluorescein isothiocyanate; RFP = red fluorescent protein; PIN1a-YFP = pZmPIN1a::ZmPIN1a:YFP; DR5-RFP = DR5rev::mRFPer; TCS-Tomato = TCSv2::NLS-tdTomato; GAR2-YFP = pZmGAR2::ZmGAR2:YFP; mDII-Venus = pZmUbi:mDII:YFP-NLS. Please click here to view a larger version of this figure.
Whole-mount analysis of maize leaf primordia
We followed protocol section 3 to visualize and analyze FP expression in whole-leaf mounts of maize leaf primordia (Figure 5). By visualizing vein patterns with PIN1a-YFP, we found that the formation of veins occurs in the entire primordium during early plastochrons, but this process becomes restricted in the proximal regions later in development (Figure 5A)24. Complementary to the transverse section analyses, the whole-leaf mount analyses have revealed tissue- and stage-specific patterns of hormone response during vein formation24. One example is the expression pattern of the CK response reporter TCSv2::NLS-tdTomato37 (TCS-Tomato), relative to PIN1a-YFP expression (Figure 5B)24. By following protocol section 3, we were able to perform both qualitative and quantitative analyses of FP expression in the leaf primordia (Figure 5D-F; unpublished data). We examined the expression patterns of an auxin response reporter, DR5rev::mRFPer41 (DR5-RFP), and a GA-responsive marker, pZmGAR2::ZmGAR2:YFP39 (GAR2-YFP), in whole-leaf mounts of single and double mutants of vt2 and dwarf plant3 (d3), a GA-deficient mutant56 (Figure 5D). We also compared the relative levels of cell proliferation between normal and vt2 leaf primordia using pZmCyclin-D2B::ZmCyclin-D2B:YFP42 (Cyclin-D2B-YFP), which is a marker for the G1/S transition in the cell cycle57 (Figure 5E,F). While there was no significant difference between normal and vt2, Cyclin-D2B-YFP expression was consistent with the known cell proliferation profile of early plastochrons31. We conclude that protocol section 3 is an effective method for analyzing whole mounts of maize leaf primordia, which are difficult to image due to their rolled morphology.
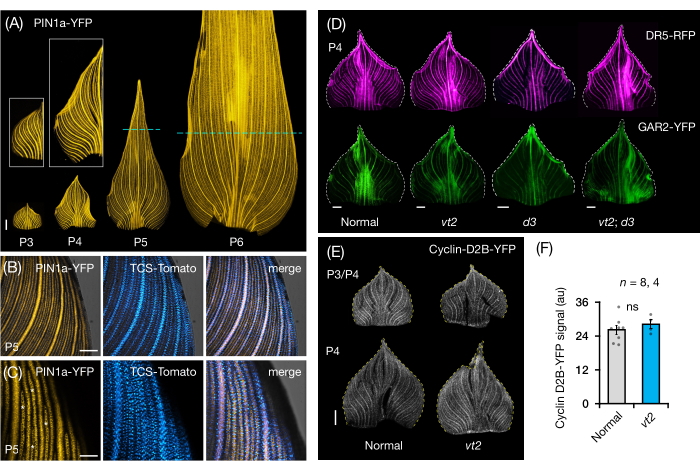
Figure 5: Representative results for whole-mount analysis of maize leaf primordia. (A) Representative fluorescence images of leaf primordia of 7 DAG maize seedlings showing developing veins and procambial strands, as marked by the auxin efflux protein marker PIN1a-YFP. P3-P6 primordia were excised from the base, unrolled, and flattened with the adaxial side up. Insets show close-ups of P3 and P4. In P5 and P6, dashed lines demarcate the distal end of the proliferative zones, where the majority of procambial strands are still developing and extending. (B,C) Representative confocal images showing the expression of PIN1a-YFP and cytokinin response reporter, TCS-Tomato, in the proximal marginal region of a P5 primordium. (D) Representative fluorescence images of P4 primordia showing the expression of auxin response reporter, DR5-RFP, and gibberellic acid-responsive marker, GAR2-YFP in normal and single and double mutants of vanishing tassel2 and dwarf plant3. (E) Representative confocal images of P3 and/or P4 showing the expression of Cyclin-D2B-YFP, a reporter for the G1/S transition in the cell cycle. (F) Relative amounts of cell proliferation in P3/P4 leaf primordia of normal and vt2, quantified by measuring the integrated density of the Cyclin-D2B-YFP signal over the area of the primordium using ImageJ/FIJI. The bars represent the mean measurements ± standard error of the mean. Scale bar = 500 µm (A,D,E); 200 µm (B); 100 µm (C). Figure 4A-C has been modified and reproduced with permission from Robil and McSteen24, while Figure 4D-F is unpublished data from the authors. Abbreviations: DAG = days after germination; P = plastochron; vt2 = vanishing tassel2; d3 = dwarf plant3; YFP = yellow fluorescent protein; RFP = red fluorescent protein; PIN1a-YFP = pZmPIN1a::ZmPIN1a:YFP; TCS-Tomato = TCSv2::NLS-tdTomato; DR5-RFP = DR5rev::mRFPer; GAR2-YFP = pZmGAR2::ZmGAR2:YFP; Cyclin-D2B-YFP = pZmCyclin-D2B::ZmCyclin-D2B:YFP; ns = no significant difference; au = arbitrary unit. Please click here to view a larger version of this figure.
Supplementary File 1: Examples of leaf development staging in maize seedlings. Please click here to download this File.
Supplementary Figure S1: Plant samples and materials used in the protocols. (A,B) A maize seedling 7-8 DAG with the upper leaf whorl removed using a wire stripper. (B) The inset shows a close-up of the shoot with the exposed P6 primordium. (C-G) Shoots of maize seedlings with the upper whorls removed for transverse section analysis (C,D) and surrounding leaves fully removed for whole-mount analysis (E-G). (H) A roll of polyurethane gel clear double-sided nano tape. (I,J) P6 leaf primordium (I) and proximal region of P8 leaf (J) unrolled and mounted on glass slides with the nano tape. (K) A glass slide with an unrolled leaf primordium mounted on the stage of an epifluorescence microscope. Abbreviation: DAG = days after germination. Please click here to download this File.
Discussion
We present two methods for preparing maize leaf primordia for cellular imaging. The first method (protocol section 2) allows measurement of the primordium for transverse section analysis, while the second method (protocol section 3) enables unrolling and flattening of the primordium for whole-mount analysis. These methods facilitate the cellular imaging of FPs in maize leaf primordia24 (as shown in Figure 4 and Figure 5) and provide simple solutions to the challenges of imaging developing maize leaves. Protocol section 2 reduces dissection time and improves sampling accuracy by measuring the primordia prior to sectioning rather than relying solely on staging parameters9,16. With commercially available nano tape, protocol section 3 solves the long-standing problem of imaging whole-leaf primordia in maize. This protocol improves on the previous method, which used dialysis tubing31, and is a much cheaper alternative to CT and MRI11,32,33. However, when it comes to visualizing leaf anatomical traits and producing optimal results, both protocols have some limitations, which are outlined in Table 2 and are discussed in more detail below.
In protocol section 2, we encountered difficulty visualizing cell outlines in thick transverse sections of the leaf primordia, and counterstaining with cell wall- or plasma membrane-binding fluorescent dyes did not provide satisfactory results. For instance, FM 4-64 produced suboptimal results compared to the plasma membrane FP marker, pZmPIP2-1::ZmPIP2-1:CFP39 (PIP2-1-CFP; Figure 3A-D). To overcome this limitation, we recommend using a vibratome to produce thinner tissue sections (~0.1 mm)58 which will allow for a vivid bright-field imaging of cell outlines or optimizing the counterstaining protocol47,59.
In protocol section 3, the main limitation is the difficulty of mounting the leaf without tearing, damage, or air bubbles, as detailed in protocol steps 3.2.5-3.2.6 (Figure 3E-K). Because the maize leaf is bilaterally symmetrical, a half-leaf mount rather than a whole-leaf mount may be sufficient for visualization9. To do this, the primordium can be cut with a razor blade along the longitudinal axis after unrolling it up to the midrib, allowing only half of the leaf to be mounted. Another limitation of protocol section 3 is that the thickness of the leaf can limit the optical resolution of the fluorophore signal during deep imaging. To address this issue, it is possible to employ a tissue clearing technique60. However, we found that ClearSee61, a commonly used clearing reagent for imaging plant tissues, is not compatible with the protocol because it causes the sample and coverslip to detach from the nano tape. A potential solution to this problem could be to apply a semipermeable membrane31 over the leaf sample, allowing it to be treated with the clearing solution while being held in place by the nano tape. Such a method that allows liquid solutions to be applied to the unrolled leaf could also be used for whole-mount RNA in situ hybridization and immunolocalization techniques, which have previously been optimized for developing maize inflorescences but not for whole-leaf primordia62,63.
We described protocols for maize, which has large leaf primordia even at the seedling stage. Other grass species with much smaller leaf primordia, such as rice, barley, wheat, Setaria, and Brachypodium16,23,64,65,66, may require the use of additional precision tools to effectively apply these protocols. Furthermore, these protocols were not intended for live cell imaging, which captures real-time dynamic processes of tissue formation and cellular responses. However, as fluorescent probes, imaging technologies, and computing capabilities continue to advance in live cell imaging for plants67, future research could build on these protocols to develop live cell imaging strategies tailored to the unique features of grass leaf primordia.
Acknowledgements
The authors would like to thank the Maize Genetics Cooperation, the Maize Cell Genomics Project, Dave Jackson (Cold Spring Harbor Laboratory, NY), Anne W. Sylvester (Marine Biological Laboratory, University of Chicago, IL), Andrea Gallavotti (Rutgers University, NJ), and Carolyn G. Rasmussen (University of California, Riverside) for providing the mutant and transgenic stocks, as well as Robert F. Baker and Alexander Jurkevich of the Advanced Light Microscopy Core at the University of Missouri-Columbia for their assistance with confocal microscopy. JMR was supported by the J. William Fulbright Fellowship, The Diane P. and Robert E. Sharp Fund, and the National Science Foundation's Plant Genome Research Program (IOS-1546873) to PM. CDTC, CMRV, EDCDP, and RJRR are supported by the Ateneo College Scholarship Program. CDTC, EDCDP, and RJRR are supported by the DOST-SEI S&T Undergraduate Scholarship. DODL is supported by Fr. Thomas Steinbugler SJ Academic Scholarship. RJRR is supported by Aiducation International–Pathways to Higher Education Scholarship. This work was supported by the School of Science and Engineering and the Rizal Library, Ateneo de Manila University.
Materials
| Name | Company | Catalog Number | Comments |
| Acrylic Gel Clear Double Sided Nano Tape 16.5 ft x 1.2 in, 2 mm thick | EZlifego Store (Amazon) | B07YB1ZXG6 | 1 roll |
| Bellucci Pick Curved micro probe 16.8 cm, 6.6 in | Bausch & Lomb | N1692 9 | 1 pc |
| Clayman guide microprobe Sinskey hook angled shaft, 11.6 cm, 4.6 in | Storz Opthalmic Instruments | E0542 | 1 pc |
| Dental Probe, Bent Needle, 14 cm (5.5 in) | Ted Pella | 13553 | 1 pc |
| DOWELL 10-22 AWG Wire Stripper | Dowell Store (Amazon) | 10-22 AWG | 1 pc |
| Feather Double Edge Carbon Steel Blades | Ted Pella | 121-9 | pkg/10; for fine sectioning |
| Frosted End Glass Microscope Slides, 75 mm x 25 mm x 1-1.2 mm | Ted Pella | 260442 | pkg/144 |
| GEM Single Edge, Stainless Steel Uncoated Blades | Ted Pella | 121-1 | box/200; for general cutting/sectioning |
| Glycerol | Thermo Scientific | PI17904 | 1 liter |
| ImageJ/FIJI with EDF plugin (version 17.05.2021) and Grid/Collection Stitching plugin | National Institutes of Health (NIH) USA | version 2.9.0/1.54s | The EDF plugin was developed by Alex Prudencio, Jesse Berent, and Daniel Sage for the Biomedical Imaging Group, École Polytechnique Fédérale de Lausanne (EPFL; http://bigwww.epfl.ch/demo/edf/). The grid/collection stitching software was developed by Stephan Preibisch for the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG). |
| Kimwipes Ex-L Small 111.76 mm x 213.36 mm | Kimtech Science | 34155 | box/280 ply |
| Micro Cover Glasses, 22 mm x 22 mm x 0.13 - 0.16 mm thick | Ted Pella | 260140 | 1 ounce |
| PU Gel Clear Double Sided Nano Tape 29.5 ft x 1.18 in, 1 mm thick | Yecaye Store (Amazon) | L354 W1.18 | 2 rolls |
| Superslip Cover Glasses, 24 mm x 50 mm x 0.13 - 0.16 mm thick | Ted Pella | 260166 | 1 ounce |
| Superslip Cover Glasses, 24 mm x 60 mm x 0.13 - 0.16 mm thick | Ted Pella | 260168 | 1 ounce |
| Tempered Glass Cutting Board | Hacaroa (Amazon) | B09XMXBT5S | 4 pc |
References
- McSteen, P., Kellogg, E. A. Molecular, cellular, and developmental foundations of grass diversity. Science. 377 (6606), 599-602 (2022).
- Wang, P., Vlad, D., Langdale, J. A. Finding the genes to build C4 rice. Current Opinion in Plant Biology. 31, 44-50 (2016).
- Wang, P., et al. Candidate regulators of early leaf development in maize perturb hormone signalling and secondary cell wall formation when constitutively expressed in rice. Scientific Reports. 7 (1), 4535 (2017).
- Perico, C., Tan, S., Langdale, J. A. Developmental regulation of leaf venation patterns: Monocot versus eudicots and the role of auxin. New Phytologist. 234 (3), 783-803 (2022).
- Wang, P., Kelly, S., Fouracre, J. P., Langdale, J. A. Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. The Plant Journal. 75 (4), 656-670 (2013).
- Liu, W. Y., et al. Regulators of early maize leaf development inferred from transcriptomes of laser capture microdissection (LCM)-isolated embryonic leaf cells. Proceedings of the National Academy of Sciences. 119 (35), e2208795119 (2022).
- Liu, W. Y., et al. Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination. Proceedings of the National Academy of Sciences. 110 (10), 3979-3984 (2013).
- Strable, J., Nelissen, H. The dynamics of maize leaf development: Patterned to grow while growing a pattern. Current Opinion in Plant Biology. 63, 102038 (2021).
- Reynolds, J. O., Eisses, J. F., Sylvester, A. W. Balancing division and expansion during maize leaf morphogenesis: Analysis of the mutant, warty-1. Development. 125 (2), 259-268 (1998).
- Richardson, A. E., et al. Evolution of the grass leaf by primordium extension and petiole-lamina remodeling. Science. 374 (6573), 1377-1381 (2021).
- Johnston, R., Leiboff, S., Scanlon, M. J. Ontogeny of the sheathing leaf base in maize (Zea mays). New Phytologist. 205 (1), 306-315 (2015).
- Sharman, B. C. Developmental anatomy of the shoot of Zea mays L. Annals of Botany. 6 (22), 245-282 (1942).
- Johnston, R., et al. Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. The Plant Cell. 26 (12), 4718-4732 (2014).
- Sharman, B. C. Leaf and bud initiation in the Gramineae. Botanical Gazette. 106 (3), 269-289 (1945).
- Nelissen, H., et al. A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Current Biology. 22 (13), 1183-1187 (2012).
- Itoh, J., et al. Rice plant development: From zygote to spikelet. Plant and Cell Physiology. 46 (1), 23-47 (2005).
- Ritchie, S. W., Hanway, J. J., Benson, G. O. How a corn plant develops. Iowa State University of Science and Technology. , (1986).
- Freeling, M., Walbot, V. . The Maize Handbook. , (2013).
- Freeling, M., Hake, S. Developmental genetics of mutants that specify knotted leaves in maize. Genetics. 111 (3), 617-634 (1985).
- Durbak, A. R., et al. Transport of boron by the tassel-less1 aquaporin is critical for vegetative and reproductive development in maize. The Plant Cell. 26 (7), 2978-2995 (2014).
- Langdale, J. A., Lane, B., Freeling, M., Nelson, T. Cell lineage analysis of maize bundle sheath and mesophyll cells. Developmental Biology. 133 (1), 128-139 (1989).
- Bosabalidis, A. M., Evert, R. F., Russin, W. A. Ontogeny of the vascular bundles and contiguous tissues in the maize leaf blade. American Journal of Botany. 81 (6), 745-752 (1994).
- Junqueira, N. E. G., et al. Anatomy and ultrastructure of embryonic leaves of the C4 species Setaria viridis. Annals of Botany. 121 (6), 1163-1172 (2018).
- Robil, J. M., McSteen, P. Hormonal control of medial-lateral growth and vein formation in the maize leaf. New Phytologist. 238 (1), 125-141 (2023).
- Donnelly, P. M., Bonetta, D., Tsukaya, H., Dengler, R. E., Dengler, N. G. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Developmental Biology. 215 (2), 407-419 (1999).
- Govindaraju, P., Verna, C., Zhu, T., Scarpella, E. Vein patterning by tissue-specific auxin transport. Development. 147 (13), (2020).
- Linh, N. M., Scarpella, E. Leaf vein patterning is regulated by the aperture of plasmodesmata intercellular channels. PLoS Biology. 20 (9), e3001781 (2022).
- Verna, C., Ravichandran, S. J., Sawchuk, M. G., Linh, N. M., Scarpella, E. Coordination of tissue cell polarity by auxin transport and signaling. eLife. 8, e51061 (2019).
- Sawchuk, M. G., Head, P., Donner, T. J., Scarpella, E. Time-lapse imaging of Arabidopsis leaf development shows dynamic patterns of procambium formation. New Phytologist. 176 (3), 560-571 (2007).
- Linh, N. M., Scarpella, E. Confocal imaging of developing leaves. Current Protocols. 2 (1), e349 (2022).
- Poethig, R. S., Szymkowiak, E. J. Clonal analysis of leaf development in maize. Maydica. 40, 67-76 (1995).
- Sprangers, K., Thys, S., van Dusschoten, D., Beemster, G. T. S. Gibberellin enhances the anisotropy of cell expansion in the growth zone of the maize leaf. Frontiers in Plant Science. 11, 1163 (2020).
- Tsuda, K., et al. KNOTTED1 cofactors, BLH12 and BLH14, regulate internode patterning and vein anastomosis in maize. The Plant Cell. 29 (5), 1105-1118 (2017).
- Schindelin, J., et al. FIJI: An open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Plant Care Protocols - Maize. The Donald Danforth Plant Science Center's Plant Growth Facility Available from: https://www.danforthcenter.org/our-work/core-facilities/plant-growth/ (2019)
- Mir, R., et al. A DII domain-based auxin reporter uncovers low auxin signaling during telophase and early G1. Plant Physiology. 173 (1), 863-871 (2017).
- DeBlasio, S. L., Sylvester, A. W., Jackson, D. Illuminating plant biology: Using fluorescent proteins for high-throughput analysis of protein localization and function in plants. Briefings in Functional Genomics. 9 (2), 129-138 (2010).
- Wu, Q., Luo, A., Zadrozny, T., Sylvester, A., Jackson, D. Fluorescent protein marker lines in maize: Generation and applications. The International Journal of Developmental Biology. 57 (6-8), 535-543 (2013).
- Mohanty, A., et al. Advancing cell biology and functional genomics in maize using fluorescent protein-tagged lines. Plant Physiology. 149 (2), 601-605 (2009).
- Baker, R. F., et al. Sucrose transporter ZmSut1 expression and localization uncover new insights into sucrose phloem loading. Plant Physiology. 172 (3), 1876-1898 (2016).
- Gallavotti, A., Yang, Y., Schmidt, R. J., Jackson, D. The relationship between auxin transport and maize branching. Plant Physiology. 147 (4), 1913-1923 (2008).
- Krishnakumar, V., et al. A maize database resource that captures tissue-specific and subcellular-localized gene expression, via fluorescent tags and confocal imaging (Maize Cell Genomics Database). Plant and Cell Physiology. 56 (1), 12 (2015).
- Snapp, E. L. Fluorescent proteins: A cell biologist's user guide. Trends in Cell Biology. 19 (11), 649-655 (2009).
- Geng, Y., Zhou, Y. Confocal live imaging of shoot apical meristems from different plant species. Journal of Visualized Experiments. (145), e59369 (2019).
- Stanislas, T., Hamant, O., Traas, J. In-vivo analysis of morphogenesis in plants. Methods in Cell Biology. 139, 203-223 (2017).
- Fodor, E., Ayaydin, F. Fluorescent probes and live imaging of plant cells. Advances in Plant Ecophysiology Techniques. , 241-251 (2018).
- Grandjean, O., et al. In vivo analysis of cell division, cell growth, and differentiation at the shoot apical meristem in Arabidopsis. The Plant Cell. 16 (1), 74-87 (2004).
- Conklin, P. A., Johnston, R., Conlon, B. R., Shimizu, R., Scanlon, M. J. Plant homeodomain proteins provide a mechanism for how leaves grow wide. Development. 147 (20), (2020).
- Shaw, S. L. Imaging the live plant cell. The Plant Journal. 45 (4), 573-598 (2006).
- Klaus, A. V., Schawaroch, V., Frischmann, K. J. Confocal imaging and three-dimensional visualization of thick autofluorescent specimens. Methods in Molecular Biology. 1075, 213-225 (2014).
- Forster, B., Van De Ville, D., Berent, J., Sage, D., Unser, M. Extended depth-of-focus for multi-channel microscopy images: a complex wavelet approach. 2004 2nd IEEE International Symposium on Biomedical Imaging: Nano to Macro (IEEE Cat No. 04EX821). IEEE. , 660-663 (2004).
- Preibisch, S., Saalfeld, S., Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 25 (11), 1463-1465 (2009).
- Phillips, K. A., et al. vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. The Plant Cell. 23 (2), 550-566 (2011).
- Robil, J. M., et al. GrasVIQ: An image analysis framework for automatically quantifying vein number and morphology in grass leaves. The Plant Journal. 107 (2), 629-648 (2021).
- Helliwell, C. A., Chandler, P. M., Poole, A., Dennis, E. S., Peacock, W. J. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proceedings of the National Academy of Sciences. 98 (4), 2065-2070 (2001).
- Gutierrez, R., Quiroz-Figueroa, F., Vazquez-Ramos, J. M. Maize cyclin D2 expression, associated kinase activity and effect of phytohormones during germination. Plant and Cell Physiology. 46 (1), 166-173 (2005).
- Atkinson, J. A., Wells, D. M. An updated protocol for high throughput plant tissue sectioning. Frontiers in Plant Science. 8, 1721 (2017).
- Lux, A., Morita, S., Abe, J., Ito, K. An improved method for clearing and staining free-hand sections and whole-mount samples. Annals of Botany. 96 (6), 989-996 (2005).
- Heriche, M., Arnould, C., Wipf, D., Courty, P. E. Imaging plant tissues: Advances and promising clearing practices. Trends in Plant Science. 27 (6), 601-615 (2022).
- Kurihara, D., Mizuta, Y., Sato, Y., Higashiyama, T. ClearSee: A rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 142 (23), 4168-4179 (2015).
- Chuck, G., Muszynski, M., Kellogg, E., Hake, S., Schmidt, R. J. The control of spikelet meristem identity by the branched silkless1 gene in maize. Science. 298 (5596), 1238-1241 (2002).
- Tran, T. M., et al. An optimized whole-mount immunofluorescence method for shoot apices. Current Protocols. 1 (4), e101 (2021).
- O'Connor, D. L. PINs Lost and PINs Gained: Auxin-Transport Mediated Patterning in the Grasses. University of California, Berkeley. , (2012).
- Sharman, B. C., Hitch, P. A. Initiation of procambial strands in leaf primordia of bread wheat, Triticum aestivum L. Annals of Botany. 31 (2), 229-243 (1967).
- Serra, L., Tan, S., Robinson, S., Langdale, J. A. Flip-flap: A simple dual-view imaging method for 3D reconstruction of thick plant samples. Plants. 11 (4), 506 (2022).
- Colin, L., et al. Imaging the living plant cell: From probes to quantification. The Plant Cell. 34 (1), 247-272 (2022).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved