Application of Ha-CoV-2 Pseudovirus for Rapid Quantification of SARS-CoV-2 Variants and Neutralizing Antibodies
In This Article
Summary
This protocol describes the application of a novel hybrid alphavirus-SARS-CoV-2 pseudovirus (Ha-CoV-2) as a platform for rapid quantification of infectivity of SARS-CoV-2 variants and their sensitivity to neutralizing antibodies.
Abstract
The coronavirus disease 2019 pandemic (COVID-19) has highlighted the need for rapid assays to accurately measure the infectivity of emerging SARS-CoV-2 variants and the effectiveness of vaccine-induced neutralizing antibodies against viral variants. These assays are essential for pandemic surveillance and validating vaccines and variant-specific boosters. This manuscript demonstrates the application of a novel hybrid alphavirus-SARS-CoV-2 pseudovirus (Ha-CoV-2) for quick quantification of SARS-CoV-2 variant infectivity and vaccine-induced neutralizing antibodies to viral variants. Ha-CoV-2 is a SARS-CoV-2 virus-like particle consisting of viral structural proteins (S, M, N, and E) and a fast-expressing RNA genome derived from an alphavirus, Semliki Forest Virus (SFV). Ha-CoV-2 also contains both green fluorescent protein (GFP) and luciferase reporter genes that allow for quick quantification of viral infectivity. As an example, the infectivity of the SARS-CoV-2 Delta (B.1.617.2) and the Omicron (B.1.1.529) variants are quantified, and their sensitivities to a neutralizing antibody (27VB) are also measured. These examples demonstrate the great potential of Ha-CoV-2 as a robust platform for rapid quantification of SARS-CoV-2 variants and their susceptibility to neutralizing antibodies.
Introduction
As of May 2023, there have now been more than 766 million COVID-19 cases1. Despite worldwide vaccination campaigns, SARS-CoV-2 continuously circulates and infects people, largely due to the emergence of new variants such as Delta (B.1.617.2) and Omicron (B.1.1.529) that drive new waves of infection2,3,4. Given that SARS-CoV-2 is constantly evolving, it is important to develop quick assays that can accurately measure the infectivity of emerging variants and the effectiveness of vaccine-induced neutralizing antibodies against these variants. These assays are essential for pandemic surveillance and for determining the efficacy of vaccines and their variant-specific boosters.
Due to the highly contagious nature of SARS-CoV-2, the Center for Disease Control and Prevention (CDC) requires that the study of SARS-CoV-2 and its variants is conducted in biosafety level (BSL) 3 facilities5,6. This BSL-3 requirement limits the use of live viruses for quantifying the infectivity of viral variants and their neutralizing antibodies in common research and clinical laboratories. In addition, traditional SARS-CoV-2 neutralization assays, such as the plaque- or cytopathic effect-based assays using replication-competent live viruses, are time-consuming and require long incubation periods7. Several spike (S) protein-pseudotyped SARS-CoV-2 pseudoviruses have been developed to quantify the effectiveness of neutralizing antibodies8,9,10,11,12. In SARS-CoV-2, the S protein is the major protein that mediates viral entry13, and is the main antigen used in SARS-CoV-2 vaccines9,10,14,15,16. The S protein-pseudotyped virions, such as those of the vesicular stomatitis virus (VSV-G) or lentivirus, have been used for the quantification of neutralizing antibodies17,18,19. Nevertheless, the lentivirus-based pseudovirus normally requires 2 to 3 days of infection in order to quantify reporter signals. VSV-based pseudovirus systems often contain residual VSV viruses, which can result in high rates of false-positive results and typically require 24 h of infection20.
A novel SARS-CoV-2 pseudovirus system, the hybrid alphavirus-SARS-CoV-2 pseudovirus (Ha-CoV-2), has been recently developed by Hetrick et al12. Ha-CoV-2 provides a new tool for the rapid quantification of virus infectivity and virus sensitivity to neutralizing antibodies in common BSL-2 laboratories. Structurally, Ha-CoV-2 resembles the SARS-CoV-2 virion particle, consisting of SARS-CoV-2 structural proteins including the S protein (S), the membrane (M), the nucleocapsid (N), and the envelope (E), and there is no structural protein from other viruses. Additionally, Ha-CoV-2 particle contains a rapid-expressing RNA genome from an alphavirus for fast reporter expression in cells. Ha- CoV-2 has been shown to quickly measure the neutralizing activity of antibodies in the sera of vaccinated and convalescent individuals12. As demonstrated by Hetrick et al., when compared with lentivirus-based SARS-CoV-2 pseudovirus in a time course assay, Ha-CoV-2 expressed the Luc reporter as early as 2-4 h post-infection while the lentivirus-pseudovirus expressed Luc after 24 h12. In addition, the potential application of Ha-CoV-2 variants for quantifying neutralizing antibodies is further demonstrated by using a standard monoclonal neutralizing antibody, 27BV (See Supplementary Figure 1)12. This work details the use of the Ha-CoV-2 platform for rapid quantification of the infectivity of SARS-CoV-2 variants, using the Delta (B.1.617.2) and the Omicron (B.1.1.529) variants as examples. In addition, the potential application of Ha-CoV-2 variants for quantifying neutralizing antibodies is further demonstrated by using a standard monoclonal neutralizing antibody, 27BV12.
Protocol
1. Virus and viral particle assembly
- Vectors: Purchase expression vectors for SARS-CoV-2 M, E, or N as well as the SARS-CoV-2 S (Wild-type,Wt), Delta (B.1.617.2), and Omicron (B.1.1.529) commercially.
NOTE: Protein sequences of the expression vectors are provided in Supplementary File 1. The vendor of these expression vectors can also be found under the Table of Materials. - Cells and cell culture: Maintain HEK293T cells in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS), 50 units/mL of penicillin, and 50 µg/mL of streptomycin.
NOTE: All cell culture work needs to be done in a laminar airflow biosafety cabinet. The polyethyleneimine (PEI) transfection typically requires 5-6 h incubation. Start times need to be considered carefully beforehand. - Virus assembly and PEI-based cotransfection: Assemble Ha-CoV-2 particles by cotransfection of HEK293T cells. Seed cells in a 10 cm cell culture Petri dish (4-5 x 106 cells per dish) in 10 mL of complete DMEM medium the day prior to cotransfection. For this study, three Petri dishes are used to seed cells for the assembly of Ha-CoV-2 wild-type, Delta, and Omicron, respectively.
- Incubate Petri dishes overnight in a CO2 incubator at 37 °C. Check the dishes next morning to ensure that cells are 80% confluent. Remove complete medium and replace it with 9 mL of DMEM serum-free media.
- For each dish, prepare a cotransfection mixture with 2.5 µg of each of the SARS-CoV-2 structural protein expression vectors (N, E, M), 10 µg pAlphaPro-Luc-GFP-PreΨ (HaCoV2 Genome), and 2.5 µg of the S protein expression vector, either the Delta or the Omicron S variants and 45 µL of PEI-based transfection reagent. Allow the cotransfection mixture to form complexes by incubating for 13 min (do not incubate for longer than 30 min).
- After incubation, add the cotransfection mixture to each Petri dish slowly, drop by drop. Place the dishes inside a CO2 incubator at 37 °C for 6 h. After 6 h, remove serum-free DMEM and replace it with complete DMEM medium. Harvest virus at 48-60 h post-cotransfection.
- Virus harvesting and storage: Harvest particles at 48 h post cotransfection. Detach cells by pipetting repeatedly over the surface of the monolayer and collect cells from each dish, put into 15 mL centrifuge tubes, and centrifuge at 400 x g for 5 min. Collect the supernatant and pass it through a 0.22 µM filter. Store the Ha-CoV-2 pseudovirus at -80 °C.
2. Viral infectivity assay
- Cells and cell culture: Maintain HEK293T(ACE2/TMPRSS2) cells in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated FBS, 50 units/mL of penicillin and 50 µg/mL of streptomycin.
- Seeding HEK293T(ACE2/TMPRSS2) cells: The day before viral infectivity assay, seed HEK293T(ACE2/TMPRSS2) cells in a 96-well plate in 50 µL of complete DMEM media. For each 96-well plate, seed 2.5 x 104 cells into each well, and a total of 2.5 x 106 cells are needed for one plate. Place the 96 well plate in a CO2 incubator at 37 °C overnight.
- Infection of HEK293T(ACE2/TMPRSS2): Use Ha-CoV-2 variants particles to infect HEK293T(ACE2/TMPRSS2) cells. On the morning of infection, remove 50 µL of DMEM from pre-seeded 96 well plate. Replace media with 50 µL of either Ha-CoV-2 Wild-type, Delta, or Omicron for 18 h at 37 °C.
NOTE: The protocol can be stopped here until the infection has reached 18 h of incubation and the plate is ready to be analyzed via Luciferase Assay. The success of the infection is determined by the Luciferase assay resulting from the Luciferase reporter gene expressed in the infected cells, therefore the more signal is produced, the more successful the infection of the Ha-CoV-2 variant is. - Luciferase assay: After 18 h of incubation, add 7.5 µL of cell lysis buffer directly to each well and mix by orbital shaking for 2 min. Lyse cells in lysis buffer for at least 5 min at room temperature.
- Prepare the Firefly luciferase assay solution by mixing the D-luciferin solution with the Firefly luciferase assay solution in a 1:50 ratio. For a whole 96-well plate, combine 3 mL of the luciferase substrate solution with 60 µL of the D-luciferin solution with 2940 µL of the Firefly luciferase buffer solution.
- Add 25 μL of the Firefly luciferase assay solution to the cell lysates and mix the plate by orbital shaking for 1 min. Analyze the luciferase activity by using a commercial luciferase microplate reader.
3. RNA extraction of Ha-CoV-2 and quantitative reverse transcriptase PCR (RT-qPCR)
- Viral RNA extraction: Extract the viral RNA from Ha-CoV-2 Wild-type and the Ha-CoV-2 Delta and Omicron variant particles using a commercial viral RNA extraction kit, following the manufacturer's instructions. Store the extracted viral RNA at -80 °C or use it immediately for RT-qPCR.
- RT-qPCR: Perform RT-qPCR on viral RNA using a one-step master mix. Perform the reaction in a commercial PCR machine. Please note that the target of amplification is the genomic RNA of Ha-CoV-2. Use Ha-CoV-2 vector DNA as a standard for creating a standard curve and calculating the RNA copy number of each variant.
4. Neutralizing antibody assay
- Seeding HEK293T(ACE2/TMPRSS2) cells: The day before the assay, seed HEK293T(ACE2/TMPRSS2) cells in a 96 well plate in 50 µL of complete DMEM media. HEK293T(ACE2/TMPRSS2) cells are commercially purchased.
- For counting cells, obtain 20 µL cells from a T75 flask containing HEK293T(ACE2/TMPRSS2) cells, and mix with 20 µL of trypan blue solution. Add 20 µL of this mixture to cell counting chamber and count the number of cells per mL. To seed a 96 well plate for infection, use 2.5 x 104 cells per well, and 2.5 x 106 cells will be needed in total. Place 96 well plate in a CO2 incubator at 37 °C overnight.
- Neutralizing antibody assay: In a sterile polypropylene 96-well plate, prepare the standard 27BV neutralizing antibody and Ha-CoV-2 mixture. Add 8 µL of 27BV (45 mg/mL) to the plate and perform serial dilutions of the antibody with 6 µL of serum-free DMEM.
NOTE: Be sure to exchange pipette tips between well transfers and ensure the antibody and serum-free medium are mixed thoroughly to produce accurate results. - To the serially diluted antibody, add 54 µL of Ha-CoV-2 particle and mix the virus and the antibody. Pre-incubate Ha-CoV-2 particles with serially diluted 27BV for 1 h at 37 °C at 5% CO2. After the 1 h incubation, apply 50 µL of the antibody and Ha-CoV-2 mixture to the 96-well plate containing the HEK293T(ACE2/TMPRSS2) cells (2.5 x 104 cells per well) seeded the day before.
- For controls, leave at least three wells containing only the HEK293T(ACE2/TMPRSS2) cells. Add 50 µL of complete medium to these wells to serve as non-infected wells for background signal of the luciferase assay readings.
NOTE: The protocol can be stopped here until the infection has reached 18 h of incubation at 37 °C, and then the plate is ready to be analyzed via luciferase assay. - Luciferase assay: After 18 h of incubation, add 7.5 µL of lysis buffer directly to each well and mix by orbital shaking for 2 min. Lyse cells in lysis buffer for at least 5 min at room temperature.
- Prepare the Firefly luciferase assay solution by mixing the D-luciferin solution with the Firefly luciferase assay solution in a 1:50 ratio. For a whole 96-well plate, combine 3 mL of the luciferase substrate solution with 60 µL of the D-luciferin solution with 2940 µL of the firefly luciferase buffer solution.
- Add 25 μL of the Firefly luciferase assay solution to the cell lysates and mix the plate by orbital shaking for 1 min. Analyze the luciferase activity by using a commercial luciferase microplate reader.
5. Quantification and statistical analysis
- Data collection: Perform infection and luciferase assays in triplicate as indicated (Figure 1). Quantify luciferase expression with luciferase assay readings. The mean is the average value of the three luciferase assay readings. Background signal readings from non-infected wells are subtracted from this mean value. Standard deviation (SD) is determined from the average value of the luciferase assay readings.
- Data analysis: Plot antibody neutralization activity and calculate the ID50 (50% inhibition dosage) values using commercial graphing software. The ID50 value is defined as the antibody inhibitory dilutions at which a 50% reduction in the infection of HEK293T(ACE2/TMPRSS2) cells (based on luciferase assay readings) is attained.
Representative Results
Ha-CoV-2 particles were assembled using five different DNA vectors that express the Ha-CoV-2 RNA genome and the structural proteins (M, N, E, and S) of SARS-CoV-2 in HEK293T cells. The S protein vector varies depending on the S variant. The S protein from the original Ha-CoV-2 Wuhan strain (Wild-type, Wt) was used as a positive control, and it was assembled along with the S protein from each of the other two variants: the Delta (B.1.617.2) or the Omicron (B.1.1.529). The same M, N, E were used in all variants. Ha-CoV-2(Wt) and variant particles were collected 48 h post-cotransfection and then used to infect HEK293T(ACE2/TMPRSS2) cells. Infectivity was measured by expression of luciferase at 18 h post-infection. In this system, higher expression levels of luciferase signal reflect a higher infection of cells by Ha-CoV-2. The luciferase signal was normalized with genomic RNA copies by RT-qPCR for each variant. As shown in Figure 1, the Ha-CoV-2 Omicron variant generated 4- to 10-fold higher signal than the original Ha-CoV-2(Wt), suggesting a higher infectivity.
Furthermore, the capacity of 27BV to neutralize HaCoV-2(Wt), Delta, and Omicron variants was quantified. 27BV is a rabbit monoclonal antibody that was developed against the RBD domain of the SARS-COV-2 S1 protein. For neutralization assays, serial dilutions of 27BV were performed in a 96-well plate, pre-incubated with Ha-CoV-2, and then added to HEK293T(ACE2/TMPRSS2) target cells. The results demonstrated that 27BV had neutralizing activity against all the variants tested (Figure 2). Interestingly, the ID50 of 27VB for Omicron was approximately 10 times less potent than the ID50 for Ha-CoV-2(WT) and Ha-CoV-2(Delta; Figure 2). These results demonstrate that the Ha-COV-2 platform can be used as a rapid method for quantifying vaccine-induced neutralizing antibodies in emerging variants.
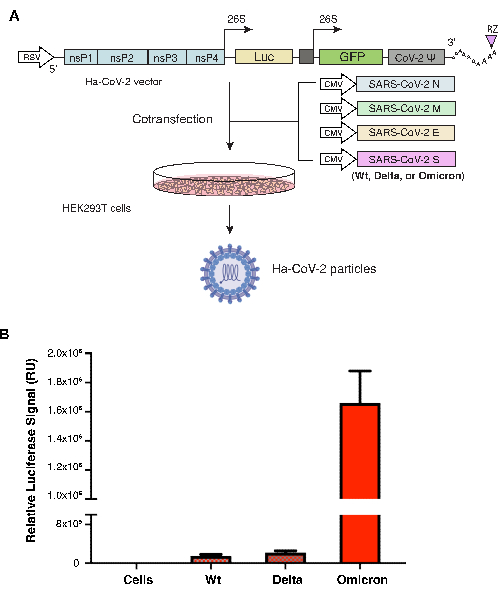
Figure 1. Assembly and quantification of Ha-CoV-2 variants. (A) Illustration of the assembly of the Ha-CoV-2 and variant particles. The vectors expressing the Ha-CoV-2 reporter genome and structural proteins (M, S, N, and E) are cotransfected in HEK293T cells. Particles were harvested 48 h post-cotransfection (the imaging of virion particle and HEK293T cells were created with Biorender.com). (B) Quantification of the infectivity of Ha-CoV-2 variants. The relative infectivity of the two variants (Delta and Omicron) is quantified and normalized using genomic RNA copies of individual Ha-CoV-2 (Luc) variants. Wild-type is the Ha-COV-2 (Wt), which is used as a control for comparison. Infection and luciferase assays were performed 3x. RU, relative unit. The mean and standard deviation (SD) are shown. Please click here to view a larger version of this figure.
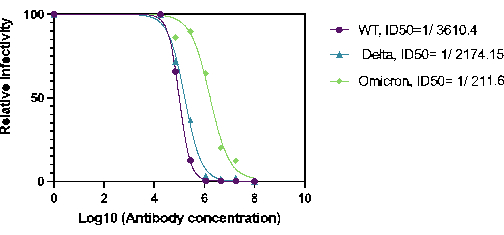
Figure 2. Quantification of 27BV neutralization activity against Ha-CoV-2(Luc) variants Neutralization activity of 27BV was analyzed 18 h post infection of HEK293T(ACE2/TMPRSS2) cells. The ID50 was calculated using relative infection rate (luciferase activity) versus 27BV concentration. Please click here to view a larger version of this figure.
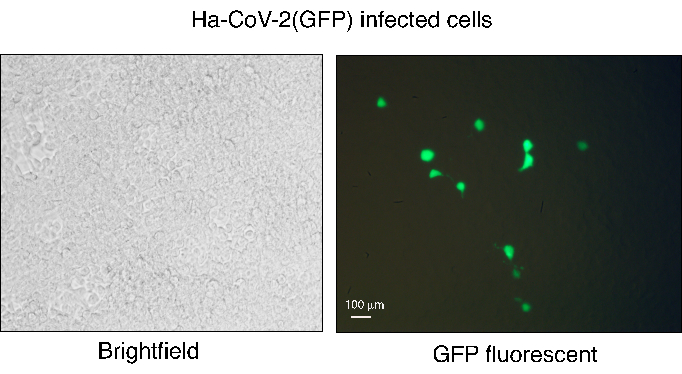
Figure 3. Infection of HEK293T(ACE2/TMPRSS2) with Ha-CoV-2(GFP). Ha-CoV-2(GFP) particles were assembled and then used to infect HEK293T(ACE2/TMPRSS2) cells. GFP expression was observed at 48 h post-infection using fluorescent microscopy. The white field of infected cells is shown on the left, and GFP imaging is shown on the right. The white bar represents 100 µm. Please click here to view a larger version of this figure.
Supplementary Figure 1. Graphical Abstract. The Ha-CoV-2 pseudovirus structure and application. Image created with Biorender.com. Please click here to download this File.
Supplementary File 1. Protein Sequences. List of the sequences of SARS-CoV-2 S, M, N, and E proteins. The S protein sequences also include the SARS-CoV-2 variants Omicron (B.1.1.529) and Delta (B.1.617.2). Please click here to download this File.
Discussion
The Ha-CoV-2 platform provides a rapid, robust, and simple workflow to quantify viral variants and neutralize antibodies. However, there are a few critical steps that need attention. The production of The Ha-CoV-2 pseudovirus should be performed using HEK293T cells with high viability. The cotransfection efficiency can be monitored 24 h post-transfection using the GFP reporter gene from the Ha-CoV-2 genome. The Ha-CoV-2 genome can contain two reporters (GFP and Luc), and GFP can be expressed during cotransfection and following Ha-CoV-2 infection of target cells12. The GFP+ cells from infection are normally at a low percentage (1% to 5%), but each infected cell expresses strong GFP signals (Figure 3). This low GFP percentage may limit the use of GFP as a robust readout for quantifying antibody neutralization, as compared with the Luc reporter, which quantifies the whole population of infected cells.
When performing the neutralization assay, it is essential to change pipette tips between well transfers and to ensure the antibody and serum-free medium are mixed thoroughly to produce accurate results. Additionally, when conducting the luciferase assay protocol, cells must be fully lysed for at least 3 min to ensure complete lysis of cells and the release of the luciferase enzyme. This will ensure the accuracy of the assay. Additionally, once the Firefly luciferase assay solution is added to the optical white-walled 96 well plates, the plate must be analyzed within 10 min as the initial light emission is high but decreases over time as the ATP is depleted21.
As more SARS-CoV-2 variants continue to evolve, there is an increased need for platforms like Ha-CoV-2 to rapidly screen for variant infectivity and variant sensitivity to vaccine-induced neutralizing antibodies. The Ha-CoV-2 platform offers faster speed, a higher signal-to-noise ratio, and a simple protocol compared to existing pseudovirus-based neutralization assays8,9,10,11. The Ha-CoV-2 platform also offers the advantage that it can be used in BSL-2 laboratories and does not require the use of BSL-3 facilities. This allows SARS-CoV-2 to research to be pursued in common research and clinical laboratories. Furthermore, the Ha-CoV-2 platform produces rapid results in comparison with other systems. For instance, the study of neutralizing antibodies against infectious SARS-CoV-2 virus often makes use of the plaque reduction neutralization test (PRINT)22. Although PRINT produces reliable results, manual counting of plaque-forming units (PFUs) is slow and requires 3-5 days to obtain results23,24. Other pseudotype systems, such as the lentivirus-pseudovirus need 24-72 h to produce a detectable reporter signal12. In comparison, the Ha-CoV-2 neutralization assay can generate results within 18 h. The Ha-CoV-2 provides a convenient tool for rapid screening and quantification of viral variants and neutralizing antibodies for pandemic monitoring.
Monitoring the infectivity of SARS-CoV-2 is essential as more variants of concern (VOCs) continue to emerge. Ha-CoV-2 offers the advantage of quickly determining the infectivity of VOCs. Previous studies have used artificial intelligence (AI)-based modeling to quantitatively analyze the infectivity of the Omicron subvariant and the other SARS-CoV-2 variants, such as the Delta variant25. These studies have shown that the Omicron variant is more contagious than the original virus, and more likely to escape neutralizing antibodies25. In these studies, using Ha-CoV-2, similar phenotypes were observed. Additionally, in the antibody neutralization assays, the Omicron variant is ten times less likely to be neutralized by 27BV than the Wuhan and Delta strains. These results are also consistent with the reported higher transmissibility of the Omicron variant, which has at least 15 mutations on its receptor binding domain (RBD), likely enhancing viral binding affinity to the ACE2 receptor for higher transmissibility and greater immune escape26.
Acknowledgements
This work was supported by George Mason University internal research fund.
Materials
| Name | Company | Catalog Number | Comments |
| 27VB1 20 µg SARS-CoV-2 Standard Neutralizing Antibody | Virongy Biosciences | 27VBI-01 | |
| 500 mL - US Origin FBS | Neuromics | FBS001 | |
| AB Mixing Plate: Olympus 96-Well PCR Plate, Non-Skirted UltraThin Wall, Natural, 25 Plates/Unit | Genesee Scientific | Cat# 24-300 | |
| Allegra 6R Centrifuge | Beckman Coulter | 2043-30-1158 | |
| DMEM (1x) | ThermoFisher | 11995-073 | |
| GenClone 25-209, TC Treated Flasks, 250ml, Vent Growth Area: 75.0cm2, 5 per Sleeve, 100 Flasks/Unit | Genesee Scientfic | 25-209 | |
| GlowMax Discover Microplate reader | Promega | GM3000 | |
| Ha-CoV-2 E Vector | Virongy Biosciences | pCoV2_E | |
| Ha-CoV-2 M Vector | Virongy Biosciences | pCoV2_M | |
| Ha-CoV-2 N Vector | Virongy Biosciences | pCoV2_N | |
| Ha-CoV-2 WT S Vector | Virongy Biosciences | pCoV2_WT S | |
| Hek293T cells | ATCC | CRL-3214 | |
| Illumination Firefly Luciferase Enhanced Assay Kit 1000 assays | Gold Bio | I-930-1000 | |
| Infection Plate: 96-Well Tissue Culture Plate, Greiner Bio-One (With Lid, μClear White Flat Round, Chimney) | VWR | Cat# 82050-758 | |
| pAlphaPro-Luc-GFP-PreΨ (Ha-CoV-2 Genome) Vector | In house | ||
| PEI-based Transfection Reagent | Virongy Biosciences | Transfectin | |
| Penicillin-Streptomycin-Glutamine (100X) | Invitrogen | 10378016 | |
| Polyethylenimine, branched | Millipore Sigma | 408727-100ML | |
| QuantStudio 7 Pro Real-Time PCR System | ThermoFisher | A43163 | |
| Ready to use (HEK293T)(ACE2/TMPRSS2) Cells | Virongy Biosciences | Ready-To-Use-Cells | |
| SARS-CoV-2 S Omicron (B.1.1.529) Vector | Virongy Biosciences | pCoV2-B.1.1.529 | |
| SARS-CoV-2 S Delta (B.1.617.2) Vector | Virongy Biosciences | pCoV2- B.1.617.2 | |
| Syringe Filters, PES, 0.22µm | Genesee Scientfic | 25-244 | |
| TaqMan Fast Virus 1-Step Master Mix | ThermoFisher | 4444432 | |
| Trypan Blue Solution, 0.4% | ThermoFisher | 15250061 |
References
- World Health Organization. . Weekly epidemiological update on COVID-19. 143, 1 (2023).
- Dhama, K., et al. Global emerging Omicron variant of SARS-CoV-2: Impacts, challenges and strategies. Journal of Infection and Public Health. 16 (1), 4-14 (2023).
- Dhama, K., et al. Coronavirus Disease 2019-COVID-19. Clinical Microbiology Reviews. 33 (4), e00028-e00020 (2020).
- El-Shabasy, R. M., et al. Three waves changes, new variant strains, and vaccination effect against COVID-19 pandemic. International Journal of Biological Macromolecules. 204, 161-168 (2022).
- CDC. . Interim laboratory biosafety guidelines for handling and processing specimens associated with coronavirus disease 2019 (COVID-19). , (2021).
- Kaufer, A., Theis, T., Lau, K., Gray, J., Rawlinson, W. Laboratory biosafety measures involving SARS-CoV-2 and the classification as a Risk Group 3 biological agent. Pathology. 52 (7), 790-795 (2020).
- Vanderheiden, A., et al. Development of a Rapid Focus Reduction Neutralization Test Assay for Measuring SARS-CoV-2 Neutralizing Antibodies. Current Protocols in Immunology. 131 (1), e116 (2020).
- Dieterle, M. E., et al. A Replication-Competent Vesicular Stomatitis Virus for Studies of SARS-CoV-2 Spike-Mediated Cell Entry and Its Inhibition. Cell Host & Microbe. 28 (3), 486-496 (2020).
- Nie, J., et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nature Protocols. 15 (11), 3699-3715 (2020).
- Nie, J., et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerging Microbes & Infections. 9 (1), 680-686 (2020).
- Yang, R., et al. Development and effectiveness of pseudotyped SARS-CoV-2 system as determined by neutralizing efficiency and entry inhibition test in vitro. Biosafety and Health. 2 (4), 226-231 (2020).
- Hetrick, B., et al. Development of a hybrid alphavirus-SARS-CoV-2 pseudovirion for rapid quantification of neutralization antibodies and antiviral drugs. Cell reports methods. 2 (3), 100181 (2022).
- Jackson, C., Farzan, M., Chen, B., Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nature Reviews. Molecular Cell Biology. 23 (1), 3-20 (2022).
- Jackson, L. A., et al. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. The New England Journal of Medicine. 383 (20), 1920-1931 (2020).
- Polack, F. P., et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. The New England Journal of Medicine. 383 (27), 2603-2615 (2020).
- Li, F. Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology. 3 (1), 237-261 (2016).
- Donofrio, G. A Simplified SARS-CoV-2 Pseudovirus Neutralization Assay. Vaccines. 9 (4), 389 (2021).
- Crawford, K. H. D., et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 12 (5), 513 (2020).
- Zettl, F., et al. Rapid Quantification of SARS-CoV-2-Neutralizing Antibodies Using Propagation-Defective Vesicular Stomatitis Virus Pseudotypes. Vaccines. 8 (3), 386 (2020).
- Li, Q., Liu, Q., Huang, W., Li, X., Wang, Y. Current status on the development of pseudoviruses for enveloped viruses. Reviews in Medical Virology. 28 (1), e1963 (2018).
- Lundin, A. Optimization of the firefly luciferase reaction for analytical purposes. Advances in Biochemical Engineering/Biotechnology. 145, 31-62 (2014).
- Deshpande, G. R., et al. Neutralizing antibody responses to SARS-CoV-2 in COVID-19 patients. The Indian Journal of Medical Research. 152 (1 & 2), 82-87 (2020).
- Chen, C., et al. Research progress in methods for detecting neutralizing antibodies against SARS-CoV-2. Analytical Biochemistry. 673, 115199 (2023).
- Manischewitz, J., et al. Development of a novel vaccinia-neutralization assay based on reporter-gene expression. The Journal of Infectious Diseases. 188 (3), 440-448 (2003).
- Chen, J., Wang, R., Gilby, N., Wei, G. Omicron Variant (B.1.1.529): Infectivity, Vaccine Breakthrough, and Antibody Resistance. Journal of Chemical Information and Modeling. 62 (2), 412-422 (2022).
- Saxena, S. K., et al. Characterization of the novel SARS-CoV-2 Omicron (B.1.1.529) variant of concern and its global perspective. Journal of Medical Virology. 94 (4), 1738-1744 (2022).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved