Three-Dimensional Bioprinting of Human iPSC-Derived Neuron-Astrocyte Cocultures for Screening Applications
In This Article
Summary
Here, we present a protocol to produce 3D-bioprinted cocultures of iPSC-derived neurons and astrocytes. This coculture model, generated within a hydrogel scaffold in 96- or 384-well formats, demonstrates high post-print viability and neurite outgrowth within 7 days and shows the expression of maturity markers for both cell types.
Abstract
For a cell model to be viable for drug screening, the system must meet throughput and homogeneity requirements alongside having an efficient development time. However, many published 3D models do not satisfy these criteria. This therefore, limits their usefulness in early drug discovery applications. Three-dimensional (3D) bioprinting is a novel technology that can be applied to the development of 3D models to expedite development time, increase standardization, and increase throughput. Here, we present a protocol to develop 3D bioprinted coculture models of human induced pluripotent stem cell (iPSC)-derived glutamatergic neurons and astrocytes. These cocultures are embedded within a hydrogel matrix of bioactive peptides, full-length extracellular matrix (ECM) proteins, and with a physiological stiffness of 1.1 kPa. The model can be rapidly established in 96-well and 384-well formats and produces an average post-print viability of 72%. The astrocyte-to-neuron ratio in this model is shown to be 1:1.5, which is within the physiological range for the human brain. These 3D bioprinted cell populations also show expression of mature neural cell type markers and growth of neurite and astrocyte projections within 7 days of culture. As a result, this model is suitable for analysis using cell dyes and immunostaining techniques alongside neurite outgrowth assays. The ability to produce these physiologically representative models at scale makes them ideal for use in medium-to-high throughput screening assays for neuroscience targets.
Introduction
Research into central nervous system (CNS) diseases in the drug discovery industry is expanding1. However, many prevalent CNS diseases such as epilepsy, schizophrenia, and Alzheimer's disease still have no curative treatments2,3,4. The lack of effective therapeutics across CNS diseases can, at least in part, be attributed to a lack of accurate in vitro models of the brain5. This has resulted in a translational gap between current in vitro models and in vivo data and a subsequent bottleneck in research efforts.
Driven by this translational gap, there has been a significant increase in the development of novel 3D cell models within recent years, including neural organoids, neurospheroids, and scaffold-based models6. The 3D structure of these models aids in recapitulating the neural microenvironment, including biomechanical stresses, cell-cell contacts, and the brain extracellular matrix (ECM)7. The brain ECM is a dynamic element of neurophysiology that occupies the space between neural cell types, including neurons, astrocytes, oligodendrocytes, and the neurovascular unit7. Recapitulation of the brain ECM has been shown to affect neuronal morphology and neuronal firing, and many complex 3D models of the brain have demonstrated deposition of ECM proteins by neural cell types8,9,10,11. Scaffold-based models consist of mature neural cocultures suspended in a porous synthetic or biological hydrogel matrix which represents the brain ECM12. Unlike organoid and spheroid systems, scaffold-based 3D models allow the customization of ECM proteins present and have the added benefit of tunability of hydrogel stiffness to mimic biomechanical stresses13,14.
Although an overwhelming majority of 3D neural models demonstrate an increased recapitulation of the brain microenvironment, not all models are well suited for implementing drug discovery applications15. For a 3D model to be implemented into industrial processes, the system must meet throughput requirements for screening applications and have a relatively short development time16. 3D Bioprinting is a novel technology that offers the potential to create 3D scaffold-based neural models with rapid development time, increased throughput, and higher levels of precision control, alongside the removal of variability caused by human error17. This protocol presents a 3D coculture model of human iPSC-derived glutamatergic neurons and astrocytes in a hydrogel scaffold. This hydrogel scaffold contains physiologically representative bioactive peptides (RGD, IKVAV, YIGSR) and ECM proteins within a mimetic biomechanical stiffness. These full-length ECM proteins include full-length laminin-211 and hyaluronic acid, abundant in the human cortex, with a stiffness of 1.1 kPa in line with in vivo measurements18. This model is designed with practicality for drug discovery, and is created using a 3D bioprinter in a 96-well or 384-well plate format suitable for screening analysis using imaging techniques with cell dyes and antibodies, alongside neurite outgrowth assays. Cells show expression of neural cell type markers and growth of neurite and astrocytic projections within 7 days of culture. Thus, this protocol will present the methodology to develop a high-throughput 3D neural coculture model for use in drug discovery applications.
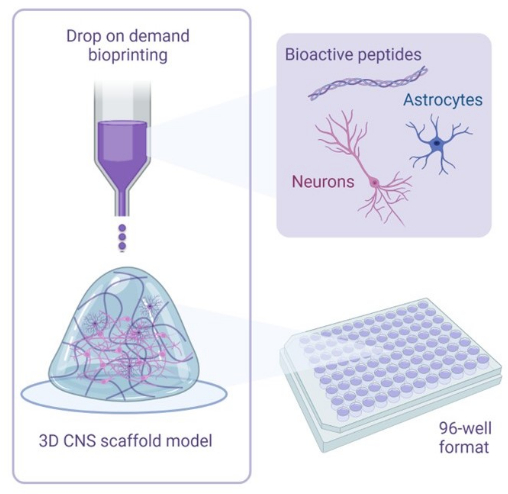
Figure 1: Illustrative overview of methodology used to 3D bioprint cocultures. Human iPSC-derived neurons and astrocytes are combined with activator and bioink solutions containing bioactive peptides and are bioprinted to hydrogel scaffolds in 96-well or 384-well formats using drop-on-demand bioprinting technology. Please click here to view a larger version of this figure.
Protocol
1. Bioprinting of 3D models
- Generating plate map and print file
- Prior to model generation, generate a plate map, protocol, and print file using the plate map software. Open the plate map software and select the Culture Cells in 3D option.
- Select the model type to be bioprinted from the available options on the drop-down menu; this model has been validated in the Imaging Model and HTP model formats. Select Imaging Model for 96-well plates and HTP for 384-well plates.
- In the Matrix Selection window, select Hydrogel Px02.53, containing Laminin-211 and hyaluronic acid proteins, with IKVAV, RGD, and YIGSR peptides.
- In the pop-up window, enter the cell types as Glutamatergic Neurons and Astrocytes and enter the cell density as 20 million cells/mL.
- Use the software to design the desired plate map by highlighting wells on the on-screen plate. Include at least 1 row of 2D control on plastic in the design. 2D control on plastic wells consists of cells deposited directly into the well without any hydrogel scaffold; coat these wells as per step 1.2.
- Ensure that the plate model listed at the top of the window is changed to the intended plate model for use (see Table of Materials).
- Download the protocol and print file for the plate map using the download buttons and save them to the bioprinter-linked computer.
- Coating of 2D control on plastic wells
- At least 24 h before printing, coat the wells for the 2D control models for cell adherence and growth. 3D model wells do not require any pre-coating. All processes are to be undertaken in a biosafety cabinet unless otherwise stated. Plates can be coated and prepared up to 7 days before printing.
- Make 50 mL of 1x borate buffer by diluting 2.5 mL of 20x borate stock solution with 47.5 mL of sterile dH2O.
- Add 100 µL of 50% (w/v) polyethyleneimine (PEI) to 50 mL of 1x borate buffer to create a 0.1% (w/v) PEI solution and sterile filter through a 0.22 µm filter.
- Add 0.1% (w/v) PEI solution to each 2D control well, as indicated by the plate map generated in step 1.1. Add 100 µL/well if using a 96-well plate or 25 µL/well if using a 384-well plate.
- Incubate the plate for 2 h at 37 °C before aspirating the 0.1% (w/v) PEI solution and rinsing the wells 5x with PBS. Let the wells dry completely in the biosafety cabinet.
- Dilute 100 µL of 1 mg/mL laminin solution in 5 mL of neurobasal medium. Add 100 µL to each 2D control well if using a 96-well plate or 25 µL if using a 384-well plate. Incubate for 4 h at 37 °C.
- If storing plates, leave the laminin solution in place after incubation and store at 4 °C. Prewarm the plates for 1 h at 37 °C before use.
- 3D bioprinting of coculture models
- Always maintain a sterile technique when using the bioprinter, and wipe gloves, cartridges, and culture plates with 70% EtOH before being put inside the printer. Unless otherwise stated, perform all processes outside of the bioprinter in a biosafety cabinet.
- Switch on the compressor for the bioprinter and initialize the printer by selecting the Initialize button on the bioprinter software. Once the airflow in the printer has started, wipe down surfaces inside the printer with a 70% EtOH wipe.
- On the day of printing, undertake the guided Start of Day Greenlighting process to ensure the correct nozzle status for the print run (minimum nozzle status 2A2B).
- Take a bioprinter cartridge from the sterile packaging, add 20 mL of sterile dH2O in compartment A, and add 6 mL of sterile-filtered 70% EtOH to compartments B1-4. Insert the cartridge into the lefthand cartridge holder inside the bioprinter and place the plate lid into the dedicated lid holder.
- Select the Greenlighting button on the software home screen and confirm on the software that the cartridge is in place within the bioprinter and that the lid has been removed. Initiate the greenlighting process.
- When prompted by the software, confirm that no consistent drips can be seen from either the left or right needles. Subsequently, when prompted by the software, confirm that water is present in the B7 and B8 compartments.
- After the bioprinter has completed the self-guided greenlighting step, insert an uncoated sterile 96-well flat bottom plate (separate from the coated plate for bioprinting cocultures) and insert this new plate into the righthand plate holder section of the bioprinter when prompted by the software. Ensure the plate holder is set to High Profile Plate, and the lid is removed and placed in the dedicated lid holder before commencing the next step.
- The bioprinter will deposit water droplets into each well of the 96-well plate. Once complete, place the plate from the bioprinter and confirm that water droplets are present in each well of the plate.
- Dispose of the 96-well plate used for greenlighting and allow the bioprinter to finish the greenlighting process. Once complete, the software will state the nozzle status of the bioprinter and confirm that the bioprinter is ready to print models. Place the lid onto the print cartridge and remove it to the biosafety cabinet for the next steps.
- Using the bioprinter software, load the print file (generated in step 1.1) into the software using the Print Run software button and open the print protocol pdf.
- Retrieve bioink and activator fluids, as indicated on the print protocol pdf, from -20 °C and thaw at room temperature (RT) for 40 min. For hydrogel matrix Px02.53 this will include: 1x vial F32, 1xvial F3, 1x vial F261, 1x vial F299. Do not thaw bioink and activator fluids in hands or water baths.
- Bring 50 mL of complete media with doxycycline and ROCK inhibitor (complete media +DOX/ROCKi) (see step 2.2) to RT while bioink and activator fluids are thawing.
- Once bioinks and activators are thawed, prepare the printing cartridge as instructed on the final two pages of the generated print protocol pdf. The greenlighting printer cartridge will be reused for the model printing stages.
- Ensure that there are 40 mL of dH2O in compartment A1 and 8 mL of 70% EtOH in compartments B1 and B2. Add 1.2 mL of activator F32 to C1, 1.2 mL of F3 to C2, and 200 µL of F261 to C4.
- Retrieve the PEI and laminin-coated plate from the incubator and place it inside the printer in the righthand plate holder compartment. Do not remove the laminin-media solution from the wells at this point. Ensure the plate holder compartment is set to Low Profile Plate and the lid is removed and in the lid holder.
- Place the print cartridge into the bioprinter, ensuring the lid is removed and placed in the holder, and start the print run on the software by selecting the Print Inert Base button on the bioprinter software.
- While the inert base is printing, retrieve the vials of glutamatergic neurons and astrocytes from liquid nitrogen storage, thaw, and resuspend as per instructions in section 2.
- Once the cells are thawed, and 8 mL of complete media +DOX/ROCKi has been added to each cell type separately (as per section 2), centrifuge the cells at 300 x g for 5 min at RT.
- Aspirate the supernatant and resuspend both cell types separately in 1 mL of complete media +DOX/ROCKi.
- Add 20 µL of cell suspension to 20 µL of trypan blue and mix before counting cells to determine viable cell concentration per mL for each cell type.
- Combine a total of 3 million glutamatergic neurons with 1 million astrocytes in a 15 mL tube. Add media to create a total volume of 8 mL.
- Centrifuge the cells at 300 x g for 5 min at RT.
- Aspirate as much of the supernatant as possible without disturbing the cell pellet and resuspend the cell pellet in 200 µL of activator fluid F299.
- Note that the activator fluid is viscous; pipette up and down to fully resuspend the pellet. The cell types are delicate; minimize shearing and bubbles by using a wide bore pipette tip and the reverse pipetting technique. Pipette up and down firmly no more than 3 times to prevent cell loss.
- Once the inert base stage is finished printing, place the lids on the cartridge and culture plate, remove both from the bioprinter and place in the biosafety cabinet.
- Add the 200 µL of cell suspension in F299 to well C3 of the print cartridge, reinsert the cartridge into the bioprinter, remove the lid, and place it into the lid holder. Do not yet reinsert the culture plate.
- Initiate the Print Models stage of the print run. While the bioprinter is priming fluids, remove the laminin-media solution from the 2D control wells of the plate and replace it with 150 µL of complete media +DOX/ROCKi in each 2D control well if using a 96-well plate and 50 µL of complete media +DOX/ROCKi if using a 384-well plate.
- After the liquids have been primed, insert the bioprinting targeting plate into the bioprinter, remove the lid, and place it in the lid holder.
- Begin the bioprinter targeting process.
- When prompted by the bioprinting software, remove the targeting plate from the bioprinter.
- Use the guide to select where droplets can be observed on the plate. Reinsert the targeting plate into the bioprinter and repeat the droplet printing and selection process.
- When prompted, again remove the targeting plate, then replace it with the cell culture plate (containing media in the 2D control wells as per step 1.3.25). Ensure the culture plate lid is off and placed in the holder. The bioprinter will finish model printing.
- Once model printing is complete, follow the cell culture methods in section 2 (step 2.8) to add media to the wells.
- Commence the bioprinter cleaning process and, once complete, dispose of cartridges and remaining liquids according to lab protocols.
2. Cell culture
- Models are generated using 1x vial of glutamatergic neurons (>5 million cells per vial) and 1x vial of astrocytes (>1 million cells per vial).
- Preheat the water bath to 37 °C.
- Transport both vials of cells to the lab on dry ice and immersed in a water bath immediately after removal from dry ice. Thaw both vials simultaneously.
- Remove the vials from the water bath when only a small ice crystal remains; this will take approximately 3 min after immersion. Do not swirl or mix the cells in the water bath.
- Spray the vials with 70% EtOH and transfer them into the biosafety cabinet.
- Add 500 µL of complete media +DOX/ROCKi dropwise into each vial before transferring each suspension into separate 15 mL tubes.
- Top up the media in each tube to 8 mL and proceed with bioprinting steps as per step 1.3.
- Following bioprinting models (step 1.3), immediately add complete media +DOX/ROCKi to all 3D coculture wells and place the models into an incubator at 37 oC and 5% CO2. If using a 96-well plate, add 150 µL of media per well; if using a 384-well plate, use 50 µL of media per well.
- No media changes are required for the first 48 h of culture.
- When performing media changes on 2D controls and models, care should be taken to prevent inducing mechanical stress, which could detach 2D cultures or cause deformation of the hydrogel. Perform media aspiration and addition slowly using a micropipette pointing down the side of the well.
- After 48 h, perform a 90% media change on all wells and remove ROCKi from the media composition (see step 2.14).
- After 96-h, perform a further 90% media change on all wells to remove DOX from the media composition (see step 2.14).
- Following the two 90% media changes at 48 h and 96 h, carry out 50% media changes every 48 h, where 50% of media is aspirated and replaced with fresh complete media
- Media composition:
- Complete media: Neurobasal media with 1x GlutaMAX, 1x B27, 12.5 nM 2-mercaptoethanol, 10 ng/mL NT3, 5 ng/µL BDNF.
- Complete media plus doxycycline (DOX): Neurobasal media with 1x GlutaMAX, 1x B27, 12.5 nM 2-mercaptoethanol, 10 ng/mL neurotrophin-3 (NT3), 5 ng/µL brain-derived neurotrophic factor (BDNF), and 1 µg/mL doxycycline.
- Complete media plus DOX/ROCK inhibitor (ROCKi): Neurobasal media with 1x GlutaMAX, 1x B27, 12.5 nM 2-mercaptoethanol, 10 ng/mL NT3, 5 ng/µL BDNF, 1 µg/mL doxycycline, and 10 µM ROCK inhibitor.
3. Neurite growth analysis
- Place cells in a live cell microscope for brightfield imaging immediately after the addition of media post-bioprinting.
- Use microscope software to schedule cells to be imaged at 4x magnification in every well, including 2D controls, every 12 h for at least 7 days.
- Carry out media changes every 48 h, as detailed in section 2, placing the cells back in the microscope after media changes each time.
- After 7 days of data, export the images from the software in .jpg format
- Import all .jpg images into ImageJ software and convert files to 8-bit format. Load the NeuronJ plugin and trace neurite growth in images, including branch points, using the tracing tool.
- Use the neurite tracing data generated from NeuronJ to plot neurite outgrowth over time.
4. Cell viability analysis
- At every 24-h time point in culture, stain 3 or more wells for cell viability analysis using a live/dead viability kit. Repeat this step at 24 h or 48-h time points for the duration of the study.
- Prepare live/dead reagent media by suspending live cell reagent (1x Calcein-AM) and dead cell reagent (1x ethidium homodimer-1) and 1x live cell nuclear stain (Hoechst 33342) in 10 mL of reduced serum media without phenol red (Opti-MEM) 30 min before imaging. Let live/dead reagent media come to RT. Store live/dead reagent media out of direct light due to fluorescence bleaching.
- Remove media from 3 wells containing cell models/2D controls, carefully preventing gel disturbance. Wash the cell models once with RT sterile PBS. Add 100 µL of live/dead reagent media to each well for a 96-well plate or 25 µL for a 384-well plate.
- Incubate the models for 30 min at 37 °C and 5% CO2.
- After incubation, models are ready for imaging. Perform imaging on any standard microscope with red (647 nm, dead cells), green (488 nm, live cells), and blue (405 nm, nuclear stain) excitation channels. For best results in 3D models, image models using a high-content confocal microscope and carry out imaging using a Z-stack function at 4x or 10x magnification.
NOTE: In this study, cell models were imaged using the INCell Analyzer 6500HS (high-content imaging system), and analysis was carried out using Signals Image Artist (image analysis platform, see step 4.7). - After imaging is complete, return cell models to the cell culture incubator at 37 °C and 5% CO2. However, omit cell models used for viability analysis from further studies because of the live/dead reagent media on long-term viability.
- Analysis of images on the image analysis platform
- Select each plane of the Z-stack image in the software. Combine plane images as a maximum-intensity projection image.
- Create a new analysis by selecting the bioprinted model as a region of interest (ROI) by identifying fluorescence from the live cell channel at 488 nm (ensure all cells are selected under ROI).
- Within the region of interest, use the imaging software to identify the number of cells in the ROI through the nuclear stain channel (405 nm), the number of live cells in the ROI using the 488 nm channel, and the number of dead cells in the ROI using the 647 nm channel.
- Run the analysis on all live/dead treated cell model wells and export the data table containing ROI area, total cell number, number of live cells, and number of dead cells.
- Calculate the number of live and dead cells as a percentage of total cell number per analysis day.
5. Immunostaining and cell population analysis
- Before fixation, remove media from the cell models and wash the models once in PBS.
- Fix cells with 4% (v/v) paraformaldehyde in PBS at RT for 20 min.
CAUTION: All work with paraformaldehyde must be carried out according to laboratory procedures. - Aspirate 4% (v/v) paraformaldehyde solution from the models and wash the models four times in PBS to completely remove paraformaldehyde.
- Permeabilize the cell models with 0.2% triton-X for 30 min at RT and wash three times with PBS.
- Block the cell models using 10% (v/v) normal donkey serum (NDS) for 3 h at RT.
- Remove the blocking solution and add primary antibodies (diluted in 1% v/v NDS in PBS) to the models. If performing cell population ratio analysis (see step 5.11), ensure to include cell models that are co-stained for Β-III tubulin (with red AF647 conjugation) and GFAP (with green AF488 conjugation). Incubate models with primary antibodies for 24 h at 4 °C.
- After primary incubation, wash the models stained with conjugated primary antibodies three times in PBS and counterstain with 20 µM Hoechst for 30 min. Image as detailed in step 5.10.
- Wash the models stained with unconjugated primary antibodies three times in PBS and add secondary antibodies (diluted in 1% v/v NDS in PBS). Incubate for 24 h at 4 °C.
- Remove secondary antibodies by washing three times in PBS, and counterstain models with 20 µM Hoechst for 30 min before imaging.
- Perform imaging on standard microscopes with red (647 nm) and green (488 nm) excitation channels. However, for best results in 3D models, image models using a high-content confocal microscope and carry out imaging using a Z-stack function at 4x or 10x magnification.
- Analysis of cell population ratios on the image analysis platform
- Obtain an analysis of cell populations using models co-stained for GFAP (with AF488 conjugation) and Β-III tubulin (with AF647 conjugation).
- Select each plane of the Z-stack image in the software and combine the plane images as a maximum-intensity projection image.
- Create a new analysis by selecting the bioprinted model as an ROI by identifying fluorescence from the Β-III tubulin channel at 647 nm (ensure all cell-containing area is selected under ROI).
- Within the ROI, use the software to identify the number of cells in the Hoechst channel (405 nm), the number of GFAP+ astrocytes in the ROI using the 488 nm channel, and number of Β-III tubulin+ cells in the ROI using the 647 nm channel.
- Run the analysis on all co-stained cell model wells and export the data table containing ROI area, total cell number, number of GFAP+ cells, and number of Β-III tubulin+ cells.
- Calculate the number of GFAP+ and Β-III tubulin+ cells as a percentage of total cell number.
Representative Results
Neurite growth analysis
In this protocol, iPSC-derived glutamatergic neurons and astrocytes were bioprinted in coculture into a hydrogel matrix using the 3D bioprinter. Over the first 7 days post-printing, cells were imaged every 12 h using a live cell microscope. Post-bioprinting, cells should have a rounded morphology and should be dispersed throughout the hydrogel matrix, gradually changing to form smaller cell clusters with few protrusions over the first few days of culture (see Supplementary Video 1 for representative healthy cell growth). By day 4, healthy cells will migrate throughout the gel to form larger clusters, which are connected through neurite outgrowths. By day 7, almost no single cells should remain, the interconnecting bundles of neurites and astrocytic projections should appear fortified, and many smaller neurite outgrowths can be seen forming from the clusters (Figure 2A). Using a series of live cell brightfield images taken over the 7-day growth period, an analysis of neurite outgrowth was performed as detailed in section 3. This analysis demonstrated that neurite outgrowth increases in a near linear fashion (R2 value = 0.84) between 12 h and 156 h (Figure 2B). During this period of neurite outgrowth, cell body clusters also increase in size (see Supplementary Video 1), which is indicative of cell migration throughout the hydrogel.
Cell viability and population ratio
In this protocol, a concentration of 20 million cells/mL, comprising 15 million neurons/mL and 5 million astrocytes/mL, is used for bioprinting the cell models. Using live cell staining with calcein-AM (live cells), ethidium homodimer-1 (dead cells) and a nuclear stain, the number of cells surviving over a 7-day period can be calculated as per section 4 (Figure 3A). Cell viability results for representative cultures are shown for day 4, where 72% ± 1% (mean ± SEM) of total cells are live and show staining for Calcein-AM, while 29% ± 2% (mean ± SEM) total cells are dead and show staining with ethidium homodimer-1 (Figure 3B). Representative images of cell staining with Calcein-AM and ethidium homodimer-1 can be seen in Supplementary Figure 1. It should be noted that cell survival values for 3D cultures cannot be directly compared to 2D cultures, as dead cells are retained in the hydrogel and will not be removed during cell feeding processes.
Using the immunofluorescent staining for Β-III tubulin and GFAP, as described in section 5 and in Figure 4, image analysis can be carried out to determine cell population ratios between neurons and astrocytes (Figure 3A). Of total cells per model in representative cultures, Β-III tubulin positive neurons represent 49% ± 3% (mean ± SEM), while GFAP positive astrocytes represent 30% ± 4% (mean ± SEM). This gives a ratio of 1:1.5, astrocytes to neurons, respectively. This leaves a remainder of 21% total cells per model, which do not stain for either cell marker. As cell viability analysis demonstrated that a mean value of 29% of cells are not viable at day 4, it is likely that these cells are dead within the hydrogel.
Expression of cell markers
Morphology of the bioprinted neurons and astrocytes was assessed through immunostaining for neuronal cell type marker (Β-III tubulin) and astrocytic marker (GFAP). In the representative cultures shown, immunostaining is localized to individual cell types, showing healthy cell morphology, with both cell types exhibiting outgrowth of cellular protrusions (Figure 4A,B). As the hydrogel and cell structures are three-dimensional, each image represents only one slice through the structure in Figure 4A,B. Figure 4C shows a merged stack of images throughout the hydrogel, demonstrating views of cell localization in the X, Y, and Z planes. Figure 4D shows immunostaining for Β-III tubulin only; highlighting finer neurite outgrowths from the cell body clusters. To further examine the phenotype of the glutamatergic neurons, immunostaining for the glutamatergic ionic receptor marker, GluR2, can be carried out. Within Figure 4E, area 4E.1 (inset) has been highlighted to show higher resolution punctate staining along the neurite bundles. This therefore, confirms that neurons in this coculture have a glutamatergic phenotype. Across all immunostaining images, immunofluorescent stained non-cell structures can be observed surrounding the cell clusters and neurites. It is likely that these structures represent debris retained within the hydrogel in combination with minor amounts of non-specific antibody binding to the hydrogel. This is expected in bioprinted cultures, as within 3D scaffold models, dead cells and debris are not removed during cell feeding. A representative negative control immunostaining image is shown in Supplementary Figure 2 for a demonstration of hydrogel non-specific binding of secondary antibodies.
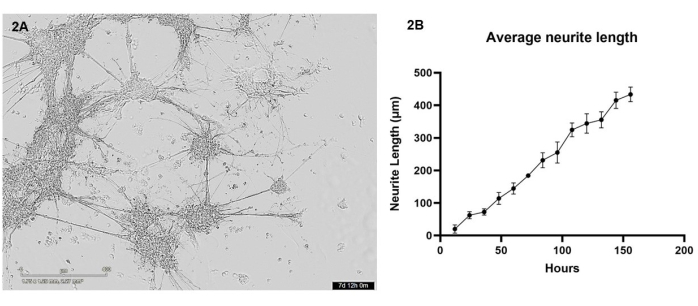
Figure 2: Glutamatergic neurons and astrocytes were bioprinted into the hydrogel matrix using the bioprinter and were imaged every 12 h using a brightfield microscope. (A) An example brightfield image taken of cell cultures during analysis. The image represents time point 156 h, and the scale bar represents 400 µm. (B) The average length of neurite outgrowths (µm) from the cultures measured using the NeuronJ package for ImageJ. Each data point is n = 3 neurites, and data is shown as mean ± SEM. Please click here to view a larger version of this figure.
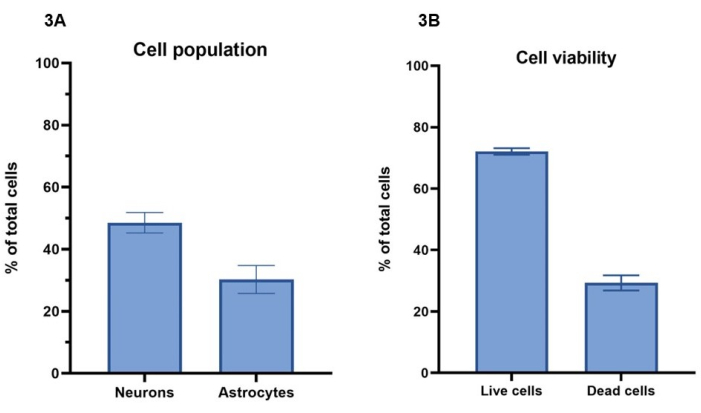
Figure 3: A concentration of 20 million cells/mL of activator solution was used for bioprinting the cell models. (A) Cell viability was calculated using live/dead cell dyes (Calcein-AM and ethidium homodimer-1, respectively). Values show 72% ± 1% (mean ± SEM, n = 3) of total cells per well are live and 29% ± 2% (mean ± SEM, n = 3) of cells are dead of total cell population per well at Day 4. Values shown represent mean ± SEM. (B) Cell populations percentage of neurons and astrocytes per well were calculated through image analysis of staining shown in Figure 4. Neurons represent the percentage of cells staining positive for Β-III tubulin at Day 7 (49% ± 3%, mean ± SEM, n = 3), while astrocytes represent the percentage of cells staining positive for GFAP at Day 7 (30% ± 4%, mean ± SEM, n = 3). Values shown represent mean ± SEM. All imaging for calculations shown in Figure 3 was performed on a confocal imaging system, and all analyses were performed on the image analysis platform and GraphPad Prism as per methods. Please click here to view a larger version of this figure.
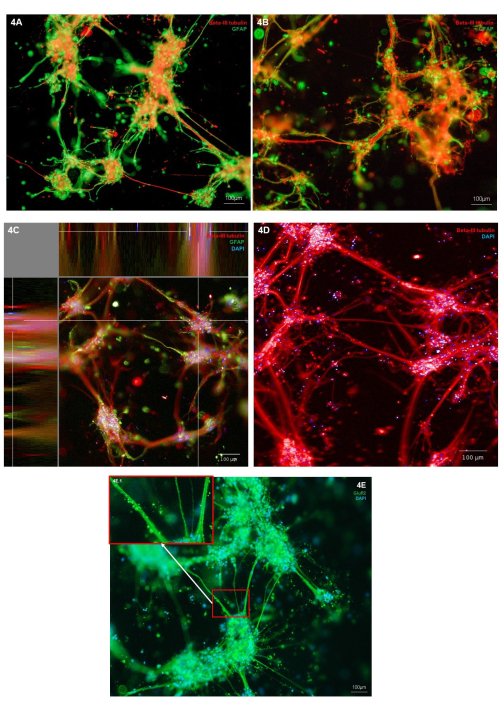
Figure 4: Expression of neural cell type markers in 3D bioprinted cocultures of glutamatergic neurons and astrocytes at day 7. (A,B) Immunofluorescent staining of neuronal marker Β-III tubulin and astrocyte marker GFAP, imaged on an inverted microscope platform at 10x magnification. Scale bars represent 100 µm. (C) Immunofluorescent staining of neuronal marker β-III tubulin and astrocyte marker GFAP co-stained with Hoechst, shown in XYZ plane view, imaged on a confocal imaging system at 10x magnification. Created on the image analysis platform. The scale bar represents 100 µm. (D) Immunofluorescent staining of neuronal marker β-III tubulin co-stained with Hoechst, imaged on a confocal imaging system at 20x magnification. Scale bar represents 100 µm. (E) Immunofluorescent staining of glutamatergic marker GluR2 co-stained with Hoechst, imaged on an inverted microscope platform at 10x magnification. Box 3E.1 shows highlighted areas of GluR2 staining. The scale bar represents 100 µm. Please click here to view a larger version of this figure.
Supplementary Video 1: Glutamatergic neurons and astrocytes were bioprinted into the hydrogel matrix using the bioprinter and were imaged every 12 h using a brightfield microscope. Video of brightfield images taken of cell cultures during analysis, time points are indicated in the bottom right corner, and scale bars represent 400 µm. Please click here to download this File.
Supplementary Figure 1: Example images of live/dead analysis of bioprinted neurons and astrocytes on Day 4. Calcein-AM stain shown in green (488 nm), and ethidium homodimer stain shown in red (647 nm). The image is shown in XYZ plane view, created on the image analysis platform . The scale bar represents 100 µm. (A) Imaging was carried out using a confocal imaging system at 4x magnification. (B) Imaging was carried out using a confocal imaging system at 10x magnification Please click here to download this File.
Supplementary Figure 2: Example of negative control image after immunofluorescent staining. Primary antibodies were omitted, and green (488 nm) and red (647 nm) secondary antibodies were used as per immunostaining protocols. The image is shown in XYZ plane view, created on the image analysis platform. The scale bar represents 100 µm. Imaging was carried out using a confocal imaging system at 10x magnification. Please click here to download this File.
Discussion
The need for accurate models of the CNS has never been higher, and limitations of two-dimensional (2D) traditional cell culture models have driven a generation of complex CNS models in recent years19. However, many complex 3D models that represent interactions between neural cell types and the ECM have limitations that would prevent the application of these models in industrial processes6,20,21. In this protocol, we develop a 3D coculture model of human iPSC-derived neurons and astrocytes, which aims to resolve some of these limitations using 3D bioprinting technology to create a bioactive hydrogel scaffold in 96-well and 384-well formats.
The methodology for developing these models has been simplified through the plate map design software, auto-generated print protocols, and guided print process from the bioprinter. However, due to the sensitive nature of the sensitive iPSC-derived cell types used in this protocol care should be taken with the following critical steps in thawing and culture. Firstly, the inclusion of ROCK inhibitor (ROCKi) has several benefits throughout the bioprinting process and during early culture. Cell thawing is a critical point in which the neurons can experience a stress response, and improper thawing protocols can decrease the chances of survival22. It is typically recommended to thaw cells, add media, and raise cells to incubator temperature as efficiently as possible23. However, during the bioprinting process described in this protocol, it is necessary that neurons and astrocytes are resuspended in an activator solution rather than media, and cells will not be raised above room temperature until the end of the print run (up to 30 minutes post-thaw). Thus, the addition of ROCKi to the media immediately after thawing and including this during the two centrifugation steps (steps 2.1--2.7 and 1.3.15-1.3.20) is imperative to inhibit cell stress pathways, which would result in lower viability levels24. Furthermore, ROCKi has been shown to promote neurite outgrowth and improve neuronal maturation25. Thus, ROCKi supplementation is continued for 48 h post-bioprinting. However, it is imperative to remove ROCKi supplementation after 48 h to ensure complete wash-off during the subsequent media changes before cells are used for assay.
A further step that requires critical attention is during post-print media addition and media changes (steps 2.8-2.13). The bioprinted hydrogel scaffold has an equivalent biomechanical stiffness of only 1.1 kPa, equivalent to grey matter. As described in step 2.10, it is critical to pipette gently into the side of the well during media addition and aspiration to prevent disturbance. This is of particular relevance to 384-well plates, where the gel level takes up a higher proportion of total well volume. This method should also be used in 2D control wells to prevent edge lifting of cells and shearing of neurite outgrowths. The authors would also like to highlight the importance of sterile technique within the bioprinter, which should be treated with equivalent caution to that of a biosafety cabinet used for iPSC-derived cell cultures. This includes sterile filtering 70% EtOH and dH2O used in the greenlighting and printing procedures, keeping lids on the cartridges and plates while moving hands in and out of the bioprinter, and decontaminating surfaces inside the bioprinter with 70% ethanol wipes before and after printing.
The bioprinted hydrogel scaffold, formed from bioink and activator solutions, selected to develop this model is selected from a range of bioinks and activator solutions developed by Inventia Life Science for use within the RASTRUM bioprinter. Laminin and hyaluronic acid were identified as molecules of relevance to iPSC-derived neuronal maturation due to their role in axonal guidance, synapse formation, and formation of the perineuronal net26,27. Furthermore, a biomechanical stiffness of 1.1 kPa was selected, as lower-density hydrogels have been shown to enable better neurite outgrowth from neurons12. If modifications are made to the protocol by using neurons and astrocytes that have been differentiated in-house or from a different commercial supplier, it would be recommended to do a matrix selection test to determine the most supportive hydrogel scaffold15. Furthermore, cell density may also need to be optimized if changes to cell sources are made to ensure optimal viability and prevent hydrogel overcrowding. For all modifications and troubleshooting related to the bioprinter function, the authors recommend contacting manufacturers and referencing manufacturer protocols.
The CNS contains a broad range of neuronal subtypes and glial cells, all of which exist in different brain niches and have specific roles contributing to neural function28. Within the context of this broad scope, this model represents only the two most abundant cell types (astrocytes and excitatory glutamatergic neurons). Important cell types such as microglia, oligodendrocytes, and blood-brain barrier-forming endothelial cells are omitted from this system. The inclusion of microglia could be of relevance in focus on neuroimmune interactions, and oligodendrocytes could be of interest in diseases that affect central myelination. In addition to their role in pathology, cells such as blood-brain barrier-forming endothelial cells excrete drug-metabolizing enzymes, which could affect the use of this model for pharmacokinetic assays29. A further limitation of the model may be the ratio of astrocytes to neurons; the ratio of astrocytes to neurons varies greatly between brain regions, with suggested values of between 1:1 and 1:330,31. This model has an approximate ratio of 1:1.5 astrocytes to neurons; thus, this model might not be of relevance to model brain areas where astrocytes are more abundant, such as in white matter areas30.
Other protocols to develop 3D bioprinted coculture models have been published in recent years. A publication by Sullivan et al., 2021, presented a 3D bioprinted neural model using iPSC-derived neural progenitor cells, which demonstrates high post-print viability and enhancement of neuronal function compared to 2D cultures32. However, in this protocol, neural progenitor cells were used as a cell source and were maintained in culture for 4 weeks. In this protocol, commercially available iPSC-derived glutamatergic neurons and astrocytes were used. This allows a 3D network of co-cultured cells to be established in as little as 7 days; as demonstrated by neurite growth analysis, neurite outgrowth begins within 24 h and continues in a linear fashion throughout the 156 h period for which cell growth was monitored. The rapid establishment of these networks can be partly attributed to the use of glutamatergic neurons that use optimized doxycycline-inducible gene expression of NGN2, which shows expression of mature neuronal subtype markers within 7 days, even in 2D culture33. The shortening of this growth period using this technique is important to implementing models within the biopharmaceutical industry, as assay development requires rapid turnaround and development of cell models15.
In conclusion, this model shows potential for a 3D model of neurons and astrocytes, which is established quickly and conveniently for screening purposes. Future applications for this model type could be for drug discovery efforts across different CNS diseases, with the opportunity to expand to different diseases using patient or gene-edited disease iPSC lines. Furthermore, the use of doxycycline-inducible NGN2 expression iPSC-derived glutamatergic neurons allows cells to reach maturity in less time, which can be utilized for developing models of the aging brain for neurodegeneration research. This system could also be expanded through the use of additional cell types in coculture, including microglia and oligodendrocytes.
Acknowledgements
The authors would like to thank Alex Volkerling, Martin Engel, and Rachel Bleach for their assistance in developing the protocol and feedback on the manuscript.
Materials
| Name | Company | Catalog Number | Comments |
| 2-mercaptoethanol | Thermofisher | 31350010 | |
| 384-well plate | PerkinElmer | 6057300 | |
| 96-well plate | PerkinElmer | 6055300 | |
| Activator fluid F299 | Inventia Life Science | N/A | |
| Activator fluid F3 | Inventia Life Science | N/A | |
| B27 (50x) minus Vit A | Thermofisher | 12587010 | |
| Bioink fluid F261 | Inventia Life Science | N/A | |
| Bioink fluid F32 | Inventia Life Science | N/A | |
| Doxycycline hyclate | Sigma Aldrich | D5207 | |
| GlutaMAX (100x) | Thermofisher | 35050061 | |
| Goat anti-mouse IgG H&L Alexa Fluor 647 | Abcam | ab150115 | |
| Goat anti-rabbit IgG H&L Alexa Fluor 488 | Abcam | ab150077 | |
| Hoechst | Abcam | ab228551 | |
| Human BDNF Recombinant Protein | Thermofisher | PHC7074 | |
| Human NT3 Recombinant Protein | Thermofisher | PHC7036 | |
| iCell Astrocytes | Fujifilm CDI | 1434 | |
| INCell Analyser 6500HS | Molecular Devices | N/A | high content imaging system |
| Incucyte S3 | Sartorius | N/A | |
| ioGlutamatergic Neurons (Large vial) | Bit.bio | e001 | |
| Laminin (1 mg/mL) | Sigma Aldrich | L2020 | |
| Live/dead kit (Calcein-AM, Ethidium homo-dimer-1) | Invitrogen | L3224 | |
| Mouse anti-BIII tubulin NL637 conjugated | R&D systems | SC024 | |
| Neurobasal media | Thermofisher | 21103049 | |
| Normal Donkey Serum | Abcam | ab7475 | |
| NucBlue Live (Hoechst 33342) | Thermofisher | R37605 | |
| Opti-MEM | Thermofisher | 11058021 | |
| Paraformaldehyde | Sigma Aldrich | P6148 | |
| PEI 50% in H2O | Sigma Aldrich | 181978 | |
| Pierce Borate Buffer 20x | Thermofisher | 28341 | |
| Prism | GraphPad | Data analysis software | |
| Rabbit anti-ionotropic glutamatre receptor 2 (GluR2) | Abcam | ab206293 | |
| RASTRUM(TM) Bioprinter | Inventia Life Science | N/A | Bioprinter |
| RASTRUM(TM) Bioprinter Cartridges | Inventia Life Science | N/A | Bioprinter Cartridges |
| RASTRUM(TM) Targeting plate | Inventia Life Science | N/A | Targeting plate |
| Rho kinase (ROCK) inhibitor | Abcam | ab120129 | |
| Sheep anti-GFAP NL493 conjugated | R&D systems | SC024 | |
| Signals Image Artist | PerkinElmer | N/A | Image analysis platform |
| Triton X-100 | Thermofisher | HFH10 | |
| Zeiss Axio Observer | Zeiss | N/A | Inverted microscope platform |
References
- Jung, Y. L., Hwang, J., Yoo, H. S. Disease burden metrics the innovations of leading pharmaceutical companies: a global and regional comparative study. Globalization and Health. 16 (1), 80-80 (2020).
- Potkin, S. G., et al. The neurobiology of treatment-resistant schizophrenia: paths to antipsychotic resistance and a roadmap for future research. npj Schizophrenia. 6, 1 (2020).
- Zahra, W., et al., Keswani, C., et al. The Global Economic Impact of Neurodegenerative Diseases: Opportunities and Challenges. Bioeconomy for Sustainable Development. , (2019).
- Perucca, E. The pharmacological treatment of epilepsy: recent advances and future perspectives. Acta Epileptologica. 3 (1), 22 (2021).
- Nikolakopoulou, P., et al. Recent progress in translational engineered in vitro models of the central nervous system. Brain. 143 (11), 3181-3213 (2020).
- Whitehouse, C., Corbett, N., Brownlees, J. 3D models of neurodegeneration: implementation in drug discovery. Trends in Pharmacological Sciences. 44 (4), 208-221 (2023).
- Rauti, R., Renous, N., Maoz, B. M. Mimicking the brain extracellular matrix in vitro: A review of current methodologies and challenges. Israel Journal of Chemistry. 60 (12), 1141-1151 (2020).
- Fawcett, J. W., Oohashi, T., Pizzorusso, T. The roles of perineuronal nets and the perinodal extracellular matrix in neuronal function. Nature Reviews Neuroscience. 20 (8), 451-465 (2019).
- Lam, D., et al. Tissue-specific extracellular matrix accelerates the formation of neural networks and communities in a neuron-glia co-culture on a multi-electrode array. Scientific Reports. 9, 4159 (2019).
- Roll, L., Lessmann, K., Brüstle, O., Faissner, A. Cerebral organoids maintain the expression of neural stem cell-associated glycoepitopes and extracellular matrix. Cells. 11 (5), 760 (2022).
- Yan, Y., Bejoy, J., Marzano, M., Li, Y. The use of pluripotent stem cell-derived organoids to study extracellular matrix development during neural degeneration. Cells. 8 (3), 242 (2019).
- Ma, L., et al. 3D bioprinted hyaluronic acid-based cell-laden scaffold for brain microenvironment simulation. Bio-Design and Manufacturing. 3 (3), 164-174 (2020).
- Liaw, C. -. Y., Ji, S., Guvendiren, M. Engineering 3D hydrogels for personalized in vitro human tissue models. Advanced Healthcare Materials. 7 (4), 1701165 (2018).
- Ma, J., Huang, C. Composition and mechanism of three-dimensional hydrogel system in regulating stem cell fate. Tissue Engineering Part B: Reviews. 26 (6), 498-518 (2020).
- Belfiore, L., et al. Generation and analysis of 3D cell culture models for drug discovery. European Journal of Pharmaceutical Sciences. 163, 105876 (2021).
- Langhans, S. A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Frontiers in Pharmacology. 9, 6 (2018).
- Engel, M., Belfiore, L., Aghaei, B., Sutija, M. Enabling high throughput drug discovery in 3D cell cultures through a novel bioprinting workflow. SLAS Technology. 27 (1), 32-38 (2022).
- Takamura, T., et al. Influence of age on global and regional brain stiffness in young and middle-aged adults. Journal of Magnetic Resonance Imaging. 51 (3), 727-733 (2020).
- Slanzi, A., Iannoto, G., Rossi, B., Zenaro, E., Constantin, G. In vitro models of neurodegenerative diseases. Frontiers in Cell and Developmental Biology. 8, 328 (2020).
- de Souza, N. Organoid variability examined. Nature Methods. 14 (7), 655-655 (2017).
- Hernández, D., et al. Culture variabilities of human iPSC-derived cerebral organoids are a major issue for the modelling of phenotypes observed in Alzheimer's disease. Stem Cell Review and Reports. 18 (2), 718-731 (2022).
- Li, R., et al. Differentiation of human iPS cells into sensory neurons exhibits developmental stage-specific cryopreservation challenges. Frontiers in Cell and Developmental Biology. 9, 796960 (2021).
- Nishiyama, Y., et al. Safe and efficient method for cryopreservation of human induced pluripotent stem cell-derived neural stem and progenitor cells by a programmed freezer with a magnetic field. Neuroscience Research. 107, 20-29 (2016).
- Uhrig, M., Ezquer, F., Ezquer, M. Improving cell recovery: Freezing and thawing optimization of induced pluripotent stem cells. Cells. 11 (5), 799 (2022).
- Harbom, L. J., et al. The effect of rho kinase inhibition on morphological and electrophysiological maturity in iPSC-derived neurons. Cell and Tissue Research. 375 (3), 641-654 (2019).
- Koh, H. S., Yong, T., Chan, C. K., Ramakrishna, S. Enhancement of neurite outgrowth using nano-structured scaffolds coupled with laminin. Biomaterials. 29 (26), 3574-3582 (2008).
- Tarus, D., et al. Design of hyaluronic acid hydrogels to promote neurite outgrowth in three dimensions. ACS Applied Materials & Interfaces. 8 (38), 25051-25059 (2016).
- Brain Initiative Cell Census Network (BICCN). Initiative Cell Census Network (BICCN). A multimodal cell census and atlas of the mammalian primary motor cortex. Nature. 598 (7879), 86-102 (2021).
- Dauchy, S., et al. Expression and transcriptional regulation of ABC transporters and cytochromes P450 in hCMEC/D3 human cerebral microvascular endothelial cells. Biochemical Pharmacology. 77 (5), 897-909 (2009).
- Herculano-Houzel, S. The glia/neuron ratio: How it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia. 62 (9), 1377-1391 (2014).
- von Bartheld, C. S., Bahney, J., Herculano-Houzel, S. The search for true numbers of neurons and glial cells in the human brain: A review of 150 years of cell counting. The Journal of Comparative Neurology. 524 (18), 3865-3895 (2016).
- Sullivan, M. A., et al. 3D bioprinting of stem cell-derived central nervous system cells enables astrocyte growth, vasculogenesis and enhances neural differentiation/function. bioRxiv. , (2022).
- Pawlowski, M., et al. Inducible and deterministic forward programming of human pluripotent stem cells into neurons, skeletal myocytes, and oligodendrocytes. Stem Cell Reports. 8 (4), 803-812 (2017).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved