A subscription to JoVE is required to view this content. Sign in or start your free trial.
DeepOmicsAE: Representing Signaling Modules in Alzheimer's Disease with Deep Learning Analysis of Proteomics, Metabolomics, and Clinical Data
In This Article
Summary
DeepOmicsAE is a workflow centered on the application of a deep learning method (i.e., an autoencoder) to reduce the dimensionality of multi-omics data, providing a foundation for predictive models and signaling modules representing multiple layers of omics data.
Abstract
Large omics datasets are becoming increasingly available for research into human health. This paper presents DeepOmicsAE, a workflow optimized for the analysis of multi-omics datasets, including proteomics, metabolomics, and clinical data. This workflow employs a type of neural network called autoencoder, to extract a concise set of features from the high-dimensional multi-omics input data. Furthermore, the workflow provides a method to optimize the key parameters needed to implement the autoencoder. To showcase this workflow, clinical data were analyzed from a cohort of 142 individuals who were either healthy or diagnosed with Alzheimer's disease, along with the proteome and metabolome of their postmortem brain samples. The features extracted from the latent layer of the autoencoder retain the biological information that separates healthy and diseased patients. In addition, the individual extracted features represent distinct molecular signaling modules, each of which interacts uniquely with the individuals' clinical features, providing for a mean to integrate the proteomics, metabolomics, and clinical data.
Introduction
An increasingly large proportion of the population is aging and the burden of age-related diseases, such as neurodegeneration, is expected to sharply increase in the coming decades1. Alzheimer's disease is the most common type of neurodegenerative disease2. Progress in finding a treatment has been slow given our poor understanding of the fundamental molecular mechanisms driving the onset and progress of the disease. The majority of information on Alzheimer's disease is gained postmortem from the examination of brain tissue, which has made distinguishing causes and consequences a difficult task3. The Religious Orders Study/Memory and Aging Project (ROSMAP) is an ambitious effort to gain a broader understanding of neurodegeneration, which involves the study of thousands of individuals who have committed to undergo medical and psychological examinations yearly and to contribute their brains for research after their demise4. The study focuses on the transition from the normal functioning of the brain to Alzheimer's disease2. Within the project, postmortem brain samples were analyzed with a plethora of omics approaches, including genomics, epigenomics, transcriptomics, proteomics5, and metabolomics.
Omics technologies that offer functional readouts of cellular states (i.e., proteomics and metabolomics)6,7 are key to interpreting disease8,9,10,11,12, due to the direct relationship between protein and metabolite abundance and cellular activities. Proteins are the primary executors of cellular processes, while metabolites are the substrates and products for biochemical reactions. Multi-omics data analysis offers the possibility to understand the complex relationships between proteomics and metabolomics data instead of appreciating them in isolation. Multi-omics is a discipline that studies multiple layers of high-dimensional biological data, including molecular data (genome sequence and mutations, transcriptome, proteome, metabolome), clinical imaging data, and clinical features. Particularly, multi-omics data analysis aims to integrate such layers of biological data, understand their reciprocal regulation and interaction dynamics, and deliver a holistic understanding of disease onset and progression. However, methods to integrate multi-omics data remain in the early stages of development13.
Autoencoders, a type of unsupervised neural network14, are a powerful tool for multi-omics data integration. Unlike supervised neural networks, autoencoders do not map samples to specific target values (such as healthy or diseased), nor are they used to predict outcomes. One of their primary applications lies in dimensionality reduction. However, autoencoders offer several advantages over simpler dimensionality reduction methods such as principal component analysis (PCA), t-distributed stochastic neighbor embedding (tSNE), or uniform manifold approximation and projection (UMAP). Unlike PCA, autoencoders can capture non-linear relationships within the data. Unlike tSNE and UMAP, they can detect hierarchical and multi-modal relationships within the data since they rely on multiple layers of computational units each containing non linear activation functions. Therefore, they represent attractive models to capture the complexity of multi-omics data. Finally, while the primary application of PCA, tSNE, and UMAP is that of clustering the data, autoencoders compress the input data into extracted features that are well-suited for downstream predictive tasks15,16.
Briefly, neural networks comprise several layers, each containing multiple computational units or "neurons." The first and last layers are referred to as the input and output layers, respectively. Autoencoders are neural networks with an hourglass structure, consisting of an input layer, followed by one to three hidden layers and a small "latent" layer typically containing between two and six neurons. This structure's first half is known as the encoder and is combined with a decoder mirroring the encoder. The decoder ends with an output layer containing the same number of neurons as the input layer. Autoencoders take the input through the bottleneck and reconstruct it in the output layer, with the goal of generating an output that mirrors the original information as closely as possible. This is achieved by mathematically minimizing a parameter termed "reconstruction loss." The input consists of a set of features, which in the application showcased herein will be protein and metabolite abundances, and clinical characteristics (i.e., sex, education, and age at death). The latent layer contains a compressed and information-rich representation of the input, which can be used for subsequent applications such as predictive models17,18.
This protocol presents a workflow, DeepOmicsAE, which involves: 1) preprocessing of proteomics, metabolomics, and clinical data (i.e., normalization, scaling, outlier removal) to obtain data with a consistent scale for machine learning analysis; 2) selecting appropriate autoencoder input features, since feature overload may obscure relevant disease patterns; 3) optimizing and training the autoencoder, including determining the optimal number of proteins and metabolites to select, and of neurons for the latent layer; 4) extracting features from the latent layer; and 5) utilizing the extracted features for biological interpretation by identifying molecular signaling modules and their relationship with clinical features.
This protocol aims to be simple and applicable by biologists with limited computational experience who have a basic understanding of programming with Python. The protocol focuses on analyzing multi-omics data, including proteomics, metabolomics, and clinical features, but its use can be extended to other types of molecular expression data, including transcriptomics. One important novel application introduced by this protocol is mapping the importance scores of original features onto individual neurons in the latent layer. As a result, each neuron in the latent layer represents a signaling module, detailing the interactions between specific molecular alterations and the patients' clinical characteristics. Biological interpretation of the molecular signaling modules is obtained by using MetaboAnalyst, a publicly available tool that integrates gene/protein and metabolite data to derive enriched metabolic and cell signaling pathways17.
Protocol
NOTE: The data used here were ROSMAP data downloaded from the AD Knowledge portal. Informed consent is not needed to download and reuse the data. The protocol presented herein utilizes deep learning to analyze multi-omics data and identify signaling modules that distinguish specific patient or sample groups based, for example, on their diagnosis. The protocol also delivers a small set of extracted features that summarize the original large-scale data and can be used for further analysis such as training a predictive model using machine learning algorithms (Figure 1). Refer to Supplemental File 1 and the Table of Materials for information regarding accessing the code and setting up the computational environment prior to performing the protocol. The methods should be performed following the order specified below.
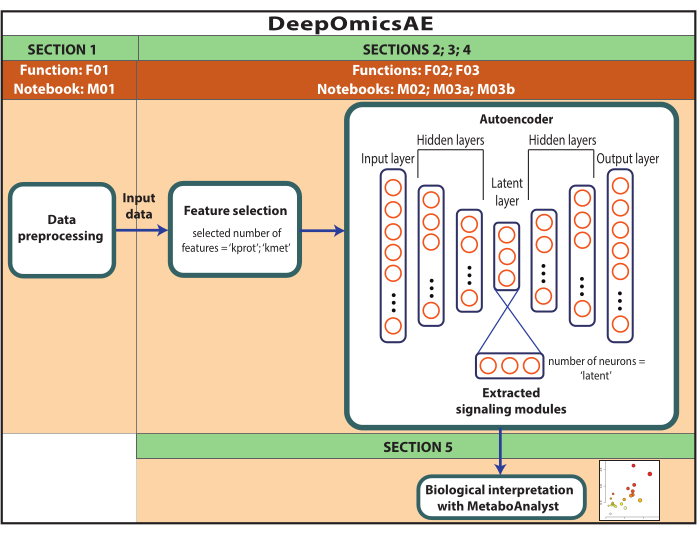
Figure 1: Schematic of the DeepOmicsAE workflow. Schematic representation of the workflow for analyzing multi-omics data using the workflow. In the autoencoder depiction, rectangles represent layers of the neural network and circles represent neurons within layers. Please click here to view a larger version of this figure.
1. Data preprocessing
NOTE: The goal of this section is to preprocess the data, including handling missing data; normalizing and scaling proteomic, metabolomic expression, and clinical data; and removing outliers. The protocol is designed for a dataset that includes proteomics data expressed as log2(ratio); metabolomics data expressed as fold change; and clinical features including continuous and categorical features. The patients or samples should be grouped based on diagnosis or other similar parameters. Samples or patients should be across the rows and features across the columns.
- To start a new instance of Jupyter Notebook in the browser, open a new terminal window, type the following and press Enter.
jupyter notebook - In the Jupyter home page on the browser, click on the notebook M01 - expression data pre-processing.ipynb to open it in a new tab (Supplemental File 2, Step 1.1).
- In the second cell of the notebook, type the name of the dataset file in place of your_dataset_name.csv.
- In the last cell of the notebook, type the desired name of the output data file in place of M01_output_data.csv.
- In the fifth cell of the notebook, specify the position of the columns for each data type as follows: proteomics data (cols_prot), metabolomics data (cols_met), continuous clinical data (e.g., age) (cols_clin_con), binary clinical data (e.g., sex) (cols_clin_bin). Enter the first column index for each data type in place of col_start and the last columns index in place of col_end; for example: cols_prot = slice(0, 8817). Ensure that the values specified in the slice objects correspond to the first and last columns indexes corresponding to each data type. Use the command in the fourth cell of the same notebook (df.iloc[:, :]) to determine the start and end position for each data type (Supplemental File 2, Step 1.2).
- Select Cell | Run all from the menu bar in Jupyter to create the output data file in the specified folder (Supplemental File 2, Step 1.3).
NOTE: These data will be used as input for the protocols described in sections 2, 3, or 4.
2. Custom optimization of the workflow (optional)
NOTE: Section 2 is optional because it is computer-intensive. Users should skip directly to section 4 if they decide to not perform section 2. This protocol will guide the user through optimizing the workflow in an automated manner. Specifically, the method identifies the parameters that deliver the best performance of the autoencoder in terms of generating extracted features that separate the sample groups well. The optimized parameters generated as an output include the number of features to use for feature selection (k_prot and k_met) and the number of neurons in the autoencoder latent layer (latent). These parameters can then be used in the protocol described in section 3 to generate the model.
- On the Jupyter home page on the browser, click on the notebook M02 - DeepOmicsAE model optimization.ipynb to open it in a new tab (Supplemental File 2, Step 2.1).
- In the second cell of the notebook, type the name of the input file in place of M01_output_data.csv. The input to this function is the output data from section 1.
- In the fifth cell of the notebook, specify the position of the columns for each data type as follows: proteomics data (cols_X_prot), metabolomics data (cols_X_met), clinical data (cols_clin; includes all the clinical data), all molecular expression data, including proteomics and metabolomics data (cols_X_expr). Enter the first column index for each data type in place of col_start and the last columns index in place of col_end; for example, cols_prot = slice(0, 8817). Ensure that the values specified in the slice objects correspond to the first and last columns index corresponding to each data type, and use the commands in the third and fourth cells of the notebook to explore the data and determine the start and end positions for each data type. Specify the name of the column containing the target variable in place of y_column_name as y_label (Supplemental File 2, Step 2.2).
NOTE: The values of the indexes specified in cols_X_prot, cols_X_met, cols_clin, and cols_X_expr will be different from those used in section 1 due to the reshaping of the dataframe occurring during data preprocessing. - In the sixth cell of the notebook, specify how many optimization rounds to perform by assigning a value to n_comb. Times for processing are approximately 4-5 min for 10 rounds; 20 min for 50 rounds, and 40 min for 100 rounds (Supplemental File 2, Step 2.3).
- Select Cell | Run all from the menu bar in Jupyter.
NOTE: The output variables kprot, kmet, and latent will be stored and can be accessed from the other notebooks, which will be used to continue the analytical workflow. The plot AE_optimization_plot.pdf will be generated and saved in the local folder (Figure 2).
3. Workflow implementation with custom-optimized parameters
NOTE: Perform this protocol only following method optimization (section 2). If users choose to not perform method optimization, skip directly to section 4. This protocol will guide the user through generating a model using the custom-optimized parameters derived from section 2. The autoencoder will 1) generate a set of extracted features that recapitulate the original data and 2) identify the important features driving each neuron in the latent layer, effectively representing unique signaling modules. The signaling modules will be interpreted using the protocol provided in section 5.
- On the Jupyter home page on the browser, click on the notebook M03a - DeepOmicsAE implementation with custom-optimized parameters.ipynb to open it in a new tab (Supplemental File 2, Step 3.1).
- In the second cell of the notebook, type the name of the input file in place of M01_output_data.csv. The input to this function is the output data from section 1.
- In the fifth cell of the notebook, specify the position of the columns for each data type as follows: proteomics data (cols_prot), metabolomics data (cols_met), clinical data (cols_clin; includes all of the clinical data). Enter the first column index for each data type in place of col_start and the last columns index in place of col_end; for example: cols_prot = slice(0, 8817). Ensure that the values specified in the slice objects correspond to the first and last columns indexes corresponding to each data type, and use the commands in the third and fourth cells of the notebook to explore the data and determine the start and end positions for each data type. Specify the name of the column containing the target variable (e.g., 0 or 1, corresponding to healthy or diseased) in place of y_column_name as y_label.
NOTE: The value of the indexes specified in cols_X_prot, cols_X_met, cols_clin, and cols_X_expr will be different from those used in section 1 due to the reshaping of the dataframe occurring during data preprocessing. - Select Cell | Run all from the menu bar in Jupyter to generate and save the plots PCA_initial_data.pdf, PCA_extracted_features.pdf, and distribution_important_feature_scores.pdf in the local folder (Figure 3 and Supplemental Figure S1). Additionally, lists of important features for each identified signaling module will be stored in text files in the local folder, named module_n.txt, where n will be substituted by the module number.
4. Workflow implementation with preset parameters
- Refer to section 3 for detailed instructions on how to run this method (Supplemental File 2, Step 4.1). The only difference between these two protocols is that the parameters kprot, kmet, and latent (in the seventh cell of the notebook) are mathematically derived based on the results of the optimization performed as shown in Figure 2.
NOTE: If section 4 delivers a poor separation of the sample groups, indicating suboptimal model performance, it is recommended to execute model optimization (section 2) using at least 15 iterations, and if possible, up to 50.
5. Biological interpretation using MetaboAnalyst
- Open the browser and navigate to the link below to access the Joint Pathway Analysis functionality on the MetaboAnalyst website: https://www.metaboanalyst.ca/MetaboAnalyst/upload/JointUploadView.xhtml.
- Access the folder where the output files from Method 3 or Method 4 were saved and open the text files module_n.txt for each signaling module n generated by Method 3 or by Method 4.
- Locate the proteins in the text files and copy them.
- Paste the list of proteins into the window Genes/proteins with optional fold changes in the MetaboAnalyst web page.
- Repeat the above step for metabolites and paste them into the window Compound list with optional fold changes on the same web page.
- Select the appropriate organism and ID type, then click Submit at the bottom of the page (Supplemental File 2, Step 5.1).
NOTE: Ensure that the identifiers are recognized by MetaboAnalyst. Recognized identifiers include Entrez ID, official gene symbols, and Uniprot ID for proteins; compound name, HMDB ID, and KEGG ID for metabolites. If the identifiers are other than these types, appropriate conversion is necessary prior to the analysis. - On the following page, check the ID mapping before clicking Proceed to verify that the identifiers are being recognized.
- In the Parameter Setting page, select Metabolic pathways (integrated) or All pathways (integrated) to visualize respectively the contribution of the input to metabolic pathways only or to all signaling pathways (Supplemental File 2, Step 5.2). In the Algorithm selection panel, choose Enrichment analysis: Hypergeometric test, Topology measure: Degree centrality, and Integration method: Combine p values (pathway-level). Click on Submit at the bottom of the page.
- The last page is the Result View, which presents the results of the enrichment analysis. Enriched pathways are plotted based on their impact and significance, and the list of pathways is also provided in tabular format.
Results
To showcase the protocol, we analyzed a dataset comprising the proteome, metabolome, and clinical information derived from postmortem brains of 142 individuals who were either healthy or diagnosed with Alzheimer's disease.
After performing the protocol section 1 to preprocess the data, the dataset included 6,497 proteins, 443 metabolites, and three clinical features (sex, age at death, and education). The target feature is clinical consensus diagnosis of cognitive status at ti...
Discussion
The structure of the dataset is critical to the success of the protocol and should be carefully checked. The data should be formatted as indicated in protocol section 1. The correct assignment of column positions is also critical to the success of the method. Proteomics and metabolomics data are preprocessed differently and feature selection is conducted separately due to the different nature of the data. Therefore, it is critical to assign column positions correctly in protocol steps 1.5, 2.3, and 3.3.
Disclosures
The author declares that they have no conflicts of interest.
Acknowledgements
This work was supported by NIH grant CA201402 and the Cornell Center for Vertebrate Genomics (CVG) Distinguished Scholar Award. The results published here are in whole or in part based on data obtained from the AD Knowledge Portal (https://adknowledgeportal.org). Study data were provided through the Accelerating Medicine Partnership for AD (U01AG046161 and U01AG061357) based on samples provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago. Data collection was supported through funding by NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG30146, R01AG36836, U01AG32984, U01AG46152, the Illinois Department of Public Health, and the Translational Genomics Research Institute. The metabolomics dataset was generated at Metabolon and preprocessed by the ADMC.
Materials
| Name | Company | Catalog Number | Comments |
| Computer | Apple | Mac Studio | Apple M1 Ultra with 20-core CPU, 48-core GPU, 32-core Neural Engine; 64 GB unified memory |
| Conda v23.3.1 | Anaconda, Inc. | N/A | package management system and environment manager |
| conda environment DeepOmicsAE | N/A | DeepOmicsAE_env.yml | contains packages necessary to run the worflow |
| github repository DeepOmicsAE | Microsoft | https://github.com/elepan84/DeepOmicsAE/ | provides scripts, Jupyter notebooks, and the conda environment file |
| Jupyter notebook v6.5.4 | Project Jupyter | N/A | a platform for interactive data science and scientific computing |
| DT01-metabolomics data | N/A | ROSMAP_Metabolon_HD4_Brain 514_assay_data.csv | This data was used to generate the Results reported in the article. Specifically, DT01-DT04 were merged by matching them based on the individualID. The column final consensus diagnosis (cogdx) was filtered to keep only patients classified as healthy or AD. Climnical features were filtered to keep the following: age at death, sex and education. Finally, age reported as 90+ was set to 91, then the age column was transformed to float64. The data is available at https://adknowledgeportal.synapse.org |
| DT02-TMT proteomics data | N/A | C2.median_polish_corrected_log2 (abundanceRatioCenteredOn MedianOfBatchMediansPer Protein)-8817x400.csv | |
| DT03-clinical data | N/A | ROSMAP_clinical.csv | |
| DT04-biospecimen metadata | N/A | ROSMAP_biospecimen_metadata .csv | |
| Python 3.11.3 | Python Software Foundation | N/A | programming language |
References
- Hou, Y., et al. Ageing as a risk factor for neurodegenerative disease. Nature Reviews Neurology. 15 (10), 565-581 (2019).
- Scheltens, P., et al. Alzheimer’s disease. The Lancet. 397 (10284), 1577-1590 (2021).
- Breijyeh, Z., Karaman, R. Comprehensive review on Alzheimer’s disease: causes and treatment. Molecules. 25 (24), 5789 (2020).
- Bennett, D. A., et al. Religious Orders Study and Rush Memory and Aging Project. Journal of Alzheimer’s Disease. 64 (s1), S161-S189 (2018).
- Higginbotham, L., et al. Integrated proteomics reveals brain-based cerebrospinal fluid biomarkers in asymptomatic and symptomatic Alzheimer’s disease. Science Advances. 6 (43), eaaz9360 (2020).
- Aebersold, R., et al. How many human proteoforms are there. Nature Chemical Biology. 14 (3), 206-214 (2018).
- Nusinow, D. P., et al. Quantitative proteomics of the cancer cell line encyclopedia. Cell. 180 (2), 387-402.e16 (2020).
- Johnson, E. C. B., et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nature Medicine. 26 (5), 769-780 (2020).
- Geyer, P. E., et al. Plasma proteome profiling to assess human health and disease. Cell Systems. 2 (3), 185-195 (2016).
- Akbani, R., et al. A pan-cancer proteomic perspective on the cancer genome atlas. Nature Communications. 5, 3887 (2014).
- Panizza, E., et al. Proteomic analysis reveals microvesicles containing NAMPT as mediators of radioresistance in glioma. Life Science Alliance. 6 (6), e202201680 (2023).
- Li, Z., Vacanti, N. M. A tale of three proteomes: visualizing protein and transcript abundance relationships in the Breast Cancer Proteome Portal. Journal of Proteome Research. 22 (8), 2727-2733 (2023).
- Subramanian, I., Verma, S., Kumar, S., Jere, A., Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinformatics and Biology Insights. 14, 1177932219899051 (2020).
- Wang, Y., Yao, H., Zhao, S. Auto-encoder based dimensionality reduction. Neurocomputing. 184, 232-242 (2016).
- Mulla, F. R., Gupta, A. K. A review paper on dimensionality reduction techniques. Journal of Pharmaceutical Negative Results. 13, 1263-1272 (2022).
- Shrestha, A., Mahmood, A. Review of deep learning algorithms and architectures. IEEE Access. 7, 53040-53065 (2019).
- Pang, Z., et al. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Research. 49 (W1), W388-W396 (2021).
- Hinton, G. E., Salakhutdinov, R. R. Reducing the dimensionality of data with neural networks. Science. 313 (5786), 504-507 (2006).
- Altmann, A., Toloşi, L., Sander, O., Lengauer, T. Permutation importance: a corrected feature importance measure. Bioinformatics. 26 (10), 1340-1347 (2010).
- Lundberg, S. M., Allen, P. G., Lee, S. -. I. A unified approach to interpreting model predictions. , (2017).
- Wang, Q., et al. Deep learning-based brain transcriptomic signatures associated with the neuropathological and clinical severity of Alzheimer’s disease. Brain Communications. 4 (1), (2021).
- Beebe-Wang, N., et al. Unified AI framework to uncover deep interrelationships between gene expression and Alzheimer’s disease neuropathologies. Nature Communications. 12 (1), 5369 (2021).
- Camandola, S., Mattson, M. P. Brain metabolism in health, aging, and neurodegeneration. The EMBO Journal. 36 (11), 1474-1492 (2017).
- Verdin, E. NAD+ in aging, metabolism, and neurodegeneration. Science. 350 (6265), 1208-1213 (2015).
- Platten, M., Nollen, E. A. A., Röhrig, U. F., Fallarino, F., Opitz, C. A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nature Reviews Drug Discovery. 18 (5), 379-401 (2019).
- Wang, R., Reddy, P. H. Role of glutamate and NMDA receptors in Alzheimer’s disease. Journal of Alzheimer’s Disease. 57 (4), 1041-1048 (2017).
- Skaper, S. D., Facci, L., Zusso, M., Giusti, P. Synaptic plasticity, dementia and Alzheimer disease. CNS & Neurological Disorders - Drug Targets. 16 (3), 220-233 (2017).
- Reisberg, B., et al. Memantine in moderate-to-severe Alzheimer’s disease. New England Journal of Medicine. 348 (14), 1333-1341 (2003).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved