Simultaneous Isolation of Principal Central Nervous System-Resident Cell Types from Adult Autoimmune Encephalomyelitis Mice
* These authors contributed equally
In This Article
Summary
To date, protocols for the simultaneous isolation of all principal central nervous system-resident cell types from the same mouse are an unmet demand. The protocol shows a procedure applicable in naïve and experimental autoimmune encephalomyelitis mice to investigate complex cellular networks during neuroinflammation and simultaneously reduce the required mice numbers.
Abstract
Experimental autoimmune encephalomyelitis (EAE) is the most common murine model for multiple sclerosis (MS) and is frequently used to further elucidate the still unknown etiology of MS in order to develop new treatment strategies. The myelin oligodendrocyte glycoprotein peptide 35-55 (MOG35-55) EAE model reproduces a self-limiting monophasic disease course with ascending paralysis within 10 days after immunization. The mice are examined daily using a clinical scoring system. MS is driven by different pathomechanisms with a specific temporal pattern, thus the investigation of the role of central nervous system (CNS)-resident cell types during disease progression is of great interest. The unique feature of this protocol is the simultaneous isolation of all principal CNS-resident cell types (microglia, oligodendrocytes, astrocytes, and neurons) applicable in adult EAE and healthy mice. The dissociation of the brain and the spinal cord from adult mice is followed by magnetic-activated cell sorting (MACS) to isolate microglia, oligodendrocytes, astrocytes, and neurons. Flow cytometry was used to perform quality analyses of the purified single-cell suspensions confirming viability after cell isolation and indicating the purity of each cell type of approximately 90%. In conclusion, this protocol offers a precise and comprehensive way to analyze complex cellular networks in healthy and EAE mice. Moreover, required mice numbers can be substantially reduced as all four cell types are isolated from the same mice.
Introduction
Multiple sclerosis (MS) is a chronic inflammatory autoimmune disease of the central nervous system (CNS) characterized by demyelination, axonal damage, gliosis, and neurodegeneration. Despite numerous research approaches in this field, the pathophysiology of MS is still not fully understood1,2,3,4. The most common animal model for investigating MS is the myelin oligodendrocyte glycoprotein peptide 35-55 (MOG35-55)-induced experimental autoimmune encephalomyelitis (EAE) which shares many of its clinical and pathophysiological features5,6,7,8,9. It is based on the response of the immune system against CNS-specific antigens leading to inflammation, demyelination, and neuroaxonal degeneration. Experimental autoimmune encephalomyelitis (EAE) is a suitable model for the investigation of neuroinflammatory pathways and signaling cascades found in MS.
Current therapy options for MS are only partially effective and focus primarily on the initial inflammatory phase of the disease. However, the neurodegenerative component of MS seems to be the major challenge for long-term therapeutic approaches. Therefore, reproducible, and precise cell isolation protocols are required to investigate molecular and cellular mechanisms in autoimmune diseases in a comprehensive way. Even if some protocols for the isolation of one single cell type exist10,11,12,13,14,15, there is an unmet need for the simultaneous isolation of several CNS-resident cell populations at once. Previous protocols for the isolation of CNS-resident cells lack in preserving cellular functionality and purity, resulting in co-cultivation with neighboring cells16,17,18 or the unsuitability for complex analyses of intracellular networks ex vivo19,20,21,22.
The aim of this protocol was to establish a reproducible and comprehensive method for the simultaneous isolation of pure viable single-cell suspensions of all principal CNS-resident cell types applicable in adult healthy and EAE mice. The different cell types were isolated using magnetic-activated cell sorting (MACS)23. The cell separation can be accomplished either by positive selection, i.e., magnetic labeling of cell-type-specific surface markers, or by negative selection via biotinylation and depletion of all undesired cells. Flow cytometry was applied to ensure a purity of above 90% and a viability of at least 80% of the isolated single-cell suspensions.
In conclusion, the main goal was to establish a protocol for the simultaneous isolation of all main CNS-resident cell types as a versatile tool for the investigation of neuroinflammatory pathways offering a comprehensive and precise analysis of complex cellular networks and biochemical signaling cascades in healthy and EAE mice.
Protocol
All EAE experiments were induced in female C57BL/6J mice at the age of 10-12 weeks and approved by local authorities (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen). The compliance with the German and EU animal protection law was also ensured at any time of the experiments. All mice were kept under individually ventilated cages animal housing conditions.
NOTE: The following reagent volumes refer to one adult murine brain and spinal cord, which are named CNS cell suspension in the following and weigh approximately about 20 mg to 500 mg. If dissociation of more than one CNS cell suspension is planned, all reagent volumes and materials have to be scaled up accordingly. It is recommended to store Dulbecco's phosphate buffered saline (D-PBS; 1x) with calcium and magnesium, supplemented with 1 g/L glucose and 36 mg/L sodium pyruvate) continuously on ice during the whole experiment. If cell cultivation is planned afterwards, perform all steps under sterile conditions by the usage of hoods. Otherwise, none of the following protocol sections need to be performed under a hood. Store the buffers on ice. Use only pre-cooled solutions and avoid vortexing throughout the whole experiment. See Figure 1 for overall workflow.
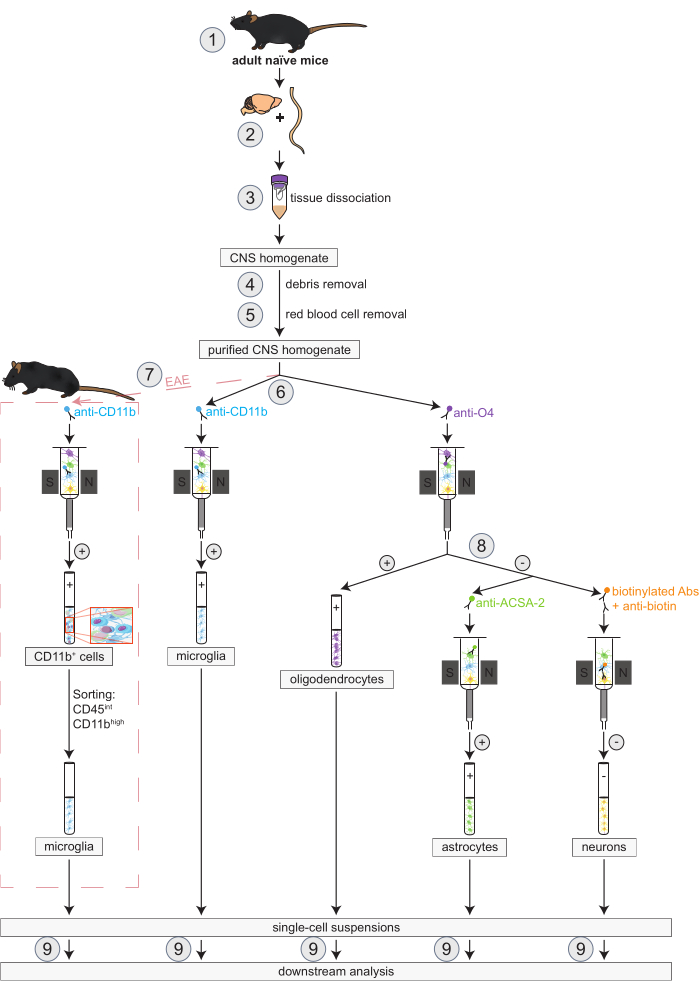
Figure 1: Workflow for the simultaneous isolation of oligodendrocytes, microglia, astrocytes and neurons in naïve and EAE mice. The first steps of the workflow are the same for both naïve and EAE mice. If working with an EAE replicate is desired, EAE induction has to be performed beforehand (1). In brief, the protocol begins with the dissection (2) and dissociation (3) of murine brain and spinal cord followed by the removal of debris (4) and red blood cells (5). Subsequently, the resulting purified CNS cell suspension is split into two fractions for the simultaneous isolation of oligodendrocytes and microglia via MACS (6). Microglia are detected via anti-CD11b micro-beads while oligodendrocytes are isolated using anti-O4 micro-beads (positive selections). From the negative flow-through of the oligodendrocytes (8), astrocytes are isolated via anti-ACSA-2 micro-beads (positive selection) and neurons by biotin labeling and depletion of all non-neuronal cells (negative selection). In EAE mice, the isolation of CD11b+ cells is followed by fluorescence-activated cell sorting of CD45intCD11bhigh cells to eliminate other CD11b+ immune cells like macrophages, dendritic cells, monocytes, granulocytes, and natural killer cells that are known to participate in neuroinflammation processes during the EAE course (7)27, 28, 48. After the isolation of the different CNS-resident cell types, purity analyses can be performed (9). Abbreviations: Abs = antibodies; ACSA-2 = astrocyte cell surface antigen-2; CD11b = cyclin-dependent kinase 11B; CD45 = receptor-type tyrosine-protein phosphatase C; CNS = central nervous system; EAE = experimental autoimmune encephalomyelitis; MACS = magnetic-activated cell sorting; O4 = oligodendrocyte marker O4. This figure has been modified from49. Please click here to view a larger version of this figure.
1. Induction of active EAE
- Preparation of reagents
- For cell separation: Prepare the PB buffer and store it at 2-8 °C for maximum 1 week. To prepare stock solution, add 475 mL of 1x PBS without supplements (pH 7.2) + 25 mL of 0.5% bovine serum albumin (BSA). Use a 1:20 dilution prepared in BSA.
- For flow cytometry and fluorescence-activated cell sorting (FACS): Prepare the FACS buffer, PBS with 2% fetal calf serum (FCS) and 2 mM EDTA and store it at 2-8 °C. To prepare, add 500 mL of 1x PBS without supplements and 10 mL of FCS + 2 mL EDTA (from 0.5 M EDTA stock)
- Perform immunization according to the protocol from Bittner et al. 5. In brief, induce EAE by subcutaneous injection of an emulsion containing 200 µg MOG35-55 peptide and 200 µL of complete Freud´s adjuvant including 200 µg Mycobacterium tuberculosis.
- Anesthetize the mouse with 2% isoflurane by using an anesthesia chamber with an isoflurane vaporizer. Use vet ointment on the animal's eyes to prevent dryness while under anesthesia.
- After 2 h, inject an intraperitoneal injection of 100 ng Pertussis toxin (PTx) dissolved in 100 µL of 1x PBS according to the protocol from Huntemann et al.24. Repeat the PTx injection on day 2 after immunization.
CAUTION: Observe each animal until it has regained sufficient consciousness to maintain sternal recumbency. Mice that have undergone the injection procedures are not returned to the company of the other mice until they have fully recovered. For Mycobacterium tuberculosis and PTx: Avoid inhalation, ingestion, and contact with the skin and eyes. Mycobacterium tuberculosis is an activator of the innate immune system. PTx has many biological effects. - Monitor EAE progression daily, performed by two blinded investigators who monitor weight and examine the mice clinically.
- For this purpose, use the following scoring system was grade 0-no clinical signs of EAE, grade 1- partial tail paresis, grade 2-complete tail paresis, grade 3-moderate hind limb weakness, grade 4-complete hind limb weakness and ataxic gait, grade 5-mild paraparesis, grade 6-paraparesis, grade 7-paraplegia, grade 8-tetraparesis, grade 9-quadriplegia, and grade 10-death.
- Use the following exclusion criteria for further participation in the experiment clinical score > 7 or a weight loss exceeding 20% of the initial body weight.
- For the dissection of the brain and the spinal cord, euthanize EAE mice on day 16 after EAE induction representing disease maximum.
2. CNS tissue preparation (Duration: approximately 10 min per mice)
- After sacrificing mice with carbon dioxide, start with the transcardial perfusion of each mouse with 20 mL of 1x PBS. Repeat perfusion again with 20 mL of 1x PBS.
- Place the mouse in the supine position and fix the limbs with cannulas. Apply 75% ethanol on the front body of the animal. Further sterility measures are not necessary at that point.
- Open the abdomen and thorax by making a longitudinal section through the skin and fascia with the help of a scissor.
- Cut the ribs laterally and fold up the thorax to gain free access to the heart. Fix the thorax folded upwards with cannulas.
- Open the right atrium using scissors. Apply 20 mL of 1x PBS into the left ventricle with a cannula to flush out the blood through the incised right atrium.
- Expose the skull by cutting the skin on top of the murine head via a longitudinal section and shift the skin around the head using a forceps. Incise the skull with help of a scissor along the sagittal suture.
- Insert the tip of a forceps along the incision line to crack open the calotte. Remove remaining parts of the calotte with forceps so that the brain is fully exposed.
- Remove the brain carefully and place it into a murine brain matrix. Cut the brain into 1 mm thick sagittal slices by using a razor blade.
- Cut the vertebral column with help of scissors just above the iliac crest so that the syringe can be inserted into the spinal canal.
NOTE: The easiest way to remove the spinal cord is to flush it out of the spinal canal with PBS. Otherwise, the vertebral arches must be opened individually with scissors and then the spinal cord can be removed. - Flush the spinal cord out of the spinal canal from caudal to cranial by using a 20 mL syringe with a 20G needle containing 1x PBS. Cut the spinal cord into 0.5 cm long segments using a scalpel.
- Store each CNS cell suspension consisting of brain and corresponding spinal cord in one separate Petri dish per mouse filled with approximately 3 mL of cold D-PBS. Store the dishes on ice until further processing.
3. CNS tissue dissociation (Duration: approximately 1-1.5 h depending on the number of CNS cell suspensions)
NOTE: Neural tissue from adult mice is dissociated by combining mechanical dissociation with enzymatic degradation of the extracellular matrix. Thereby, the structural integrity remains, and the cell suspension can be used for further cell isolation procedures.
- Prepare the appropriate volume of enzyme mix 1 consisting of 50 µL of enzyme P and 1,900 µL of buffer Z per CNS cell suspension. Both reagents belong to the adult brain dissociation kit.
- Prepare the appropriate volume of enzyme mix 2 consisting of 10 µL of enzyme A and 20 µL of buffer Y per CNS cell suspension. Both reagents belong to the adult brain dissociation kit.
- Transfer 1,950 µL of enzyme mix 1 into C tube and add the tissue pieces of one CNS cell suspension afterwards. Use one C tube per mouse.
- Add 30 µL of enzyme mix 2 to each C tube. Close the C tubes tightly and attach them upside down onto the sleeve of the cell dissociator with heaters.
- Run the appropriate program named 37C_ABDK_01 (takes 30 min). Observe at least the first 5 min of the program to ensure that all tubes turn at the same velocity. The occurrence of errors during the run is possible. Then, go on to step 6.
- In the last 2 min of the program, place one 70 µm strainer on a 50 mL tube for each dissociated CNS cell suspension. Pre-moisten these strainers with 2 mL of D-PBS.
- After termination of the program, attach the C tubes from the dissociator and place them into a centrifuge. Centrifugate the samples at 300 x g and 4 °C for 1 min to collect the sample at the bottom of the tube.
- Resuspend the sample and apply it to the pre-moistened strainer. Add 10 mL of cold D-PBS to the empty C tube and close it. Shake it gently and apply the suspension onto the corresponding strainer.
- Discard the strainers and close the 50 mL tubes. Centrifugate the cell suspension again at 300 x g and 4 °C for 10 min. Afterwards, aspirate the whole supernatant very carefully.
4. Debris removal (Duration: Approximately 1.5-2 h depending on the number of CNS cell suspensions)
NOTE: Tissue dissociation often leads to myelin and cell debris that can impair downstream analysis. By adding a debris removal solution, this debris can be efficiently removed from the CNS cell suspension.
- Resuspend the cell pellet carefully with 3,100 µL of D-PBS for each CNS cell suspension. Do not vortex.
- If working with more than one CNS cell suspension, pool maximum two CNS cell suspensions derived from one condition or experimental group in one 15 mL tube.
- Add 900 µL of the debris removal solution from the adult brain dissociation kit to one CNS cell suspension or 1,800 µL of debris removal solution to two pooled CNS cell suspensions.
- Invert the tube and mix the suspension. Afterwards, overlay it very gently with 4 mL of cold D-PBS. A clear gradient should be visible (Figure 2A).
- Centrifuge the tubes for 10 min at 3000 x g and 4 °C with full acceleration and no brake.
- If the separation occurs as intended, three phases are formed (Figure 2C). Aspirate the two top phases completely (Figure 2C-1,2) and discard them. It is important that no myelin residues are left behind (Figure 2E).
NOTE: If the gradient did not work and the cells are needed urgently, do not suck off the two top phases. Instead, fill up the 15 mL tube with cold D-PBS up to 15 mL and invert several times. Centrifugate again at 1000 x g for 10 min at 4 °C with full acceleration and no brake. Suck off the supernatant and repeat the steps 4.1- 4.4. - Fill up the tube with cold D-PBS up to 14 mL and close it. Invert the tube powerfully on the work bench until the cell pellet becomes detached from the bottom of the tube. Do not vortex.
- Centrifugate the sample again at 1000 x g and 4 °C for 10 min. Set full acceleration and full brake. Aspirate the supernatant carefully and completely.
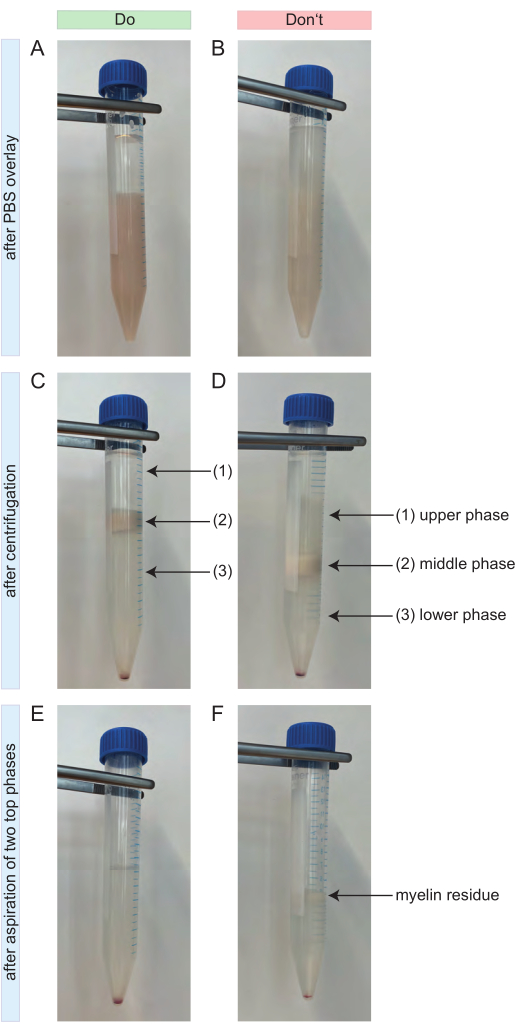
Figure 2: Do's and Dont's during debris removal. (A) Positive example for the gradient after overlaying with 4 mL of PBS. The upper phase consisting of 4 mL of PBS is clearly distinguishable from the lower phase consisting of the CNS cell suspension with the debris removal solution. (B) Negative example for the gradient after overlaying with 4 mL of PBS. The gradient lacks a clear separation between the PBS and the cell suspension below. A bit of the PBS is diffused into the cell suspension. (C) Positive example for the gradient after centrifugation. Three separate phases can be easily distinguished. No myelin residues are visible in the upper (1) or lower phase (3) of the gradient. The middle phase contains all of the myelin (2). The cell pellet is visible at the bottom of the 15 mL tube. (D) Negative example for the gradient after centrifugation. There is no accurate separation between the three phases possible. Some myelin residues are visible in the upper (1) and lower phase (3) of the gradient. (E) Positive example for the gradient after aspirating the two top phases. The resulting sample contains only the cell pellet and a clear supernatant above. No myelin residues are left behind. (F) Negative example for the gradient after aspirating the two top phases. The sample still contains some myelin residues (black arrow). Abbreviations: CNS = central nervous system; PBS = phosphate-buffered saline Please click here to view a larger version of this figure.
5. Red blood cell removal (Duration: Approximately 1 h depending on the number of CNS cell suspensions)
NOTE: This step prevents later contamination by red blood cells and ensures an optimal lysis of erythrocytes with minimal effect on the other cell types isolated from the CNS tissue. The following volumes are indicated for cell suspensions derived from 100 mg to 1 g neuronal tissue corresponding to two adult mouse brains and spinal cords. If working with more than two CNS cell suspensions, scale up all reagents and total volumes accordingly.
- Start with the preparation of red blood cell removal solution (RBCRS): per two pooled CNS cell suspensions. Dilute 100 µL of red blood cell removal stock solution (10x) from the adult brain dissociation kit in 900 µL of ddH2O to reach a final dilution of 1:10.
- Store the RBCRS at 2-8 °C until use. Discard unused remains at the end of the day.
- Resuspend the cell pellet of up to two CNS cell suspensions in 1 mL of the RBCRS. Avoid vortexing. Incubate the solution for 10 min at 4 °C.
- Add 10 mL of cold PB buffer to two pooled cell suspensions. Centrifugate the sample at 300 x g and 4 °C for 10 min and aspirate the supernatant completely afterwards.
- Resuspend each cell pellet from one CNS cell suspension in 80 µL of PB buffer by pipetting slowly up and down. Accordingly, use 160 µL to resuspend cell pellets derived from two CNS cell suspensions.
- When working with several CNS cell suspensions from the same experimental condition, pool all of these cell suspensions.
- Determine the cell count, e.g., using an improved counting chamber. The cell suspensions were usually diluted 1:50 in PB buffer, followed by a further dilution of 1:10 in 0.4% trypan blue solution.
6. Magnetic beads protocol in naïve and EAE mice (Duration: Approximately 1 h)
- Magnetically label the different CNS cell types with MicroBeads specific for their surface antigen. Then, place the cell suspension in the column and magnetically separate labeled cells retained within the column and unlabeled cells that run through.
- After removing the column from the magnetic field, flush out magnetically labeled cells from the column into a tube as the positively selected cell fraction.
NOTE: The volumes for the magnetic labeling process are calculated for up to 1 x 107 total cells. If more cells are obtained, scale up all reagent and total volumes accordingly. It is recommended to work fast and only use pre-cooled solutions to prevent the capping of antibodies on the cell surface and non-specific cell labeling as well as to ensure a high viability of the isolated cell populations. It is also important to perform the washing steps as soon as the column reservoir is empty by adding the PB buffer so that the columns do not dry out. - Divide the purified undiluted CNS cell suspension into two fractions for the following isolations of microglia and oligodendrocytes. The ratio of both fractions depends on the desired cell count of each cell type.
NOTE: Further details (duration of incubation, detailed protocol steps, volumes, reagents, and cell count method) are indicated in Table 1.
Table 1: Workflow for the simultaneous magnetic labeling and isolation of oligodendrocytes and microglia from naïve and EAE mice. Both cell types are isolated via a positive selection. Steps that are listed in the same row are indicated to be performed at once. Abbreviations: CD11b = cyclin-dependent kinase 11B; EAE = experimental autoimmune encephalomyelitis; FcR = Fc receptor-like protein; O4 =oligodendrocyte marker O4. Please click here to download this Table.
7. Protocol amendment: additional sorting for the isolation of microglia in EAE mice (Duration: Approximately 1.5-2 h)
NOTE: When working with EAE mice, it is necessary to complement the MACS-based cell isolation protocol by FACS to remove CD11b+ cell populations other than microglia (e.g., monocytes, macrophages, natural killer cells, granulocytes, or dendritic cells) from the CD11b+ cell fraction. Otherwise, this step can be ignored.
- Prepare the staining master mix containing 1x PBS supplemented by CD11b FITC (clone M1/70, 1:50) and CD45 APC/Cy7 (clone 30-F11, 1:200). Use 100 µL of the staining master mix per 5 x 106 cells. Vortex all antibodies before use.
- Centrifugate the microglia cell suspension at 300 x g and 4 °C for 10 min and aspirate the supernatant carefully.
- Resuspend the cell pellet with 100 µL of the prepared staining master mix per 5 x 106 cells. Incubate for 15 min in the dark at room temperature (RT).
- Stop the reaction by adding 500 µL of PBS and centrifugate the sample again at 300 x g and 4 °C for 10 min.
- Aspirate the supernatant carefully and resuspend the cell pellet with 1x PBS supplemented by 10 µg/mL DNAse to reach a final concentration of 1 x 107 cells per mL. Store the cells at 4 °C until sorting starts.
- Apply the cell suspension on a 100 µm strainer placed on a new FACS tube immediately before starting with sorting.
- Set the flow rate to 1000 events per second and use the 100 µm nozzle. Sort the desired cell population of CD45intCD11bhigh cells into a new 15 mL tube prepared with 1x PBS at RT.
8. Preparation of negative flow-through of oligodendrocytes for the isolation of neurons and astrocytes (Duration: Approximately 1 h)
NOTE: The negative flow-through of oligodendrocytes from step 6 is collected for further isolation of neurons and astrocytes. To this end, the cell suspension is split into two parts. Due to the previous isolation of oligodendrocytes from the CNS cell suspension, contamination by O4+ cells is minimized that would otherwise be observed.
- Centrifugate the negative flow-through of the oligodendrocytes at 300 x g and 4 °C for 10 min and aspirate the supernatant carefully.
- Resuspend the cell pellet in 80 µL of PB buffer per pooled CNS cell suspension previously used for the isolation of the oligodendrocyte positive fraction.
- Count the cells. Perform the counting of cells assumed to be O4- using an improved counting chamber after diluting the cell suspension 1:50 in PB buffer followed by a further 1:10 dilution in 0.4% trypan blue.
- Split the purified undiluted cell suspension into two fractions for the following simultaneous isolation of neurons and astrocytes. The ratio of both fractions depends on the preferred amount of each cell type.
NOTE: Further details (duration of incubation, detailed protocol steps, volumes, reagents, and cell count method) are indicated in Table 2.
Table 2: Workflow for the simultaneous magnetic labeling and isolation of neurons and astrocytes from naïve and EAE mice. Both cell types are isolated from the negative flow-through of oligodendrocytes. Astrocytes are separated as a positive selection via anti-ACSA-2 micro-beads while neurons are purified via biotinylation and depletion of all non-neuronal cells as a negative selection. Steps that are listed in the same row are indicated to be performed at once. Abbreviations: Anti-ACSA-2 = astrocyte cell surface antigen-2; EAE = experimental autoimmune encephalomyelitis; FcR = Fc receptor-like protein; MACS = magnetic-activated cell sorting. Please click here to download this Table.
9. Purity analyses of the isolated CNS-resident cell types (Duration: Approximately 2 h)
NOTE: Performing flow cytometry of all four isolated CNS-resident cell populations is recommended to measure and compare their purities and viability. Therefore, it is necessary to stain all cell types with an antibody labeled with fluorophore. Live/dead cell staining is implemented using a fixable viability dye (1:10,000).
- Purity panel - extracellular staining protocol
- Use 1 x 105 cells dissolved in 50 µL of PBS per staining.
- Prepare the staining master mix dissolved in PBS with 2% FCS/2 mM EDTA consisting of the following fluorochrome-conjugated monoclonal antibodies targeting cell-type-specific surface markers: CD11b FITC (clone 1/70, 1:100)25,26,27,28, Biotin-PE (clone Bio3-18E7, 1:200)29,30,31,32, ACSA-2 PE-Vio615 (clone REA-969, 1:200)33,34,35, O4 APC (clone REA-576, 1:400), and CD45 BV510 (clone 30-F11, 1:150)36,37. Add 1 µg of anti-CD16/32 per 1 x 106 cells to block the Fc receptor38,39. Vortex all antibodies before usage.
- Centrifugate the cell suspension for 5 min at 540 x g and 4 °C and aspirate the supernatant carefully.
- Resuspend the cell pellet in 100 µL of the respective master mix and incubate the sample for 15 min at RT in the dark.
- Wash the cells with 500 µL of 1x PBS with 2% FCS/2 mM EDTA and centrifugate the sample for 5 min at 540 x g and 4 °C.
- Aspirate the supernatant and resuspend the cell pellet with 70 µL of 1x PBS with 2% FCS/2 mM EDTA.
- Vortex the sample to dissociate the cell pellet completely. Subsequently, the sample is ready for flow cytometry analysis.
- Purity panel - intracellular staining protocol with NeuN
- Use 1 x 105 cells of each cell population for intracellular staining of NeuN which is a neuron-specific nuclear marker 40,41. This is an additional way to stain viable neurons.
- Transfer 1 x 105 cells of each cell population into a FACS tube. Add 1 mL of PBS with 2% FCS/2 mM EDTA per tube. Centrifugate the tubes at 540 x g and 4 °C for 5 min.
- In the meantime, prepare the master mix dissolved in PBS with 2% FCS/2 mM EDTA consisting of the following fluorochrome-conjugated monoclonal antibodies targeting cell-type-specific surface markers: CD11b FITC (clone M1/70, 1:100)25,26,27,28, Biotin-PE (clone Bio3-18E7, 1:200)29,30,31,32, ACSA-2 PE-Vio615 (clone REA-969, 1:200)33,34,35, and CD45 BV510 (clone 30-F11, 1:150)36,37.
- Aspirate the supernatant and resuspend cells in 100 µL of the prepared master mix and incubate the sample for 10 min at RT in the dark.
- Wash the cells with 100 µL of PBS with 2% FCS/2 mM EDTA and centrifugate them again at 540 x g and 4 °C for 5 min.
- In the meantime, prepare 200 µL of the fixation/permeabilization solution: Add 50 µL of the concentrated stock of fixation/permeabilization concentrate to 150 µL of fixation/permeabilization diluent to reach a final dilution of 1:4.
- Aspirate the supernatant and resuspend the cells in 100 µL of 1x fixation/permeabilization solution. Incubate the sample for 30 min at 4 °C.
- In the meantime, prepare 1 mL of 1x permeabilization/wash buffer by adding 100 µL of the permeabilization buffer stock to 900 µL of ddH2O to reach a final dilution of 1:10.
- Wash the cells 1x with 100 µL of 1x permeabilization/wash buffer and centrifugate the sample at 540 x g and 4 °C for 5 min.
- In the meantime, prepare another master mix in 1x permeabilization/wash buffer consisting only of NeuN (NeuN AF647, clone EPR12763, 1:200) and 1 µg of anti-CD16/32 per 106 cells to block the Fc receptor.
- Aspirate the supernatant. Resuspend the fixed and permeabilized cells in 50 µL of the second master mix and incubate for 30 min at 4 °C.
- Wash the sample with 100 µL of 1x permeabilization/wash buffer and centrifugate at 540 x g and 4 °C for 5 min.
- Discard the supernatant and resuspend the cell pellet in 70 µL of PBS with 2% FCS/2 mM EDTA. Subsequently, the sample is ready for the flow cytometric analysis.
- After setting up the panel on the flow cytometer, acquire cells for purity analysis using a flow cytometry analysis software.
10. Statistical analysis
- Perform statistical analyses and design graphs with a graphical analysis program. Data are presented as mean ± SEM.
Representative Results
The current protocol offers the possibility to simultaneously isolate all principal CNS-resident cells, i.e., microglia, oligodendrocytes, astrocytes, and neurons from one single CNS replicate. This is important for the reduction of mice numbers needed for these kinds of experiments and to ensure the comparability of molecular and biochemical analyses on cellular level. If the individual cell types are isolated from different CNS replicates, cellular interactions cannot be mapped truthfully and potential technical deviations during the isolation processes could bias further downstream analyses. In addition, molecular and biochemical findings from each cell type would not be comparable to each other as they are not derived from the same EAE context. A pre-existing MACS protocol using a commercial system/kit was adapted to enable simultaneous isolation of the above-mentioned cell types.
The isolation of microglia was performed using anti-CD11b micro-beads, oligodendrocytes were isolated via anti-O4 micro-beads (Table 1), and anti-ACSA-2 micro-beads were used to isolate astrocytes (Table 2). In contrast, the isolation of neurons represents a negative selection and was accomplished by biotinylation and magnetic labeling of all non-neuronal cells (Table 2). All non-neuronal cells (e.g., oligodendrocytes, microglia, astrocytes, endothelial cells, and fibroblasts) except for blood cells can be magnetically labeled by using a biotin-conjugated antibody, specifically directed against a surface antigen expressed on these non-neuronal cells (Table 2). By depletion of these magnetically labeled non-neuronal cells, highly pure and viable neuronal cell populations can be generated30,42,43.
Two new flow cytometry panels for the purity analyses of the generated single-cell suspensions were designed. Here, cell-type-specific surface and nuclear markers combined with live/dead cell discrimination were used.
The resulting cell yields per mouse and cell-type (Figure 3A) were analyzed and led to an average of 8. 4 x 105 ± 3 x 104 microglia, 3.23 x 106 ± 1 x 105 oligodendrocytes, 8.6 x 105 ± 2 x 104 astrocytes, and 4.5 x 105 ± 1 x 104 neurons per naïve mouse.
In the context of the aim to investigate disease models of neuroinflammation, the protocol was also applied to a mouse model of EAE. The mice were euthanized on day 16 after EAE induction representing the disease maximum. In this EAE setting, approximately 2.9 x 106 ± 6.7 x 105 oligodendrocytes, 5.2 x 105 ± 9 x 104 astrocytes and 4.4 x 105 ± 4 x 104 neurons were isolated. The microglia cell yield was decreased to approximately 1.7 x 105 ± 4 x 104 microglia per EAE mouse because of the additional cell sorting after the MACS steps (Figure 3A).
After isolation, phenotypic characterizations of the different cell populations via flow cytometry proved that viable single-cell suspensions with a purity of approximately 90% for all main CNS-resident cell types (Figure 3B) could be achieved. Microglia were gated as being CD45intCD11bhigh as defined in the literature44,45,46,47.
In EAE, microglia had to be sorted from all CD11b+ cells to differentiate them from other CD11b+ immune cells like monocytes, neutrophils, natural killer cells, granulocytes, and macrophages that immigrate into the CNS during neuroinflammation27,28,48. Therefore, the microglia were sorted as CD45intCD11high cells from the CD11b+ cell suspension. The whole microglia sorting strategy is depicted in Figure 4. In naïve mice, the microglia population was 91.16% of all live single cells (96% of the total CD11b+ population) (Figure 4A). In EAE mice, the microglia population was 46.85% of all live single cells (55% of the total CD11b+ population) (Figure 4B). Although both MACS and FACS procedures apply mechanical stress to the single cells, 75.41% ± 8.63% of the sorted purified microglia were viable (Figure 3B).
Astrocytes and neurons that were directly isolated from the initial CNS cell suspension showed relevant contamination with oligodendrocytes which led to the assumption that the simultaneous isolation of neurons and astrocytes from the negative flow-through of oligodendrocytes could prevent this contamination. Flow cytometry analyses confirmed that astrocytes isolated from the negative flow-through of oligodendrocytes had a purity of 89.23% ± 2.78% and demonstrated a viability of 80.56% ± 1.41%. Similar to these results, the purity of the neurons isolated from the O4- cell fraction was 81.25% ± 3.31% and the viability was 83.90% ± 2.61% (Figure 3B). These findings also confirm that the simultaneous isolation of these two cell types only subsequently to the isolation of oligodendrocytes has no impact on the amount of viable functional cells.
The results regarding the viability and purity of the isolated single-cell suspensions were very similar in EAE mice compared to those received in naïve mice confirming that this protocol is suitable for healthy mice as well as in the context of EAE (Figure 3B).
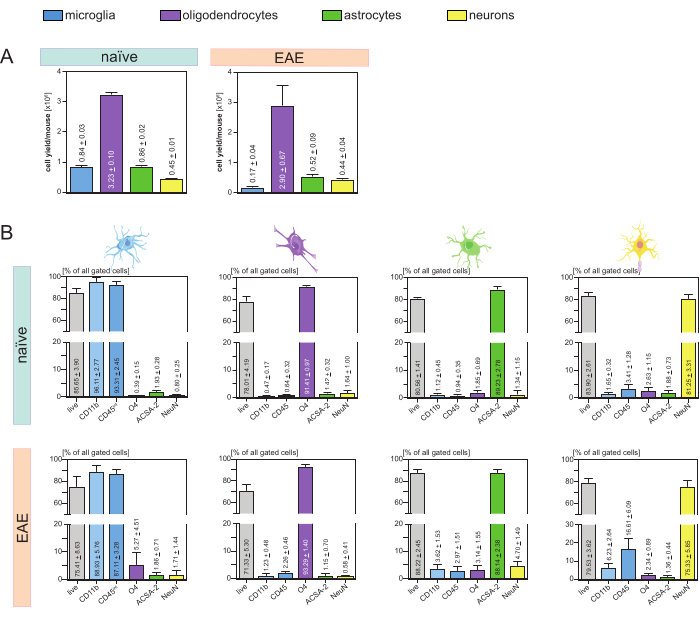
Figure 3: Cell yields and flow cytometry-based validation of isolated CNS-resident cells. (A) Cell yields per mouse and cell-type after isolation of CNS-resident cells in naïve and EAE mice. Bar graphs visualize the amount of cell yields per mouse and cell-type after the implementation of the presented protocol. Five biological replicates were processed for the results in naïve mice, and four biological replicates were analyzed in EAE mice. Respective means ± SEMs are depicted. (B) Corresponding purity and viability analyses of the purified cell fractions. Bar graphs indicate the viability and purity of the resulting single-cell suspensions based on their expression of cell-type-specific markers. NeuN was used as a cell-type specific nuclear marker for neurons. Five biological replicates were acquired and compared for each cell type for both healthy and EAE mice. Respective means ± SEMs are indicated. Abbreviations: Anti-ACSA-2 = astrocyte cell surface antigen-2; CD11b = cyclin-dependent kinase 11B; CD45 = receptor-type tyrosine-protein phosphatase C; CNS = central nervous system; EAE = experimental autoimmune encephalomyelitis; MACS = magnetic-activated cell sorting; NeuN =RNA binding protein fox-1 homolog 3; O4 = oligodendrocyte marker O4; SEM = standard error of the mean. This figure has been modified from49. Please click here to view a larger version of this figure.
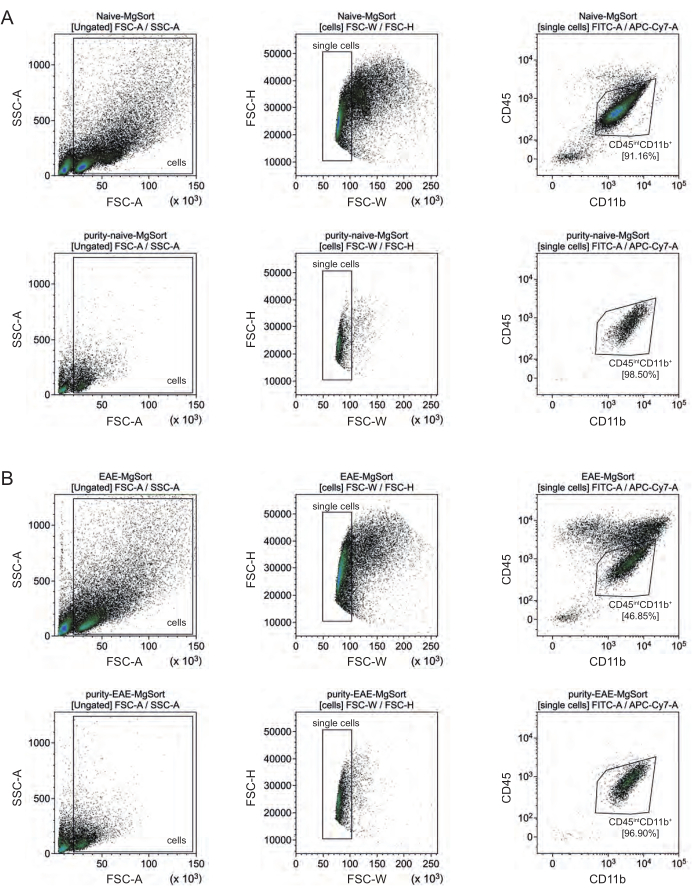
Figure 4: Gating strategy for cell sorting of microglia after isolation of CD11b+ cells. (A) Gating strategy in naïve and (B) EAE mice. The upper row of each panel shows dot plots before sorting and the lower row after sorting. After selection of live (SSC-A / FSC-A) and single cells (FCS-H / FSC-W), the population of CD45intCD11b+ cells were sorted as microglia population. Please click here to view a larger version of this figure.
Discussion
So far, methods to map CNS-resident cells ex vivo by combining mass spectrometry and RNA sequencing offer a very precise cellular profiling in health and disease but require ambitious technical knowledge and expertise in this field50,51. Further, they do not allow functional analyses and are very expensive. Besides that, microfluidic brain-on-a-chip systems provide a rapid and affordable screening for disease mechanisms and testing of new therapeutic approaches with the restriction of cell growth and migration52,53,54,55. CNS organoids could also represent an equivalent alternative in the future for the investigation of cellular modeling, inter-cellular connections, and interactions during disease courses56,57,58,59. However, fluorescence- and magnetic-activated cell sorting are currently the most effective methods to generate pure and viable single-cell suspensions ex vivo35,60,61. Even if other established manufactural protocols for the isolation of CNS-resident cell types are similar regarding the individual steps of the magnetic isolation and the prior cell dissociation, they are intended to be performed for each cell type separately. By contrast, the current protocol integrates different isolation methods for each CNS-resident cell type into a logical context so that they can be performed simultaneously at once and from one single CNS cell suspension (Table 1, Table 2). Thus, it enables multi-omics analyses from one single CNS cell suspension and, eventually, the exploration of complex neuronal networks. Even if it is not a must to pool several tissues from multiple animals to perform this protocol, this pooling ensures an adequate number of isolated cells for further downstream analysis. The usage of different mice for the isolation of the single cell types would exclude the possibility of analyzing potential cellular interactions. Besides that, combining individual isolation methods for the different CNS cell types, which all follow a prior CNS dissociation, saves material costs by using one dissociated CNS cell suspension for all following magnetic isolation steps. Additionally, a potential technical bias caused by the usage of different mice is minimized.
One limitation of the protocol could be the nearly exclusive usage of female C57BL/6J mice. The EAE immunization protocol has been designed and established for female mice, so this cell isolation protocol was implemented in female C57BL/6J mice as well. Nevertheless, naïve male mice were also used during the development of this protocol, without recognizing any effect on the resulting cell number or purities. Another restriction affects the magnetic cell isolation of neurons as there exist no specific micro-beads for the isolation of neurons in terms of a positive selection. It was assumed that a pure single-cell suspension could be obtained via biotin labeling and depletion of all non-neuronal cells (Table 2). This assumption was verified by the usage of NeuN as a specific nuclear marker for neurons, integrated in the mentioned flow cytometry purity panel. Another limitation concerns the isolation of microglia in EAE mice. Here, resulting cell yields are decreased in comparison to the other cell types because of the additional sorting step after the MACS protocol. Moreover, one could argue that sorting increases the mechanical stress of the microglia compared to the other cell populations. Individual sorting strategies may lead to different amounts of cell yields. If isolated cell number is less than expected or desired, adjusting the gating set up and/or improving the live/dead discrimination is recommended.
A critical step in the protocol represents the debris removal. The gradient must be layered very slowly and gently to create the three wanted separate phases (Figure 2A). Only if the myelin and other debris residues in the two top phases are removed totally (Figure 2E), pure single-cell suspensions can be generated, and further contamination can be reduced. If the resulting cell suspensions lack purity, this is probably the section of the protocol that should be improved first next to the assurance of the right usage of all micro-beads.
Receiving high levels in purity and viability can be challenging in this type of experiment. Some recommendations for troubleshooting are:
-Working under sterile conditions is obligatory to prevent contamination of the different micro-beads and enable repeated use, especially for subsequent cultivation.
-Labeling of each tube to prevent mix-ups is highly recommended.
-Avoid the usage of uncooled reagents/buffers. Store all cell suspensions on ice during the whole experiment to ensure high viability.
-Keep the time between the different working steps as short as possible. There is no specific part in the protocol where pausing the experiment is recommended.
-It is highly relevant to adhere to the specified incubation periods.
In conclusion, this current protocol for the simultaneous isolation of all main CNS-resident cell types from one CNS replicate offers the possibility to analyze complex neuronal networks and neuroinflammatory pathways ex vivo from one CNS cell suspension. Thus, CNS-resident cells can be investigated during different stages of disease courses, e.g., during neuroinflammation, neurodegeneration and/or remission in EAE. Furthermore, cell-cell interactions and biochemical pathways can be studied on an individual level and variability within experimental groups can be reduced. There is also the opportunity to cultivate fractions of the isolated CNS cells in monocultures for further functional assays and validation. All in one, this protocol offers significant advances potentially affecting preclinical and clinical research approaches.
Acknowledgements
Figures were created using Adobe Illustrator (version 2023) and Servier Medical Art (https://smart.servier.com). Antonia Henes was supported by Jürgen Manchot Stiftung.
Materials
| Name | Company | Catalog Number | Comments |
| 70 μm cell strainers | Corning, MA, USA | 352350 | CNS tissue dissociation |
| ACSA-2 Antibody, anti-mouse, PE-Vio 615 (clone REA-969) | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-116-244 | Flow cytometry, store at 4 °C |
| Adult Brain Dissociation Kit, mouse, and rat | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-107-677 | Tissue dissociation,contains debris and red blood cell removal solutions; prepare aliquots of enzyme A and P upon arrival and store them at -20 °C; store the remaining kit at 4 °C |
| Anti-ACSA-2 MicroBead Kit, mouse | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-097-678 | MACS of astrocytes, store at 4 °C |
| Anti-mouse CD16/32 antibody | BioLegend, London, UK | 101301 | Flow cytometry, store at 4 °C |
| Anti-O4 MicroBeads, human, mouse, rat | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-094-543 | MACS of oligodendrocytes, store at 4 °C |
| AstroMACS Separation buffer | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-091-221 | MACS of astrocytes, store at 4 °C |
| Biotin Antibody, PE (clone Bio3-18E7) | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-113-853 | Flow cytometry, store at 4 °C |
| BRAND Neubauer counting chamber | Thermo Fisher Scientific,Waltham, MA, USA | 10195580 | Cell counting |
| Brilliant Violet 510 anti-mouse CD45 Antibody (clone 30-F11) | BioLegend, London, UK | 103137 | Flow cytometry, store at 4 °C |
| CD11b MicroBeads, human, mouse | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-049-601 | MACS of microglia, store at 4 °C |
| DNAse I, recombinant, Rnase-free | Merck KGaA, Darmstadt, Germany | 4716728001 | Flow cytometry, store at -20° C |
| D-PBS with Calcium, Magnesium, Glucose, Pyruvat | Thermo Fisher Scientific,Waltham, MA, USA | 14287080 | Buffer, store at 4 °C |
| D-PBS, without calcium, without magnesium | Thermo Fisher Scientific,Waltham, MA, USA | 14190250 | Buffer, store at 4 °C |
| eBioscience Fixable Viability Dye eFluor 780 | Thermo Fisher Scientific,Waltham, MA, USA | 65-0865-14 | Flow cytometry, store at 4 °C |
| eBioscience Foxp3/Transcription factor staining buffer set | Thermo Fisher Scientific,Waltham, MA, USA | 00-5523-00 | Flow cytometry, store at 4°C |
| Falcon (15 mL) | Thermo Fisher Scientific,Waltham, MA, USA | 11507411 | Cell tube |
| Falcon (50 mL) | Thermo Fisher Scientific,Waltham, MA, USA | 10788561 | Cell tube |
| Falcon Round-Bottom Polystyrene Test Tubes with Cell Strainer Snap Cap, 5 mL | Thermo Fisher Scientific,Waltham, MA, USA | 08-771-23 | Flow cytometry |
| FcR Blocking Reagent, mouse | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-092-575 | MACS of oligodendrocytes, store at 4 °C |
| Female C57BL/6J mice | Charles River Laboratories, Sulzfeld, Germany | Active EAE induction | |
| Fetal calf serum (FCS) | Merck KGaA, Darmstadt, Germany | F2442-50ML | Flow cytometry, store at -5 to -20 °C |
| FITC Rat Anti-CD 11b (clone M1/70) | BD Biosciences, San Jose, CA, USA | 553310 | Flow cytometry, store at 4 °C |
| Freund’s Complete adjuvant | Merck KGaA, Darmstadt, Germany | AR001 | Active EAE induction, store at 4 °C |
| GentleMACS C Tubes | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-093-237 | CNS tissue dissociation |
| GentleMACS Octo Dissociator with Heaters | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-096-427 | CNS tissue dissociation |
| Graphpad Prism 8.4.3 | Graphpad by Dotmatics | Graphical Analysis | |
| Isoflurane | AbbVie, North Chicago, IL, USA | Active EAE induction, store at 4 °C | |
| Kaluza Analysis Software V2.1.1 | Beckman Coulter, Indianapolis, IN, USA | Flow cytometry analysis | |
| LS Columns | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-042-401 | MACS |
| MACS BSA Stock Solution | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-091-376 | PB-buffer |
| MACS MultiStand | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-042-303 | MACS |
| MOG35–55 peptide | Charité, Berlin, Germany; alternatives: Genosphere Biotechnologies (Paris, France) or sb-Peptide (Saint Egrève, France) | Active EAE induction, store at -20 °C | |
| Mycobacterium tuberculosis strain H37 Ra | Becton, Dickinson and Company (BD),Franklin Lakes, NJ, USA | Active EAE induction, store at 4 °C | |
| Neuron Isolation Kit, mouse | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-115-390 | MACS of neurons, store at 4 °C |
| O4 Antibody, anti-human/mouse/rat, APC, (clone REA-576) | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-119-897 | Flow cytometry, store at 4 °C |
| Pertussis toxin in glycerol | Hooke Laboratories Inc., Lawrence, MA, USA | BT-0105 | Active EAE induction; store at -20 °C |
| pluriStrainer Mini 100 μm | pluriSelect Life Science UG, Leipzig, Sachsen, Germany | 43-10100-40 | Flow cytometry |
| QuadroMACS Separator | Miltenyi Biotec, Bergisch Gladbach, NRW, Germany | 130-090-976 | MACS |
| Recombinant Alexa Fluor 647 Anti-NeuN antibody (clone EPR12763) | Abcam, Cambridge, UK | EPR12763 | Flow cytometry, store at -20 °C |
| Stainless Steel Brain Matrices, 1 mm | Ted Pella, Redding, CA, USA | 15067 | CNS tissue dissection |
| Trypan blue solution, 0.4% | Thermo Fisher Scientific,Waltham, MA, USA | 15250061 | Cell counting |
| UltraPure 0.5 M EDTA, pH 8.0 | Thermo Fisher Scientific,Waltham, MA, USA | 15575020 | Flow cytometry, store at room temperature |
References
- Trapp, B. D., Nave, K. A. Multiple Sclerosis: An Immune or Neurodegenerative Disorder. Annu Rev Neurosci. 31 (1), 247-269 (2008).
- Stys, P. K., Zamponi, G. W., van Minnen, J., Geurts, J. J. Will the real multiple sclerosis please stand up. Nat Rev Neurosci. 13 (7), 507-514 (2012).
- Korn, T. Pathophysiology of multiple sclerosis. J Neurol. 255 (Suppl 6), 2-6 (2008).
- Ward, M., Goldman, M. D. Epidemiology and Pathophysiology of Multiple Sclerosis. CONTINUUM. 28 (4), 988-1005 (2022).
- Bittner, S., Afzali, A. M., Wiendl, H., Meuth, S. G. Myelin Oligodendrocyte Glycoprotein (MOG35-55) Induced Experimental Autoimmune Encephalomyelitis (EAE) in C57BL/6 Mice. J Vis Exp. (86), 51275 (2014).
- Bittner, S., et al. The TASK1 channel inhibitor A293 shows efficacy in a mouse model of multiple sclerosis. Exp Neurol. 238 (2), 149-155 (2012).
- Göbel, K., et al. Plasma kallikrein modulates immune cell trafficking during neuroinflammation via PAR2 and bradykinin release. Proc Natl Acad Sci U S A. 116 (1), 271-276 (2019).
- Ballerini, C. Experimental Autoimmune Encephalomyelitis. Methods Mol Biol. 2285, 375-384 (2021).
- Birmpili, D., Charmarke Askar, I., Bigaut, K., Bagnard, D. The Translatability of Multiple Sclerosis Animal Models for Biomarkers Discovery and Their Clinical Use. Int J Mol Sci. 23 (19), 11532 (2022).
- Tsatas, O., Ghasemlou, N. Isolation and RNA purification of macrophages/microglia from the adult mouse spinal cord. J Immunol Methods. 477, 112678 (2020).
- Calvo, B., Rubio, F., Fernández, M., Tranque, P. Dissociation of neonatal and adult mice brain for simultaneous analysis of microglia, astrocytes and infiltrating lymphocytes by flow cytometry. IBRO Rep. 8, 36-47 (2020).
- Diaz-Amarilla, P., et al. Isolation and characterization of neurotoxic astrocytes derived from adult triple transgenic Alzheimer's disease mice. Neurochem Int. 159, 105403 (2022).
- Galatro, T. F., Vainchtein, I. D., Brouwer, N., Boddeke, E. W. G. M., Eggen, B. J. L. Isolation of Microglia and Immune Infiltrates from Mouse and Primate Central Nervous System. Methods Mol Biol. 1559, 333-342 (2017).
- Altendorfer, B., et al. Transcriptomic Profiling Identifies CD8+ T Cells in the Brain of Aged and Alzheimer's Disease Transgenic Mice as Tissue-Resident Memory T Cells. J Immunol. 209 (7), 1272-1285 (2022).
- Lanfranco, M. F., Sepulveda, J., Kopetsky, G., Rebeck, G. W. Expression and secretion of apoE isoforms in astrocytes and microglia during inflammation. Glia. 69 (6), 1478-1493 (2021).
- Swire, M., Ffrench-Constant, C. Oligodendrocyte-Neuron Myelinating Coculture. Methods Mol Biol. 1936, 111-128 (2019).
- Park, J., Koito, H., Li, J., Han, A. Microfluidic compartmentalized co-culture platform for CNS axon myelination research. Biomed Microdevices. 11 (6), 1145-1153 (2009).
- Facci, L., Barbierato, M., Skaper, S. D. Astrocyte/Microglia Cocultures as a Model to Study Neuroinflammation. Methods Mol Biol. 1727, 127-137 (2018).
- Speicher, A. M., Wiendl, H., Meuth, S. G., Pawlowski, M. Generating microglia from human pluripotent stem cells: novel in vitro models for the study of neurodegeneration. Mol Neurodegener. 14 (1), 46 (2019).
- Homayouni Moghadam, F., et al. Isolation and Culture of Embryonic Mouse Neural Stem Cells. J Vis Exp. (141), 58874 (2018).
- Santos, R., et al. Differentiation of Inflammation-Responsive Astrocytes from Glial Progenitors Generated from Human Induced Pluripotent Stem Cells. Stem Cell Reports. 8 (6), 1757-1769 (2017).
- Tcw, J., et al. An Efficient Platform for Astrocyte Differentiation from Human Induced Pluripotent Stem Cells. Stem Cell Reports. 9 (2), 600-614 (2017).
- Miltenyi, S., Müller, W., Weichel, W., Radbruch, A. High gradient magnetic cell separation with MACS. Cytometry. 11 (2), 231-238 (1990).
- Huntemann, N., et al. An optimized and validated protocol for inducing chronic experimental autoimmune encephalomyelitis in C57BL/6J mice. J Neurosci Methods. 367, 109443 (2022).
- Martin, E., El-Behi, M., Fontaine, B., Delarasse, C. Analysis of Microglia and Monocyte-derived Macrophages from the Central Nervous System by Flow Cytometry. J Vis Exp. (124), 55781 (2017).
- Sarkar, S., et al. Rapid and Refined CD11b Magnetic Isolation of Primary Microglia with Enhanced Purity and Versatility. J Vis Exp. (122), 55364 (2017).
- Rodríguez Murúa, S., Farez, M. F., Quintana, F. J. The Immune Response in Multiple Sclerosis. Annu Rev Pathol. 17, 121-139 (2021).
- Engelhardt, B., Ransohoff, R. M. Capture, crawl, cross: the T cell code to breach the blood-brain barriers. Trends Immunol. 33 (12), 579-589 (2012).
- Elia, G. Biotinylation reagents for the study of cell surface proteins. Proteomics. 8 (19), 4012-4024 (2008).
- Berl, S., et al. Enrichment and isolation of neurons from adult mouse brain for ex vivo analysis. J Neurosci Methods. 283, 15-22 (2017).
- Turvy, D. N., Blum, J. S. Biotin Labeling and Quantitation of Cell-Surface Proteins. Curr Protoc Immunol. 18 (7), (2001).
- Mao, S. Y. Biotinylation of Antibodies. Methods Mol Biol. 115, 39-41 (1999).
- Kantzer, C. G., et al. Anti-ACSA-2 defines a novel monoclonal antibody for prospective isolation of living neonatal and adult astrocytes. Glia. 65 (6), 990-1004 (2017).
- Batiuk, M. Y., et al. An immunoaffinity-based method for isolating ultrapure adult astrocytes based on ATP1B2 targeting by the ACSA-2 antibody. J Biol Chem. 292 (21), 8874-8891 (2017).
- Pan, J., Wan, J. Methodological comparison of FACS and MACS isolation of enriched microglia and astrocytes from mouse brain. J Immunol Methods. 486, 112834 (2020).
- Donovan, J. A., Koretzky, G. A. CD45 and the immune response. J Am Soc Nephrol. 4 (4), 976-985 (1993).
- Hathcock, K. S., Hirano, H., Hodes, R. J. CD45 expression by murine B cells and T cells: Alteration of CD45 isoforms in subpopulations of activated B cells. Immunol Res. 12 (1), 21-36 (1993).
- Balogh, P., Tew, J. G., Szakal, A. K. Simultaneous blockade of Fc? receptors and indirect labeling of mouse lymphocytes by the selective detection of allotype-restricted epitopes on the kappa chain of rat monoclonal antibodies. Cytometry. 47 (2), 107-110 (2002).
- Becerril-García, M. A., et al. Langerhans Cells From Mice at Birth Express Endocytic- and Pattern Recognition-Receptors, Migrate to Draining Lymph Nodes Ferrying Antigen and Activate Neonatal T Cells in vivo. Front Immunol. 11, 744 (2020).
- Dent, M. A., Segura-Anaya, E., Alva-Medina, J., Aranda-Anzaldo, A. NeuN/Fox-3 is an intrinsic component of the neuronal nuclear matrix. FEBS Lett. 584 (13), 2767-2771 (2010).
- Duan, W., et al. Novel Insights into NeuN: from Neuronal Marker to Splicing Regulator. Mol Neurobiol. 53 (3), 1637-1647 (2016).
- Monteiro, R., Sivasubramanian, M. K., Balasubramanian, P., Subramanian, M. Obesity-Induced Sympathoexcitation is Associated with Glial Senescence in the Brainstem. FASEB J. 34 (S1), 1-1 (2020).
- Li, S., Chang, L., Teissie, J. . Electroporation protocols: mircroorganism, mammalian system, and nanodevice. , (2020).
- Kettenmann, H., Hanisch, U. K., Noda, M., Verkhratsky, A. Physiology of Microglia. Physiol Rev. 91 (2), 461-553 (2011).
- Haage, V., et al. Comprehensive gene expression meta-analysis identifies signature genes that distinguish microglia from peripheral monocytes/macrophages in health and glioma. Acta Neuropathol Commun. 7 (1), 20 (2019).
- Kosior, N., Petkau, T. L., Connolly, C., Lu, G., Leavitt, B. R. Isolating cells from adult murine brain for validation of cell-type specific cre-mediated deletion. J Neurosci Methods. 328, 108422 (2019).
- Jurga, A. M., Paleczna, M., Kuter, K. Z. Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front Cell Neurosci. 14, 198 (2020).
- Man, S., Ubogu, E. E., Ransohoff, R. M. Inflammatory Cell Migration into the Central Nervous System: A Few New Twists on an Old Tale. Brain Pathol. 17 (2), 243-250 (2007).
- Schroeter, C. B., et al. One Brain-All Cells: A Comprehensive Protocol to Isolate All Principal CNS-Resident Cell Types from Brain and Spinal Cord of Adult Healthy and EAE Mice. Cells. 10 (3), 651 (2021).
- Sankowski, R., et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nate Neurosci. 22 (12), 2098-2110 (2019).
- Brennan, F. H., et al. Microglia coordinate cellular interactions during spinal cord repair in mice. Nat Commun. 13 (1), 4096 (2022).
- Enright, H. A., et al. Functional and transcriptional characterization of complex neuronal co-cultures. Sci Rep. 10 (1), 11007 (2020).
- Mofazzal Jahromi, M. A., et al. Microfluidic Brain-on-a-Chip: Perspectives for Mimicking Neural System Disorders. Mol Neurobiol. 56 (12), 8489-8512 (2019).
- Chin, E., Goh, E. Blood-brain barrier on a chip. Methods Cell Biol. 146, 159-182 (2018).
- Miccoli, B., Braeken, D., Li, Y. E. Brain-on-a-chip Devices for Drug Screening and Disease Modeling Applications. Curr Pharm Des. 24 (45), 5419-5436 (2019).
- Giandomenico, S. L., et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat Neurosci. 22 (4), 669-679 (2019).
- Pellegrini, L., et al. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science. 369 (6500), eaaz5626 (2020).
- Chhibber, T., et al. CNS organoids: an innovative tool for neurological disease modeling and drug neurotoxicity screening. Drug Discov Today. 25 (2), 456-465 (2020).
- Tang, X. Y., et al. Human organoids in basic research and clinical applications. Signal Transduct TargetTher. 7 (1), 168 (2022).
- Sutermaster, B. A., Darling, E. M. Considerations for high-yield, high-throughput cell enrichment: fluorescence versus magnetic sorting. Sci Rep. 9 (1), 227 (2019).
- Doughty, D., et al. Development of a novel purification protocol to isolate and identify brain microglia. Exp Biol Med. 247 (16), 1433-1446 (2022).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved