A Simple, Quick, and Partially Automated Protocol for the Isolation of Single Nuclei from Frozen Mammalian Tissues for Single Nucleus Sequencing
In This Article
Summary
The study describes a simple, quick, and partially automated protocol to isolate high-quality nuclei from frozen mammalian tissues for downstream single nuclei RNA sequencing.
Abstract
Single-cell and single-nucleus RNA sequencing have become common laboratory applications due to the wealth of transcriptomic information that they provide. Single nucleus RNA sequencing, particularly, is useful for investigating gene expression in difficult-to-dissociate tissues. Furthermore, this approach is also compatible with frozen (archival) material. Here, we describe a protocol to isolate high-quality single nuclei from frozen mammalian tissues for downstream single nucleus RNA sequencing in a partially-automated manner using commercially available instruments and reagents. Specifically, a robotic dissociator is used to automate and standardize tissue homogenization, followed by an optimized chemical gradient to filter the nuclei. Lastly, we accurately and automatically count the nuclei using an automated fluorescent cell counter. The performance of this protocol is demonstrated on mouse brain, rat kidney, and cynomolgus liver and spleen tissue. This protocol is straightforward, rapid, and readily adaptable to various mammalian tissues without requiring extensive optimization and provides good quality nuclei for downstream single nuclei RNA sequencing.
Introduction
Single-cell (sc) and single-nucleus (sn) RNA sequencing have become commonly used protocols in molecular and cellular biology due to the increased resolution of gene expression compared to bulk RNA sequencing. However, the isolation of good quality single cell and single nuclei preparations from solid tissues remains a challenge and is often the rate-limiting step in sc/sn-RNAseq experiments. Indeed, a plethora of protocols has been developed that use various chemical and mechanical procedures to obtain cell/nuclei suspensions1,2,3,4,5,6,7,8,9,10,11,12,13,14,15. Furthermore, strategies to clean up such preparations from debris/clumps, etc., range from flow sorting to filtration to washing. Such protocols are often manual (leading to user-related variability), can be time-consuming (leading to reduced cell/nucleus viability), and/or may require access to a flow cytometer for cell/nucleus sorting. This study focused on developing a simple, quick, and partially automated single nuclei isolation protocol from frozen mammalian tissues for downstream RNA sequencing applications. We focused specifically on nuclei isolation as opposed to cell isolation as it is compatible with the use of frozen tissues, rendering sample collection/processing more practical and enabling unbiased batching of samples, especially in time-course experiments. Furthermore, although the nuclear transcriptome does not fully reflect the cellular transcriptome, several studies have now shown that single nuclei RNA sequencing data is comparable to single cell RNA sequencing data for cell-type identification, even though the proportions of cell types can vary6,16,17,18,19.
Nuclei isolation consists of several steps: 1) mechanical or chemical disruption of the tissue to release the nuclei, 2) clean-up of debris and clumps, and 3) accurate counting of nuclei for preparation for downstream applications. In a number of protocols, step 1 frequently involves the use of a Dounce homogenizer in order to disrupt the tissue3,20. Alternatively, chemical methods can be used, although these often need to be optimized for different tissues2,5,6. We have experienced that a manual tissue disruption procedure is prone to operator-associated variability, leading to variable quality and yield of nuclei. In order to minimize technical variability and to have a more consistent and reproducible protocol that works across tissues, a protocol that uses a commercially available robotic tissue dissociator was developed21. For step 2, although buffer exchange is usually the simplest means for washing nuclei, we adopted the use of a relatively short sucrose gradient centrifugation step to have a more thorough removal of debris. For brain tissue specifically, we use a silica colloid gradient instead of a sucrose gradient for more effective myelin removal. Finally, for counting, the use of a hemocytometer is the gold standard for counting and visually inspecting the nuclei. In our protocol, this step can be reliably automated using a commercially available automated fluorescent cell counter22. This protocol has been tested and is compatible with several frozen mammalian tissues, including brain, kidney, spleen, and liver, from different mammalian species (rat, mouse, and non-human primate) and provides good quality nuclei for downstream single nuclei RNA sequencing with a droplet-based commercial platform. The protocol takes approximately 75 min from tissue preparation to the start of the single nuclei RNA sequencing workflow.
Protocol
All animal studies were conducted with the approval of the cantonal veterinary authority of Basel-Stadt in strict adherence to the Swiss federal regulations on animal protection or with the approval of the Institutional Animal Care and Use Committee in compliance with the German Animal Welfare Act.
1. Tissue and reagent/instrument preparation
- Cleaning and instrument preparation
- Clean benchtops and tweezers with 70% ethanol and RNase decontamination solution. Pre-cool the centrifuges to 4 °C.
- Pre-cool the nuclei isolation cartridges in the fridge at 4 °C for at least 30 min.
- Start the robotic dissociator and turn on the cooling by setting the slider at the top right of the screen to cool and by clicking on it to start cooling so that the slider appears orange. Check that the attached nuclei storage reagent (NSR) bottle and the nuclei isolation buffer (NIB) bottle have enough liquid left and are properly cooled.
- Prepare a polystyrene foam box filled with dry ice and pre-cool Petri dishes and scalpel blades on dry ice.
- Buffer preparation
- Prepare the 1.5 M sucrose cushion solution (SCS) as shown in Table 1. Distribute the SCS into 500 µL aliquots in 2 mL DNase/RNase tubes to obtain four 500 µL SCS aliquots per sample. Keep the aliquots on ice until further use.
- In case of processing brain tissue, prepare instead an 18% silica colloid solution as described in Table 2, by diluting the silica colloid stock solution in NSR and adding RNase inhibitor. Prepare 3 mL of 18% silica colloid solution per sample and keep it on ice.
2. Tissue homogenization and nuclei isolation
- Remove the sample from the -80 °C freezer and place it immediately on dry ice.
- Cut the sample on a pre-cooled Petri dish or metal plate on dry ice with a pre-cooled scalpel into a 15-50 mg piece (if it is not already the right size). Be sure to cut the sample in the correct direction so that the sample is still representative of the organ structures of interest.
NOTE: With this protocol, 15-50 mg is the optimal sample size for nuclear extraction. To achieve a good yield after the clean-up, a sample size of at least 25 mg is recommended. For smaller samples, special cartridges are available that are optimized for processing small inputs with the robotic dissociator. Small input nuclei isolation cartridges were employed to dissociate tissue samples weighing as little as 4 mg with sufficient yield for downstream single nuclei RNA sequencing. An example of nuclei yield using the low input cartridge from rat liver tissue is shown in Table 3. - Remove the nuclei isolation cartridge from the refrigerator, unpack it, remove the grinder, and pipette 15 µL of RNase inhibitor (40 U/µL) to the bottom of the cartridge.
NOTE: During the nuclei extraction, the robotic dissociator will add NIB and NSR to the cartridge to a total volume of 3 mL (with the low volume nuclei extraction protocol). By adding 15 µL of RNase inhibitor (40 U/µL) to the cartridge prior to the extraction, the suspension will have the desired RNAse inhibitor concentration of 0.2 U/µL. - Place the tissue sample on the bottom of the cartridge using tweezers. Do not place the sample exactly in the center of the cartridge in order to have optimal disruption efficiency.
- Select Run a Protocol on the instrument and click on the Nuclei option in the upper left corner.
- From the menu, select the Low Volume Nuclei Isolation protocol and verify that the disruption speed is set to fast by clicking on Modify. Load the cartridge to the instrument by opening the door and sliding out the stage by lifting the red knob.
- Insert the cartridge into the designated location, rotate the cartridge lock, and slide in the stage until the red knob clicks into place. Close the door and start the nuclei extraction run on the instrument by clicking Next. The run lasts approximately 7 min.
- Once the run is finished, remove the cartridge from the instrument by lifting the red knob and pulling out the stage. Immediately place the cartridge on ice.
- For all tissues except the brain, proceed to step 3.1. For brain samples, proceed directly to step 3.2.
3. Nuclei clean-up
- Sucrose gradient clean-up
NOTE: For brain tissue, skip this step and go directly to step 3.2. All the clean-up steps are performed on ice in order to minimize RNA degradation. Buffers and tubes as well as the centrifuges need to be pre-cooled. All resuspension and mixing steps are performed by careful pipetting only, as vortexing could compromise the quality and integrity of the nuclei.- Carefully pierce the round foil on the dissociator cartridge with a pipette tip.
NOTE: After dissociation, the resulting nuclei suspension has an approximate volume of 2 mL. To facilitate sucrose gradient clean-up, divide the nuclei suspension into two 900 µL aliquots, resulting in a total volume of 1.8 mL of nuclei suspension being used during clean-up. - Remove the first 900 µL aliquot of the nuclei suspension from the cartridge and add it to a 500 µL SCS aliquot previously prepared in a 2 mL tube. Mix well by pipetting until the mixture is homogeneous.
- Remove the 1400 µL of nuclei suspension - SCS mixture and carefully layer it onto a new 500 µL SCS aliquot by holding the tube at an angle and adding the mixture dropwise, creating a clearly visible phase separation (see Figure 1A).
- Carefully close the tube and place it back on ice without disturbing the phase separation.
- Repeat steps 3.1.2-3.1.4 with the second 900 µL aliquot of suspension and a new SCS aliquot to have two 2 mL tubes per sample with a clearly visible phase separation for gradient centrifugation.
- Carefully add the tubes to a pre-cooled centrifuge and spin at 13,000 x g for 15 min at 4 °C.
- In the meantime, prepare the NSR described in Table 4, by adding the RNase inhibitor to an aliquot of NSR. Prepare 1 mL of NSR per sample.
NOTE: At this point, the single cell gene expression reagent gel beads can be removed from the -80 °C freezer, allowing them to equilibrate to room temperature (RT), and the template switch oligo can be resuspended in low TE buffer. - After centrifugation, remove the supernatant from both tubes without disturbing the pellet and carefully resuspend the pellet in 50 µL of ice-cold NSR as per the manufacturer's recommendation. Pool the two pellets of the same sample in a new 1.5 mL tube and add 900 µL of ice-cold NSR to a total volume of 1 mL. Mix well by pipetting.
- Centrifuge the sample at 500 x g for 5 min at 4 °C with a swinging-bucket rotor centrifuge.
NOTE: It is highly recommended to use a swinging-bucket rotor to minimize nuclei loss, especially when nuclei yield is expected to be low or when starting with small amounts of tissue. - In the meantime, prepare 500 µL per sample of 1x PBS (without Ca2+ and Mg2+) with 0.04% bovine serum albumin (BSA) and 0.2 U/µL RNase inhibitor as described in Table 5.
- Carefully remove the supernatant without discarding the pellet and resuspend the pellet in 100 µL of PBS solution as prepared above (1x PBS + 0.04% BSA + RNase inhibitor at 0.2 U/µL).
NOTE: For small tissue samples, the nuclei concentration may be low, and therefore, it is recommended to resuspend the pellet in only 50 µL of PBS solution to ensure sufficiently high concentrations for single nuclei RNA sequencing. - Proceed directly to step 4.
- Carefully pierce the round foil on the dissociator cartridge with a pipette tip.
- Silica colloid gradient clean-up
NOTE: For brain tissue, a silica colloid gradient is more suitable than a sucrose gradient to remove myelin and debris from the nuclei suspension. All the clean-up steps are performed on ice to minimize RNA degradation. Buffers and tubes, as well as the centrifuges, need to be pre-cooled. All resuspension and mixing steps are performed by careful pipetting only, as vortexing could compromise the quality and integrity of the nuclei.- Carefully pierce the round foil on the dissociator cartridge with a pipette tip.
- Remove the nuclei suspension from the cartridge and add it to a 5 mL tube.
- Centrifuge at 500 x g for 5 min at 4 °C in a pre-cooled centrifuge.
- Carefully remove the supernatant without disturbing the pellet and resuspend the pellet in 1 mL of ice-cold 18% silica colloid solution.
- Add an additional 2 mL of 18% silica colloid solution to have a total volume of 3 mL and mix well by pipetting.
- Centrifuge the sample at 700 x g for 5 min at 4 °C in a swinging-bucket rotor with the brake set off.
- In the meantime, prepare the NSR described in Table 4 by adding the RNase inhibitor to an aliquot of NSR. Prepare 1 mL of NSR per sample.
NOTE: At this point, the single cell gene expression reagent gel beads can be removed from the -80 °C freezer, allowing it to equilibrate to RT and the template switch oligo can be resuspended in low TE buffer. - Carefully remove the sample from the centrifuge without disturbing the myelin layer floating on top.
- First, remove the myelin layer from the top and discard; then, carefully remove the entire supernatant without disturbing the pellet.
NOTE: The myelin layer can be easily removed by wrapping a sterile lint-free wipe around a 1 mL pipette tip to aspirate the myelin layer along with 1-2 mL of the supernatant. - Resuspend the pellet in 1 mL of ice-cold NSR as per the manufacturer's recommendation.
- Centrifuge the sample at 500 x g for 5 min at 4 °C in a centrifuge with a swinging-bucket rotor.
NOTE: It is highly recommended to use a swinging-bucket rotor to minimize nuclei loss, especially when nuclei yield is expected to be low or when starting with small amounts of tissue. - In the meantime, prepare 500 µL per sample of 1x PBS (without Ca2+ and Mg2+) with 0.04% bovine serum albumin (BSA) and 0.2 U/µL RNase inhibitor as described in Table 5.
- Carefully remove the supernatant without disturbing the pellet and resuspend the sample in 100 µL of PBS solution as prepared above (1x PBS + 0.04% BSA + RNase inhibitor at 0.2 U/µL).
NOTE: For small tissue samples, the nuclei concentration may be low, and therefore, it is recommended to resuspend the pellet in only 50 µL of PBS solution to ensure sufficiently high concentrations for single nuclei RNA sequencing.
4. Counting
- For each sample to be counted, dilute 10 µL of nuclei suspension in 20 µL of PBS solution to obtain a 1:3 dilution.
- For counting, add 25 µL of propidium iodide (PI) staining solution to a mixing well of the fluorescent counter counting plate. Add 25 µL of the diluted nuclei suspension and mix well by pipetting. Transfer the 50 µL stained sample from the mixing well to the loading well.
- Load the counting plate on the cell counter and start the count.
NOTE: The nuclei count is taken from the red fluorescent channel with an exposure time of 700 ms. This channel was optimized to get an accurate nuclei count by cross-comparing to manual counting with a Neubauer Chamber and Trypan Blue staining under the microscope. At high concentrations, the nuclei are very close together, making it difficult for the software to separate them. In this case, recounting the sample in a suitable dilution is recommended. The integrity of the nuclei as well as the cleanliness, can be assessed from the bright field image or under a microscope. - Dilute the samples with PBS (1x PBS + 0.04% BSA + RNase inhibitor at 0.2 U/µL) to the desired concentration for single nuclei RNA sequencing.
NOTE: Concentrations between 700-1,200 nuclei/µL are considered optimal for single nuclei RNA sequencing. Lower cell concentrations, such as 700 nuclei/µL, may result in reduced background contamination from ambient RNA.
5. Library preparation
- Perform single nuclei RNA sequencing with the single cell gene expression reagents using the manufacturer's protocol targeting a nuclei recovery of 8000-10,000 nuclei per sample.
6. Sequencing
- Sequence the libraries with the desired sequencing depth with paired-end, dual indexing, and the following sequencing reads: Read 1: 28 cycles, i7 Index: 10 cycles, i5 Index: 10 cycles and Read 2: 90 cycles.
Results
The performance and versatility of this protocol are demonstrated by performing single nuclei RNA sequencing on fresh frozen brain occipital cortex tissue from three B6 mice, fresh frozen transversally cut kidney tissue from three Wistar rats, archival (11-year-old) liver and spleen tissue from three Mauritian Cynomolgus macaques. All animals were non-perfused.
As shown in Figures 1B,C, good-quality nuclei that were free of signs of blebbing, debris, and clumping were obtained. The sucrose gradient-based filtration was optimized to remove the majority of debris by testing different densities, spin speeds, and times, and assessing the nuclear purity/integrity under a microscope as well as assessing nuclei size distribution and yield (Figure 1D). This allowed us to choose a sucrose gradient density of 1.5 M and to use a short spin time of 15 min. Next, to further assess the quality of the nuclei, the data was preprocessed using 10X Cell Ranger, and further downstream data analysis was performed using Besca23. Nuclei with >5% percent mitochondrial content (as these tend to be damaged/stressed nuclei) were filtered out, and nuclei with 500-7,000 genes (to minimize empty droplets and multiplets) were retained. We only included genes that were present in at least 30 nuclei. We targeted 8,000 nuclei per brain cortex sample and 10,000 nuclei per kidney, liver, and spleen sample. After filtration, 10,644 high-quality nuclei from the three brain samples, 14,960 high-quality nuclei from the three kidney samples, 18,795 high-quality nuclei from the three liver samples, and 13,882 high-quality nuclei from the three spleen samples were obtained. Figure 2A,D,G,J show violin plots representing the distribution of UMI counts, gene counts, and mitochondrial content in each sample. The median number of counts across all brain samples was 7,563 UMI/nucleus and 3,208 genes/nucleus. The median number of counts across all kidney samples was 3,841 UMI/nucleus and 1,915 genes/nucleus. The median number of counts across all liver samples was 2,649 UMI/nuclei and 1,676 genes/nuclei. The median number of counts across all spleen samples was 1,609 UMI/nuclei and 1,138 genes/nuclei. We then generated clusters using highly variable genes and annotated them using known marker genes17,24,25,26. As seen in Figure 2B,E,H,K, we were able to identify the expected cell types from each tissue. Furthermore, as seen in Figure 2B,E,H,K, all animals contributed to all clusters, indicating overall low technical variability introduced by the protocol. Furthermore, the cellular proportions were comparable in all three samples per tissue type, as were the UMI and gene counts (Figure 2A,C,D,F,G,I,J,L). A notable exception is the liver, where the hepatocyte populations among the three liver samples were different in proportions and profile. This is most likely due to biological differences between the animals (sex, age, metabolic status).
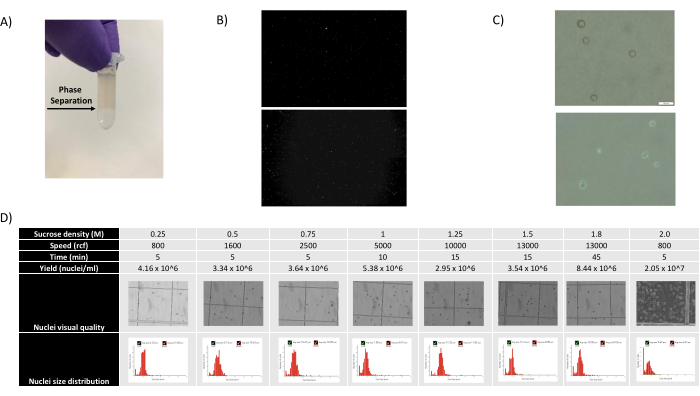
Figure 1: Assessment of nuclei quality and sucrose gradient optimization. (A) The expected phase separation during sucrose gradient centrifugation is shown with an arrow. (B) Representative fluorescent images of propidium iodide-stained rat kidney (top) and cynomolgus spleen (bottom) nuclei obtained with the protocol. (C) Representative bright field microscopy images of nuclei isolated from mouse liver (top) and mouse brain (bottom), scale bar 500 µm. Note the regular smooth surface of nuclei indicating good nuclear quality. (D) Sucrose gradient optimization. Several sucrose densities, spin speeds, and spin times were tested. Bright-field microscopy images of nuclei, nuclei size distribution, and nuclei yield are shown for each condition. Please click here to view a larger version of this figure.
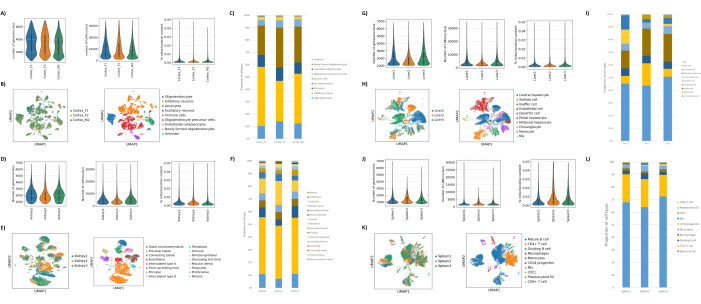
Figure 2: Representative data from snRNAseq on mouse brain occipital cortex, rat kidney (cortex and medulla), and cynomolgus macaque liver and spleen. (A) Violin plots showing the distribution of genes/nucleus, UMIs/nucleus, and percent mitochondrial content per brain sample. (B) Left panel: UMAP plot showing the contribution of each sample to the clusters identified in the brain. Right panel: UMAP showing the identities of the clusters annotated based on marker genes in brain tissue. (C) Cellular proportions observed in the 3 brain samples. (D) Violin plots showing the distribution of genes/nucleus, UMIs/nucleus, and percent mitochondrial content per kidney sample. (E) Left panel: UMAP plot showing the contribution of each sample to the clusters identified in the kidney. Right panel: UMAP showing the identities of the clusters annotated based on marker genes in kidney tissue. (F) Cellular proportions observed in the 3 kidney samples. (G) Violin plots showing the distribution of genes/nucleus, UMIs/nucleus, and percent mitochondrial content per liver sample. (H) Left panel: UMAP plot showing the contribution of each sample to the clusters identified in the liver. Right panel: UMAP showing the identities of the clusters annotated based on marker genes in liver tissue. (I) Cellular proportions observed in the 3 liver samples. (J) Violin plots showing the distribution of genes/nucleus, UMIs/nucleus, and percent mitochondrial content per spleen sample. (K) Left panel: UMAP plot showing the contribution of each sample to the clusters identified in the spleen. Right panel: UMAP showing the identities of the clusters annotated based on marker genes in spleen tissue. (L) Cellular proportions observed in the 3 spleen samples. Please click here to view a larger version of this figure.
| Components | Stock concentration | Volume per sample | Final Concentration |
| Sucrose Cushion Solution | 2 M | 1500 µL | 1.5 M |
| Sucrose Cushion Buffer | - | 500 µL | - |
| Dithiothreitol (DTT) | 1 M | 2 µL | 1 mM |
| RNAse inhibitor | 40 U/µL | 10 µL | 0.2 U/µL |
Table 1: Preparation of 1.5 M sucrose cushion solution (SCS). This solution is used for the sucrose gradient centrifugation during the clean-up in step 3.1 and should be freshly prepared each time before starting the protocol. Always keep the SCS on ice during the protocol. The solutions mentioned in this table are referenced in the Table of Materials.
| Components | Stock concentration | Volume per sample | Final Concentration |
| Silica colloid stock solution | 90% | 600 µL | 18% |
| Nuclei Storage Reagent (S2 Genomics) | - | 2400 µL | - |
| RNAse inhibitor | 40 U/µL | 15 µL | 0.2 U/µL |
Table 2: Preparation of 18% silica colloid solution. This solution is used for the silica colloid gradient centrifugation during clean-up in step 3.2 and should be freshly prepared each time before starting the protocol. Always keep the 18% silica colloid solution on ice during the protocol.
| Tissue | Sample weight | Cartridge | Yield |
| Rat liver | 25 mg | Nuclei Isolation Cartridge | 65,000 nuclei per mg of tissue |
| Rat liver | 4 mg | Small Input Nuclei Isolation Cartridge | 32,000 nuclei per mg of tissue |
Table 3: Nuclei yield from the low input nuclei isolation cartridge versus the nuclei isolation cartridge post sucrose gradient clean-up.
| Components | Stock concentration | Volume per sample | Final Concentration |
| Nuclei Storage Reagent | - | 1000 µL | - |
| RNAse inhibitor | 40 U/µL | 5 µL | 0.2 U/µL |
Table 4: Preparation of nuclei storage reagent (NSR). This solution is used during Nuclei Isolation in steps 3-5 as well as during the clean-up in step 3.1.8. It can be stored at 4 °C for up to 4 months. Prepare a fresh aliquot with RNase inhibitor during the centrifugation step in the clean-up step 6. The solutions mentioned in this table are referenced in the Table of Materials.
| 1x PBS + 0.04% BSA stock solution | |||
| Components | Stock Concentration | Volume for stock | Final Concentration |
| PBS (no Ca2+, no Mg2+) | 1x | 30 mL | - |
| Bovine Serum Albumin (BSA) | 30% | 40 µL | 0.04% |
| 1x PBS + 0.04% BSA + 0.2 U/µL RNAse inhibitor | |||
| Components | Stock Concentration | Volume per sample | Final Concentration |
| 1x PBS + 0.04% BSA Stock Solution | - | 500 µL | - |
| RNAse inhibitor | 40 U/µL | 2.5 µL | 0.2 U/µL |
Table 5: Preparation of PBS + 0.04% BSA. This solution is used at the end of the clean-up in step 3.1.10 and after counting to dilute the nuclei suspension to the desired concentration for 10X single nuclei RNA sequencing (counting step 4.4). The stock solution can be stored at 4 °C for up to 1 month. Prepare a fresh aliquot with RNase inhibitor during the centrifugation step in the clean-up step 6.
Discussion
We have developed a versatile and partially automated protocol to obtain high-quality single nuclei from frozen mammalian tissues and demonstrated the protocol on mouse brain, rat kidney, and cynomolgus liver and spleen tissue.
When comparing the performance of this protocol to that of other published protocols for single nucleus RNA sequencing in brain, kidney, spleen, and liver tissue6,7,20,24,25,26, we observe that we are able to detect a similar number of genes and UMI counts per nucleus and are able to recover the expected cell types. Compared to existing methods, there are several advantages to this protocol. First, the protocol in this study automates tissue homogenization and isolation of single nuclei. This is achieved with the use of a robotic tissue disruptor21. In most protocols, tissue is homogenized with a Dounce homogenizer in order to liberate single nuclei3,20. However, we have noticed that this manual step can lead to experimental variability in nuclei yield and integrity depending on the amount of force exerted during homogenization, compromising the reproducibility of the experiments. Here, by using an automated tissue grinder with fixed settings, good nuclei quality and yield with greater consistency were obtained across experiments. Furthermore, automating this step also reduces the hands-on time of the protocol (the tissue disruption step takes approximately 7 min), allowing the user to prepare for the subsequent steps. Second, the protocol described in this study is versatile, i.e., it is compatible with different tissues from different species. This enables us to avoid lengthy protocol optimization, e.g., to identify homogenization buffers/detergents for different tissues2,5,6. Third, this protocol does not depend on access to a flow sorter in order to obtain clean nuclei, hence making it more accessible for laboratories that do not have the required equipment/expertise for flow sorting. Instead, we optimized the sucrose gradient-based filtration to remove most of the debris. However, for brain tissue in particular, using a silica colloid gradient instead of a sucrose gradient is recommended for more efficient myelin removal. We have also found that the use of a swinging-bucket rotor at the end of the sucrose/silica colloid gradient centrifugation step minimizes nuclei loss. Hence, the use of such a rotor is highly recommended. Fourth, after testing multiple methods to count nuclei (manual counting under the microscope, use of several automated counters), the use of an automated fluorescent cell counter22 is recommended. The use of a DNA intercalating dye, such as propidium iodide, increases the accuracy of nucleus counting. Fifth, this protocol takes about 75 min from start to loading the microfluidic chip. This helps to ensure that nuclei integrity remains high when processing multiple samples. Finally, we have found the protocol to also be compatible with optimal cutting temperature compound (OCT)-embedded tissue. If using such material, the tissue can be removed from the OCT block using a scalpel before homogenization.
One frequent challenge in single nuclei RNA sequencing datasets is the presence of ambient RNA, which can be non-nuclear (e.g., mitochondrial) as well as nuclear derived27,28. In our protocol, mitochondrial RNA (a proxy for non-nuclear ambient RNA) is low even prior to filtering (0.1-1.6% for the tissues shown). However, similar to other protocols and datasets, ambient RNA contamination from highly expressed genes in the nuclei of abundant cell types (such as hepatocytes in the liver, neurons in brains, etc.) is still present27. Several bioinformatics tools, such as CellBender, SoupX, etc., exist that can remove such ambient RNA contamination prior to nuclei annotation29,30,31. Another limitation of this protocol is that although the tissue disruption and nuclei isolation steps are automated, the throughput of this step is still limiting as only one sample can be processed at a time. However, since this step takes only approximately 7 min per piece of tissue, multiple samples can still be processed in a batch. We typically process four samples per batch but have done up to six samples per batch with good results. Recent improvements in the robotic dissociator to allow the parallel processing of two samples simultaneously will enable the processing of 8-12 samples per batch, which is compatible with the throughput of the microfluidic chip that is used for single nuclei encapsulation.
Although we have not used the nuclei isolated by this protocol for other downstream applications such as ATAC-seq or snRNAseq using other platforms, based on the quality of data obtained with the gene expression reagents used here, we believe our protocol should be compatible with additional downstream applications. However, future work will involve testing this protocol with other downstream applications, such as ATAC-seq.
In conclusion, we have developed a quick, simple, and partially automated nuclei isolation protocol for downstream single nucleus RNA sequencing that has been demonstrated to be compatible with different types of frozen mammalian tissues.
Disclosures
All authors are/were employees of F. Hoffmann-La Roche during the conduct of the study.
Acknowledgements
The authors would like to thank Filip Bochner, Marion Richardson, Petra Staeuble, and Matthias Selhausen for providing the animal tissues that were analyzed in this manuscript. We would also like to thank Petra Schwalie, Klas Hatje, Roland Schmucki, and Martin Ebeling for their bioinformatics support.
Materials
| Name | Company | Catalog Number | Comments |
| 1 M DTT | Thermo Fisher Scientific | P2325 | |
| 10% Tween 20 | Bio-Rad | 1662404 | |
| 10x Magnetic Separator | 10x genomics | PN-120250 | |
| 10x Vortex Adapter | 10x genomics | PN-120251 | |
| 1x DPBS (10x), no calcium, no magnesium | Thermo Fisher Scientific | 14190144 | stored at 4°C |
| 30% Bovine Serum Albumin | Sigma-Aldrich | A9576_50ML | |
| 400 mM Tris-HCl, pH 8.0 | Thermo Fisher Scientific | 15568025 | |
| 40U/μl RNaseOUT Recombinant Ribonuclease Inhibitor | Thermo Fisher Scientific | 10777019 | Stored at -20 °C |
| Agilent High Sensitivity DNA Kit | Agilent | 5067-4626 | |
| Cellaca MX High-throughput Automated Cell Counter | Nexcelom Bioscience | CELMXSYSF2 | Automated fluorescent cell counter |
| Chromium Next GEM Chip G Single Cell Kit, 16 rxns | 10x genomics | PN-1000127 | Single cell gene expression reagent, stored at room temperature |
| Chromium Next GEM Secondary Holder | 10x genomics | PN-1000195 | |
| Chromium Next GEM Single Cell 3' Gel Bead Kit v3.1, 4 rxns | 10x genomics | PN-1000129 | Single cell gene expression reagent, stored at -80 °C |
| Chromium Next GEM Single Cell 3' GEM, Library & Gel Bead Kit v3.1, 4 rxns | 10x genomics | PN-1000128 | Single cell gene expression reagent |
| Chromium Next GEM Single Cell 3' Library Kit v3.1 4 rxns | 10x genomics | PN-1000158 | Single cell gene expression reagent, stored at -20 °C |
| Chromium Next GEM Single Cell 3'GEM Kit v3.1 4 rxns | 10x genomics | PN-1000130 | Single cell gene expression reagent, stored at -20 °C |
| Divided Polystyrene Reservoirs | VWR | 41428-958 | |
| DNA LoBind Tubes 1.5ml Eppendorf | Sigma-Aldrich | EP0030108051 | |
| DNA LoBind Tubes 2ml Eppendorf | Sigma-Aldrich | EP0030108078 | |
| Dry ice | - | - | |
| Dynabeads MyOne SILANE | 10x genomics | PN-2000048 | Single cell gene expression reagent, stored at 4 °C |
| Ethanol Pure | Sigma-Aldrich | E7023 | |
| Glycerin (Glycerol), 50% (v/v) | Ricca Chemical Company | 3290-16 | |
| Heatblock | |||
| High-Throughput Nexcelom Counting Plates | Nexcelom Bioscience | CHM24-A100-001 | Cell counter counting plate |
| Low TE Buffer (10 mM Tris-HCl pH 8.0, 0.1 mM EDTA) | Thermo Fisher Scientific | 12090015 | |
| Mini Centrifuge | - | - | |
| NovaSeq 6000 SP Reagent Kit v1.5 (100 cycles) | Illumina | 2002840 | |
| Nuclei Isolation Buffer | S2 Genomics | 100-063-396 | Stored at 4 °C |
| Nuclei Isolation Cartridge | S2 Genomics | 100-063-287 | Precooled at 4 °C before use |
| Nuclei PURE 2 M Sucrose Cushion Solution | Sigma-Aldrich | NUC201-1KT | Sucrose cushion solution |
| Nuclei PURE Sucrose Cushion Buffer | Sigma-Aldrich | NUC201-1KT | |
| Nuclei Storage Reagent | S2 Genomics | 100-063-405 | Stored at 4 °C |
| PCR Tubes 0.2 ml 8-tube strips | Eppendorf | 30124359 | |
| Percoll | GE Healthcare | 17-0891-02 | Silica colloid solution |
| PhiX Control v3 | Illumina | FC-110-3001 | |
| Qiagen Buffer EB | Qiagen | 19086 | |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Q32854 | |
| Refrigerated Centrifuge (Eppendorf 5804R) | Eppendorf | 5805000010 | |
| Refrigerated Centrifuge with Swinging-Bucket Rotor (Eppendorf 5810R) | Eppendorf | 5811000015 | |
| RNAseZap | Ambion | AM9780 | RNAse decontamination solution |
| Round cell culture petri dish | SPL | 330005 | |
| Scalpel disposable | Aesculap AG | BA210 | pre-cooled on dry ice before use |
| Single Index Kit T Set A, 96 rxns | 10x genomics | PN-1000213 | Single cell gene expression reagent, stored at -20 °C |
| Singulator 100 System | S2 Genomics | - | Commercially available robotic tissue dissociator |
| Sodium Hydroxide 1M | Sigma-Aldrich | 72068 | |
| SPRIselect Reagent Kit | Beckman Coulter | b23318 | |
| Sterile tweezers | - | - | |
| UltraPure DNase/RNase-Free Distilled Water | Thermo Fisher Scientific | 10977049 | |
| ViaStain PI Staining Solution | Nexcelom Bioscience | CS1-0109-5mL | Propidium iodide staining solution |
| Vortex Mixer+A2:D44 | VWR | - |
References
- Burja, B., et al. An Optimized Tissue Dissociation Protocol for Single-Cell RNA Sequencing Analysis of Fresh and Cultured Human Skin Biopsies. Front Cell Dev Biol. 10, 872688 (2022).
- Kimbley, L. M., et al. Comparison of optimized methodologies for isolating nuclei from esophageal tissue. Biotechniques. 72 (3), 104-109 (2022).
- Maitra, M., et al. Extraction of nuclei from archived postmortem tissues for single-nucleus sequencing applications. Nature Protocols. 16 (6), 2788-2801 (2021).
- Nadelmann, E. R., et al. Isolation of nuclei from mammalian cells and tissues for single-nucleus molecular profiling. Current Protocols. 1 (5), e132 (2021).
- Rousselle, T. V., et al. An optimized protocol for single nuclei isolation from clinical biopsies for RNA-seq. Scientific Reports. 12, 9851 (2022).
- Slyper, M., et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nature Medicine. 26 (5), 792-802 (2020).
- Leiz, J., et al. Nuclei isolation from adult mouse kidney for single-nucleus RNA-sequencing. Journal of Visualized Experiments: JoVE. (175), 62901 (2021).
- Alvarez, M., et al. Isolation of nuclei from human snap-frozen liver tissue for single-nucleus RNA sequencing. Bio-Protocol. 13 (3), e4601 (2023).
- Ayhan, F., Douglas, C., Lega, B. C., Konopka, G. Nuclei isolation from surgically resected human hippocampus. STAR Protocols. 2 (4), 100844 (2021).
- Joshi, N., Misharin, A. Single-nucleus isolation from frozen human lung tissue for single-nucleus RNA-seq. Protocols.io. , (2019).
- Martelotto, L. G., Luciano Martelotto, L. 'Frankenstein' protocol for nuclei isolation from fresh and frozen tissue for snRNAseq. Protocols.io. , (2020).
- Masilionis, I., Chaudhary, O., Chaligne, R., Mazutis, L. Nuclei extraction for single-cell RNAseq from frozen tissue using Singulator™ 100. Protocols.io. , (2022).
- Matson, K. J. E., et al. Isolation of adult spinal cord nuclei for massively parallel single-nucleus RNA sequencing. Journal of Visualized Experiments: JoVE. (140), 58413 (2018).
- Mendelev, N., et al. Multi-omics profiling of single nuclei from frozen archived postmortem human pituitary tissue. STAR Protocols. 3 (2), 101446 (2022).
- Soule, T. G., et al. A protocol for single nucleus RNA-seq from frozen skeletal muscle. Life Science Alliance. 6 (5), e202201806 (2023).
- Bakken, T. E., et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One. 13 (12), e0209648 (2018).
- Ding, J., et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nature Biotechnology. 38, 737-746 (2020).
- Hu, P., et al. Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts. Genes & Development. 32 (19-20), 1344-1357 (2018).
- Lake, B. B., et al. A comparative strategy for single-nucleus and single-cell transcriptomes confirms accuracy in predicted cell-type expression from nuclear RNA. Scientific Reports. 7, 6031 (2017).
- Narayanan, A., et al. Nuclei Isolation from Fresh Frozen Brain Tumors for Single-Nucleus RNA-seq and ATAC-seq. Journal of Visualized Experiments: JoVE. (162), 61542 (2020).
- Jovanovich, S., et al. . Automated processing of solid tissues into single cells or nuclei for genomics and cell biology applications with the Singulator™ 100 and 200 systems. , (2022).
- Bell, J., et al. Characterization of a novel high-throughput, high-speed and high-precision plate-based image cytometric cell counting method. Cell & Gene Therapy Insights. 7 (4), 427-447 (2021).
- Madler, S. C., et al. Besca, a single-cell transcriptomics analysis toolkit to accelerate translational research. NAR Genomics and Bioinformatics. 3 (4), lqab102 (2021).
- Wu, H., et al. Mapping the single-cell transcriptomic response of murine diabetic kidney disease to therapies. Cell Metabolism. 34 (7), 1064-1078 (2022).
- Han, L., et al. Cell transcriptomic atlas of the non-human primate Macaca fascicularis. Nature. 604 (7907), 723-731 (2022).
- Madissoon, E., et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biology. 21 (1), 1 (2019).
- Caglayan, E., Liu, Y., Konopka, G. Neuronal ambient RNA contamination causes misinterpreted and masked cell types in brain single-nuclei datasets. Neuron. 110 (24), 4043-4056 (2022).
- Luecken, M. D., Theis, F. J. Current best practices in single-cell RNA-seq analysis: a tutorial. Molecular Systems Biology. 15 (6), e8746 (2019).
- Fleming, S. J., et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. bioRxiv. , (2022).
- Yang, S., et al. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biology. 21 (1), 57 (2020).
- Young, M. D., Behjati, S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience. 9 (12), giaa151 (2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved