Multi-Gene Single Nucleotide Polymorphism Detection in Gastric Cancer Based on Ion Semiconductor Sequencing Platform
In This Article
Summary
This protocol proposes holistic laboratory procedures required to detect single nucleotide polymorphisms in gastric cancer samples based on an ion semiconductor sequencing platform. The target sequences, ligating adapters, library amplification and purification, and quality control criteria are also described in detail.
Abstract
Gastric cancer is a common heterogeneous tumor. Most patients have advanced gastric cancer at the time of diagnosis and often need chemotherapy. Although 5-fluorouracil (5-FU) is widely used for treatment, its therapeutic sensitivity and drug tolerance still need to be determined, which emphasizes the importance of individualized administration. Pharmacogenetics can guide the clinical implementation of individualized treatment. Single nucleotide polymorphisms (SNPs), as a genetic marker, contribute to the selection of appropriate chemotherapy regimens and dosages. Some SNPs are associated with folate metabolism, the therapeutic target of 5-FU. Methylenetetrahydrofolate reductase (MTHFR) rs1801131 and rs1801133, dihydrofolate reductase (DHFR) rs1650697 and rs442767, methionine synthase (MTR) rs1805087, gamma-glutamyl hydrolase (GGH) rs11545078 and solute carrier family 19 member 1 (SLC19A1) rs1051298 have been investigated in different kinds of cancers and antifolate antitumor drugs, which have potential forecasting and guiding significance for application of 5-FU. The ion torrent next-generation semiconductor sequencing technology can rapidly detect gastric cancer-related SNPs. Each time a base is extended in a DNA chain, an H+ will be released, causing local pH changes. The ionic sensor detects pH changes and converts chemical signals into digital signals, achieving sequencing by synthesis. This technique has low sample requirement, simple operation, low cost, and fast sequencing speed, which is beneficial for guiding individualized chemotherapy by SNPs.
Introduction
Gastric cancer is a heavy burden in the field of global public health. According to the Global Cancer Statistics 2020, published by the International Agency for Research on Cancer (IARC), gastric cancer is the fifth most diagnosed cancer and the fourth leading cause of cancer-related death. Worldwide, the incidence of age-standardized rate in East Asia is the highest in both males and females1. The occurrence of gastric cancer is insidious, which means that patients often do not have any obvious and specific symptoms in the early stage. Among all gastric cancer patients, in countries without routine screening, 80%-90% of patients are either diagnosed at an advanced stage when the tumor cannot be operated on or relapses within 5 years after the operation2.
For advanced or metastatic gastric cancer, chemotherapy is the main treatment, which can improve the survival rate and quality of life of patients. For the initial therapy of patients with metastatic gastric cancer, a platinum-fluoropyrimidine regimen is the principal choice for a first-line chemotherapeutic regimen3. Fluoropyrimidine mainly includes 5-fluorouracil (5-FU) and oral fluoropyrimidine derivatives, such as capecitabine and tegafur. The main target of 5-FU is folate metabolism-related enzymes, which inhibit DNA synthesis and slow the growth of tumor tissue. Adverse drug reactions limit their utility, with diarrhea, mucositis, myelosuppression, and hand-foot syndrome among the most frequent side effects. It has been reported that therapeutic response and adverse drug reactions are closely related to factors in the folate metabolic pathway. Notably, the homozygous mutation of rs1801131 has been identified as an indicator for hand-foot syndrome (p = 4.1 x 10-6, OR =9.99, 95% CI: 3.84-27.8)4. Although fluoropyrimidines are extensively used in anticancer chemotherapy, their chemoresistance is a common emergency, causing therapeutic failure in gastric cancer treatment. For instance, the overall response rate is only 10%-15% among patients with advanced colorectal cancer treated with 5-FU only5. Also, fluoropyrimidines have toxicity that cannot be ignored. The toxicity reactions induced by 5-FU mainly include diarrhea, hand-foot syndrome, stomatitis, neutropenia, thrombocytopenia, neurotoxicity, and even death6. Severe treatment-related toxicity occurs in 10%-30% of patients treated with fluoropyrimidines, and fatal toxicity occurs in 0.5%-1% of these patients7.
A quality-of-life study of patients with advanced gastric cancer found that the response rate was less than 50% for those who received 5-FU-based chemotherapy8. Therefore, understanding the factors related to the sensitivity of 5-FU-based chemotherapy is particularly important for precise treatment to maximize the response rate and effectiveness while minimizing toxicity. Considering that 5-FU is closely related to folate metabolism, the genetic variants of enzymes in folate metabolic pathway may be one of the factors. About 90% of human sequence variation is attributed to single base mutations in DNA, known as single nucleotide polymorphisms (SNPs)9. When SNPs change the enzyme properties of folate metabolism, it may lead to individual differences in efficacy, toxicity and chemoresistance to fluorouracil in gastric cancer patients.
Methylenetetrahydrofolate reductase (MTHFR) is mainly used to convert 5,10-methylenetetrahydrofolate (5,10-MTHF) into 5-methyltetrahydrofolate. 5-FdUMP,a metabolite of 5-FU, forms inactive ternary with 5,10-MTHF and thymidylate synthase (TS), inhibiting activity of TS and leading to deficiency of dTMP10. Accumulation of 5,10-MTHF can enhance the inhibition effect of 5-FU on TS, which is correlated with the activity of MTHFR. MTHFR rs1801131 and rs1801133 are the most common polymorphisms, which are related to lower enzyme activity (a decrease of 75% for rs1801133 and 30% for rs181131) and accumulation of 5,10-MTHF11.
Dihydrofolate reductase (DHFR) is the key enzyme in folate metabolism and DNA synthesis. DHFR reduces dihydrofolate, using NADPH, to tetrahydrofolate (THF) which is used to carry one-carbon unit. SNPs of DHFR may affect its expression, change the activity and abundance of THF, and further affect folate metabolism and sensitivity of 5-FU. DHFR rs1650697 point mutation occurs in the major promoter of DHFR gene, which increases the DHFR expression12. A study found that rs442767 is associated with the efficacy and toxicity of antifolate antitumor drugs, such as pemetrexed and methotrexate. Regarding the SNP rs442767, a GT genotype signifies the inheritance of a G allele and a T allele at the same locus on the homologous chromosomes from each parent. Similarly, the genotypes GG and TT denote the inheritance of either two G alleles or two T alleles, correspondingly. Compared with genotype GT+TT, GG is related to decreased event-free survival and increased risk13. This suggests that rs442767 may lead to certain potential influence on 5-FU.
Methionine synthase (MTR) catalyzes the re-methylation of homocysteine to methionine, which plays an important role in folate metabolism. MTR rs1805087 is the most common polymorphism of MTR gene. MTR rs1805087 substitutes glycine for aspartic acid at the potentially functional site of protein, which may decrease the activity of MTR. Subjects with the G allele had increased plasma folate level and decreased plasma homocysteine level14. On the contrary, a study showed that rs1805087 has no statistically significant association with the efficacy of 5-FU. But this study focused on colorectal cancer and the sample size was small. The relationship between rs1805087 and the efficacy of 5-FU in gastric cancer patients remains to be explored15.
Gamma-glutamyl hydrolase (GGH) is a lysosomal enzyme that regulates intracellular folate concentrations. Pteroylglutamic acid is the synonym of folic acid that is made up of pterin, p-aminomethylbenzoic acid and glutamic acid. There are two forms of folic acid in organisms, monoglutamate folate and polyglutamated folate. THF-polyglutamate is enzymatically converted to monoglutamic folate by GGH, successively releasing either mono-Glutamate (mono-Glu) or di-Glutamate (di-Glu)16. A study about GGH expression in patients with locally advanced gastric cancer, showed high GGH expression could reduce 5,10-MTHF and TS, which means that only a small dosage of 5-FU is needed to achieve the TS inhibitory effect in these patients17. GG is a prognostic biomarker in patients with locally advanced gastric cancer treated with postoperative adjuvant chemotherapy with S-1, a prodrug of 5-FU, and plays an important role in maintaining intracellular homeostasis of folic acid18. GGH rs11545078 is missense variant and alters Thr-127 to Ile-127. A study focusing on substrate specificity of GGH suggests that rs11545078, compared to wild-type, results in higher Km and lower catalytic efficiency for methotrexate and the structure is similar to folic acid19. Together, exploring the relationship between rs11545078 and clinical outcomes of 5-FU is a promising strategy to understand drug resistance.
Solute carrier family 19 member 1 (SLC19A1), also named reduced folate transporter, is a typical facilitative transmembrane protein that imports reduced folates that mammalian cells lack the ability to de novo synthesize, which is recognized to estimate the response of tumor to 5-FU20. However, only a few studies have been performed regarding the association between 5-FU and SLC19A1 polymorphism21. In patients with non-small cell lung cancer receiving pemetrexed which is a folate analog, the rs1051298 on the SLC19A1 gene contributed to increase the risk of all adverse drug reaction and decrease overall survival22,23. SLC19A1 rs1051298, a 3'untranslated region variant about folate metabolism, may help explain some of the individual differences about 5-FU therapy. Here the aim is to evaluate the association between rs1051298 and 5-FU resistance among patients with gastric cancer.
A kit, based on semiconductor sequencing, is used for qualitative gene detection in vitro (Figure 1), which can detect 7 selected SNPs in 5 genes, the rs1801131 and rs1801133 mutations of MTHFR gene, rs1650697 and rs442767 mutations of DHFR gene, rs1805087 mutation of MTR gene, rs11545078 mutation of GGH gene and rs1051298 of SLC19A1 gene in tumor tissue samples of gastric cancer patients. Firstly, the sample nucleic acid was extracted, and the target fragment was specifically amplified by PCR. A universal sequencing adapter was added at both ends of the DNA fragment to construct a library that can be used for sequencing. Then amplification of the sequencing library by PCR to form a sequencing template was done. The positive template was enriched to meet the sequencing requirements. Using the semiconductor sequencing system, by fixing the DNA strand in the tiny hole of the semiconductor chip. DNA polymerase takes the single-stranded DNA as the template and synthesizes the complementary DNA strand according to the principle of complementary base pairing. Each time a base is extended in a DNA chain, a proton will be released, causing local pH changes. An ionic sensor detects pH changes and converts chemical signals into digital signals, so that bases can be interpreted in real time, and finally the base sequence of each DNA segment can be obtained. Bioinformatics analysis was used to match these sequences to the reference map of the human genome. When gastric cancer related genes mutate, their corresponding DNA base sequences will change, so as to obtain the mutation information of related genes.
The result can display gene mutation status and provide reference for clinicians to select appropriate types and dosages of chemotherapy drugs and predict the drug resistance for gastric cancer patients. However, the test results are only for clinical reference and are not recommended to be the only basis for individualized treatment of patients. Clinicians should make a comprehensive judgment based on the patient's condition, drug indications, treatment reactions and other laboratory test indicators.
Protocol
All protocols and the tissue samples of gastric cancer (step 1), obtained from upper gastrointestinal endoscopy or surgery, used in this study were reviewed and approved by medical ethics committee on July 24th, 2023 (NFEC-2023-298). Besides, this protocol is designed exclusively to illustrate multi-gene detection in gastric cancer without engaging in cohort comparisons, therefore it imposes no specific criteria for including or excluding patients. The patients/participants provided their written informed consent to participate in this study.
1. Preparation of embedded blocks of gastric cancer
- Configure the tissue embedding system, comprising of a paraffin reservoir and dispenser, alongside warm and cold plates, following the prescribed operational guidelines.
- Extract the prepared tissue for embedding from the dehydrator and deposit it into the embedding center's storage slot.
- Select fitting embedding molds based on the tissue size, pour enough amount of melted paraffin to cover the tissue, and then position the mold on the warm plate.
- Retrieve the tissue from the cassettes and place the gastric mucosa perpendicular to the base of the embedding mold to ensure the cross-sectional plane cuts through all tissue layers. Align the tissue in the mold.
- Set the cassette on the top of the mold, followed by a secondary pouring of paraffin, filling the mold.
- Place the embedded mold onto the cold plate, and following the paraffin's solidification, snap off the embedded blocks from the molds.
2. Extraction of nucleic acid from samples
- Use nucleic acid extraction and purification kits to extract nucleic acids from paraffin-embedded tissue samples.
- Use a nucleic acid quantifier to quantify the extracted nucleic acid. Nucleic acid concentration is recommended to be greater than 2 ng/µL.
NOTE: Materials used here are available in the Table of Materials.
3. Preparation for sequencing library
- Preparation before experiment
- Turn ON the UV lamp in the ultra-clean workbench, sterilize for 30 min. Then, turn OFF the UV lamp and turn ON the fan to ventilate for 10 min.
- Take the nucleic acid out of the -20 ± 5 °C. refrigerator, check and record the sample ID, nucleic acid barcode and specific label tag number assigned to the sample. Put it on the centrifuge tube rack for dissolution at room temperature (RT) after verification, and centrifuge it for 10 s, with a fixed 2,500 x g for standby.
- PCR amplification of target fragment
- Take the fragment capture reaction solution, amplification primer of gastric cancer and fusion amplification primer from the custom-made gastric cancer multi-gene joint detection kit and put them on ice for melting. Shake and mix them after melting, centrifugate them for 10 s, and prepare nuclease-free water. Detailed primer information is shown in Table 1.
NOTE: The kit is not commercially available yet, contact authotrs for further details. - Prepare a 0.2 mL PCR reaction tube, add reagents into the tube in turn according to Supplementary Table 1, vortex and mix for 5 s, and centrifugate instantaneously for 10 s, with a fixed 2,500 x g, so that there are no obvious drops on the tube wall and cover. RNA samples shall be reverse transcribed into cDNA and then used for subsequent library construction. cDNA products shall be added into the tubes in turn according to Supplementary Table 2.
- Place each reaction tube on the thermal cycler. For the DNA samples, run the amplification program as detailed in Table 2. For cDNA products, run the amplification program as detailed in Table 3.
- Take the fragment capture reaction solution, amplification primer of gastric cancer and fusion amplification primer from the custom-made gastric cancer multi-gene joint detection kit and put them on ice for melting. Shake and mix them after melting, centrifugate them for 10 s, and prepare nuclease-free water. Detailed primer information is shown in Table 1.
- Digestion of primer sequence
- Take out the primer digestion enzyme and put it on ice for melting. After amplification, take out the above reaction tubes, add 2 µL of primer digestion enzyme to each tube, and ensure the total volume is 22 µL.
- Vortex and mix the reaction solution in the PCR tube and centrifuge instantaneously for 10 s, with a fixed 2,500 x g.
- Put each reaction tube on the thermal cycler and run the program as detailed in Table 4.
- Ligation of adapter
- Put the adapter connecting reagents on ice to dissolve. Prepare 1.5 mL microcentrifuge tube, and then mix each component according to Table 5, and mark it as adapter mixture X.
NOTE: The adapter connecting reagents include P1 adapters and specific adapters X, numbered 1 through 48. These are designed to uniquely tag various samples. When adding a specific adapter, only open one tube at a time to prevent cross contamination of the specific adapter. The diluted specific adapter mixture can be stored at -20 ± 5 °C for standby. In sequencing workflows, samples are not processed individually, rather, multiple samples, each tagged with a specific adapter, are combined into a unified library, for sequencing. This method enables subsequent differentiation of samples based on their specific adapter sequences. - Take out the digested primer product (22 µL) from the thermal cycler. Add reagents into the tube in turn according to Table 6, vortex and mix for 5 s. Centrifuge at low speed for 10 s, with a fixed 2,500 x g, so that there is no obvious drop on the wall and cover of the tube.
- Place the reaction tube on the thermal cycler. For the DNA samples, run the amplification program as detailed in Table 7. For cDNA products, run the amplification program as detailed in Table 8.
- Put the adapter connecting reagents on ice to dissolve. Prepare 1.5 mL microcentrifuge tube, and then mix each component according to Table 5, and mark it as adapter mixture X.
- Purification of ligation product and amplification
- Take the DNA purification magnetic beads out of the 2-8 °C refrigerator in advance, vortex them evenly and centrifugate them instantaneously for 10 s, with a fixed 2,500 x g. Equilibrate the magnetic beads at RT for 30 min.
- Prepare a 1.5 mL low adsorption microcentrifuge tube and transfer the product of ligation reaction to the corresponding tube.
- Add 45 µL of DNA purified magnetic beads into each tube, vortex and mix well, centrifugate instantaneously for 10 s, and incubate at RT for 5 min.
- Place the tube on a magnetic rack for 3 min. Discard the supernatant and avoid pipetting the beads out.
- Transfer 300 µL of newly prepared 75% ethanol into the microcentrifuge tube, and gently rotate the tubes 4x at 180°. After the solution is clear, quickly discard the supernatant. Avoid pipetting the beads out. Repeat the wash procedure 1x more.
- Remove the 1.5 mL microcentrifuge tubes from the magnet rack, and centrifuge briefly (10 s, with a fixed 2,500 x g). Place the tubes back into the magnet rack and pipette out the remaining liquid. Ensure that there is no residual liquid on the tube wall.
- Open the cap of each 1.5 mL microcentrifuge tube and dry the beads at RT for 5 min. Pay attention to the dry-wet condition of the beads. After the magnetic beads are dried, check for no water stains on the surface. Extend the drying time appropriately if the magnetic beads are too wet. Close the lid immediately if any cracks emerge on the beads and continue the next step to amplify and purify the library.
- Amplification and purification of library
- Put PCR related reagents on ice for dissolution in advance, vortex them, and centrifuge them for 10 s. Remove the 1.5 mL microcentrifuge tube from the magnet rack, pipette the PCR regents into the tube according to Table 9 and close the cap and vortex for 5s. Centrifuge briefly (10 s) to get no obvious liquid drop on the tube wall and cover.
- Transfer the product above into a new PCR tube. Incubate the sample on a thermal cycler. For the DNA samples, run the amplification program as detailed in Table 10. For cDNA products, run the amplification program as detailed in Table 11.
- Prepare a new 1.5 mL low adsorption microcentrifuge tube. Centrifuge the PCR tube for 10 s after incubation. Transfer the product from the PCR tube into the EP tube.
- Add 25 µL of DNA purified magnetic beads into each tube. Vortex and mix well, centrifuge at low speed and incubate at RT for 5 min.
- Place the tubes on a magnetic rack for 3 min and wait till the solution becomes clear. Transfer the supernatant into new microcentrifuge tubes. Avoid pipetting the beads out.
- Add 60 µL of DNA purified magnetic beads into each tube. Vortex and mix well, centrifuge at low speed and incubate at RT for 5 min.
- Place the tubes on a magnetic rack for 3 min and wait till the solution becomes clear. Discard the supernatant. Avoid pipetting the beads out.
- Transfer 300 µL of the newly prepared 75% ethanol into the tubes, and gently rotate the tubes 4x at a 180°. After the solution is clear, quickly pipette and discard the supernatant. Avoid pipetting the beads out. Repeat the wash procedure 1x.
- Remove the 1.5 mL microcentrifuge tubes from the magnetic rack, and centrifuge briefly (10 s). Insert the tubes back into the magnetic rack and pipette out the remaining liquid. Ensure that there is no residual liquid on the tube wall.
- Open the caps of 1.5 mL tubes and dry the beads at RT for 5 min. Pay attention to the dry-wet condition of the beads. After the magnetic beads are dried, there is no water stain on the surface. Extend the drying time appropriately if the magnetic beads are too wet. Close the lid immediately if any cracks emerge on the beads.
- Pipette 50 µL of eluent into the tubes, vortex and mix well. Centrifuge briefly (5 s, with a fixed 2,500 x g) and place the tubes at RT for 5 min.
- Place the tubes on a magnetic rack for 3 min and wait till the solution becomes clear. Carefully remove the liquid into a new tube and mark the library name.
- Store the library in a refrigerator at 2-8 °C temporarily and wait for quantitative determination or store the library in a refrigerator at -20 ± 5 °C for long-term storage.
4. Quantification of the constructed library
- Use the nucleic acid quantifier to quantify the library. If the library concentration is ≥ 0.2 ng/µL, it is qualified. Otherwise, rebuild the library.
- Mix equal volume of DNA (or RNA) and quantify the solution. According to the quantitative result, dilute the mixed solution to 100 pmol/L by using the following formula.
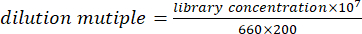
NOTE: An alternative is to dilute the library to 100 pmol/L according to the formula, and then quantify by fluorescent quantitative PCR. Mix DNA( or RNA) libraries equally according to quantitative results of PCR instrument. - Mix 100 pmol/L DNA library and RNA library in a ratio of 4:1. Use the mixed solution of DNA and RNA library for computer sequencing.
5. Sequencing
- Perform sequencing by referring to the manual of the universal kit of sequencing reaction24,25(semiconductor sequencing method).
6. Quality control of samples
- Positive quality control of gastric cancer DNA: Take the DNA positive control of gastric cancer and perform the test according to the instructions of the kit. The control is provided with the kit. The result shows that MTHFR, DHFR, MTR, GGH and SLC19A1 gene mutants are detected.
- Negative quality control of gastric cancer DNA: Take the DNA negative control of gastric cancer and perform the test according to the instructions of the kit. The control is provided with the kit. The result shows that the wild types of MTHFR, DHFR, MTR, GGH and SLC19A1 genes are detected.
NOTE: Ensure criteria are met in both the cases, otherwise, re-detection is required.
7. Data analysis
- Run the corresponding plug-in (the Variant Caller, Coverage Analysis, and Ion Reporter Software) in the Torrent Suite Software to get the analysis results of the samples. Judge the sample detection results according to the analysis results of the plug-in.
Results
The determination of test results relies on the positive judgement value, which is also recognized as the reference interval. Use semiconductor sequencing method to detect the collected clinical samples. When the mutation frequency value of MTHFR, DHFR, MTR, GGH and SLC19A1 genes is ≥ 5%, the detection result is the mutant of corresponding gene. When the mutation frequency value is < 5 %, the detection result is the wild type of the corresponding gene26.
The following criteria can be used to determine whether the detection results are credible. Firstly, if the average coverage of DNA sequencing results is ≥ 500, and the mapped reads of RNA sequencing results are ≥ 20000, the test results are credible27. Otherwise, it is recommended to retest. Secondly, the average coverage of DNA positive quality control of gastric cancer is ≥ 500, and the test result should conform to the rs1801131 and rs1801133 mutations of MTHFR gene, rs1650697 and rs442767 mutations of DHFR gene, rs1805087 mutation of MTR gene, rs11545078 mutation of GGH gene and rs1051298 mutation of SLC19A1 gene as positive. Otherwise, it is recommended to retest. Thirdly, the average coverage of DNA negative quality control of gastric cancer is ≥ 500, and the detection results should show that all the sites within the detection range of this kit are wild type. Otherwise, it is recommended to retest. Lastly, the library concentration is lower than 0.2 ng/µL, which may be due the degradation of DNA or RNA in the sample, or the failure to strictly follow the experimental process or use of expired reagents during the experiment. The above conditions may cause the sequencing quality to decline or fail. It is recommended to rebuild the library.
With this kit, a series of steps are followed to build a sequencing library from clinical samples. The library is then sequenced on an ion semiconductor sequencing platform, with ANNOVAR used for annotating the results. After sequencing, a document is used to summarize the mutant types for each sample. The absence of a mutant type indicated that the sample did not undergo that specific mutation. SupplementaryTable 3 provides a typical example of this. Table 12 includes basic information about the SNPs detected here. Analyzing each SNPs using filter-based annotation in various database, such as 1000 Genomes Project (1000g2015aug; https://www.internationalgenome.org/; Table 13) and the Exome Aggregation Consortium (ExAC; https://gnomad.broadinstitute.org/; Table 14), can reveal that different SNPs are significantly present in different populations.
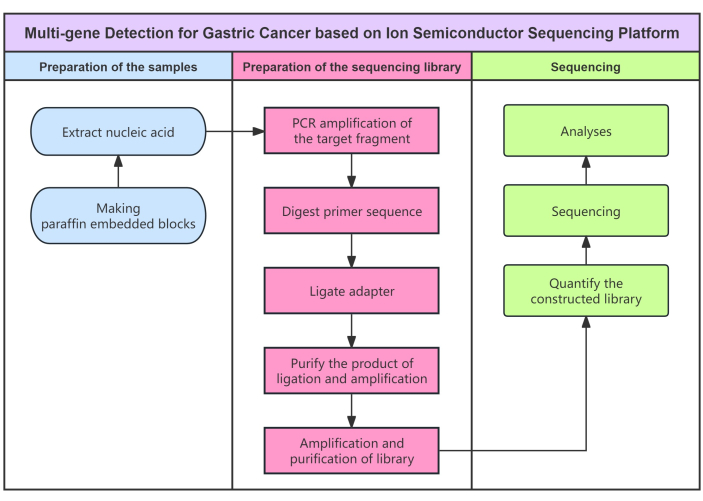
Figure 1: Flow diagram for the protocol. Flowchart of multi-gene detection for gastric cancer using semiconductor sequencing method. Please click here to view a larger version of this figure.
| Chromosome | Location | Primer design interval | Left primer | Right primer | |
| chr1 | 11854475 | chr1:11854175-11854775 | ACAGGATGGG GAAGTCACAG | AAACCGGAAT GGTCACAAAG | |
| 11856377 | chr1:11856077-11856677 | CTTCAGGTCA GCCTCAAAGC | TCCCTGTGGT CTCTTCATCC | ||
| chr5 | 79950780 | chr5:79950480-79951080 | CGCCGCACAT AGTAGGTTCT | CTTCCTCCTC CAGCCCTATC | |
| 79951495 | chr5:79951195-79951795 | CTTGGGTCAC CTGCACAGTA | ATTTTGAAGC ACCCAAGCTG | ||
| chr1 | 237048499 | chr1:237048199-237048799 | GTCAAAGGCC AGTCCCTTCT | CTCCCTTCAC CAACTGTGCT | |
| chr8 | 63938763 | chr8:63938463-63939063 | CAGTGAAGTT CAGCGGCAT | TATTTTCCTGT GTGGGGCAC | |
| chr21 | 46934825 | chr21:46934525-46935125 | CCAACCTGAG ATGGCTTTTC | TCCTTGGTGC TCTTGCTTTT | |
Table 1: Primers used for the seven selected SNPs in five genes. This sheet displays the information about the location of the primers of SNPs associated with 5-FU resistance in gastric cancer.
| Temperature | Time | No. of cycles |
| 99 °C | 2 min | 1 cycle |
| 99 °C | 15 s | 22 cycles |
| 60 °C | 4 min | |
| 10 °C | Hold | 1 cycle |
Table 2: Thermal cycle program for DNA amplification. Thermal reaction conditions such as temperature, time, and cycles are shown.
| Step | Temperature | Time | No. of cycles |
| Activate enzymes | 99 °C | 2 min | 1 cycle |
| Denature | 99 °C | 15 s | 30 cycles |
| Anneal and extend | 60 °C | 4 min | |
| Preserve | 10 °C | Hold | 1 cycle |
Table 3: Thermal cycle program for cDNA amplification. Thermal reaction conditions such as temperature, time, and cycles are shown.
| Temperature | Time | No. of cycles |
| 50 °C | 10 min | 1 cycle |
| 55 °C | 10 min | 1 cycle |
| 60 °C | 20 min | 1 cycle |
| 10 °C | Hold | 1 cycle |
Table 4: Thermal cycle program for digestion of primer sequence. Thermal reactions conditions such as temperature, time, and cycles are shown.
| Component | Volume |
| P1 adapter | 1.5 μL |
| Specific adapter X | 1.5 μL |
| Nuclease-free water | 3 μL |
| Total volume of reaction system | 6 μL |
| X: Indicates the specific adapter number |
Table 5: Sequence of adding reagents for preparation of adapter mixture. Add reagents into the tube in the order provided here.
| Component | Volume |
| Connecting buffer | 4 μL |
| Adapter mixture X | 2 μL |
| DNA ligase | 2 μL |
| Total volume of reaction system | 30 μL |
Table 6: Sequence of adding reagents into the digested primer production. Add reagents into the tube in the order provided here.
| Temperature | Time | No. of cycles |
| 22 °C | 30 min | 1 cycle |
| 72 °C | 10 min | 1 cycle |
| 10 °C | Hold | 1 cycle |
Table 7: Thermal cycle program for connecting adapter to DNA. Thermal reactions conditions such as temperature, time, and cycles are shown.
| Temperature | Time | No. of cycles |
| 22 °C | 30 min | 1 cycle |
| 68 °C | 5 min | 1 cycle |
| 72 °C | 5 min | 1 cycle |
| 10 °C | Hold | 1 cycle |
Table 8: Thermal cycle program for connecting adapter to cDNA. Thermal reactions conditions such as temperature, time, and cycles are shown.
| Component | Volume |
| Reaction solution of library amplification | 50 μL |
| Library primer mixture | 2 μL |
| Total volume of reaction system | 52 μL |
Table 9: Sequence of adding amplification reagents. Add reagents into the tube in the order provided here.
| Temperature | Time | No. of cycles |
| 98 °C | 2 min | 1 cycle |
| 98 °C | 15 s | 5 cycles |
| 60 °C | 1 min | |
| 10 °C | Hold | 1 cycle |
Table 10: Thermal cycle program for amplifying DNA library. Thermal reactions conditions such as temperature, time, and cycles are shown.
| Temperature | Time | No. of cycles |
| 98 °C | 2 min | 1 cycle |
| 98 °C | 15 s | 5 cycles |
| 64 °C | 1 min | |
| 10 °C | Hold | 1 cycle |
Table 11: Thermal cycle program for amplifying cDNA library. Thermal reactions conditions such as temperature, time, and cycles are shown.
| rs1801131 | rs1801133 | rs1650697 | rs442767 | rs1805087 | rs11545078 | rs1051298 | |
| Ref | T | G | A | G | A | G | G |
| Alt | G | A | G | T | G | A | A |
| Func.ref GeneWithVer | exonic | exonic | UTR5 | upstream | exonic | exonic | UTR3 |
| Gene.ref GeneWithVer | MTHFR | MTHFR | DHFR | DHFR | MTR | GGH | SLC19A1 |
| GeneDetail .refGeneWithVer | . | . | NG_023304.1: g.5020T>G | . | . | . | NM_001205206.1: c.*64C>T; NM_001205207.1: c.*746C>T; NM_194255.2:c. *746C>T |
| ExonicFunc .refGeneWithVer | nonsynonymous; SNV | nonsynonymous; SNV | . | . | nonsynonymous; SNV | nonsynonymous; SNV | . |
| AAChange.ref GeneWithVer | MTHFR:NM_00 1330358.1:exo n8:c.A1409C:p. E470A;MTHFR: NM_005957.4: exon8:c.A1286 C:p.E429A | MTHFR:NM_00 1330358.1:exo n5:c.C788T:p. A263V;MTHFR:N M_005957.4:e xon5:c.C665T: p.A222V | . | . | MTR:NM_0012 91939.1:exon2 5:c.A2603G:p. D868G;MTR:N M_001291940. 1:exon25:c.A1 535G:p.D512G; MTR:NM_0002 54.2:exon26:c. A2756G:p.D91 9G | GGH:NM_003878 .2:exon5:c.C452T: p.T151I | jin |
| cytoBand | 1p36.22 | 1p36.22 | 5q14.1 | 5q14.1 | 1q43 | 8q12.3 | 21q22.3 |
Table 12: Relevant information of the SNPs. Annotating the samples using ANNOVAR.
| MTHFR | DHFR | MTR | GGH | SLC19A1 | ||||
| rs1801131 | rs1801133 | rs1650697 | rs442767 | rs1805087 | rs11545078 | rs1051298 | ||
| 1000g2015aug_all | 0.249401 | 0.245407 | 0.76857 | 0.290136 | 0.218251 | 0.085463 | 0.524361 | |
| 1000g2015aug_afr | 0.1513 | 0.09 | 0.9349 | 0.0318 | 0.2844 | 0.056 | 0.5772 | |
| 1000g2015aug_amr | 0.1513 | 0.4741 | 0.755 | 0.3991 | 0.1772 | 0.0403 | 0.4323 | |
| 1000g2015aug_eas | 0.2192 | 0.2956 | 0.6607 | 0.5853 | 0.1052 | 0.0873 | 0.5635 | |
| 1000g2015aug_eur | 0.3131 | 0.3648 | 0.7555 | 0.3191 | 0.173 | 0.0924 | 0.4493 | |
| 1000g2015aug_sas | 0.4172 | 0.1186 | 0.6779 | 0.228 | 0.3211 | 0.1483 | 0.5552 | |
Table 13: Filter- based annotation of SNPs in 1000 Genomes Project.
| MTHFR | DHFR | MTR | GGH | SLC19A1 | |||
| rs1801131 | rs1801133 | rs1650697 | rs442767 | rs1805087 | rs11545078 | rs1051298 | |
| ExAC_ALL | 0.295 | 0.3037 | . | . | 0.2091 | 0.0936 | . |
| ExAC_AFR | 0.1588 | 0.1124 | . | . | 0.2666 | 0.0545 | . |
| ExAC_AMR | 0.1555 | 0.5141 | . | . | 0.1864 | 0.0361 | . |
| ExAC_EAS | 0.2148 | 0.3052 | . | . | 0.1132 | 0.0824 | . |
| ExAC_FIN | 0.3128 | 0.2227 | . | . | 0.1892 | 0.0581 | . |
| ExAC_NFE | 0.3191 | 0.345 | . | . | 0.1919 | 0.0969 | . |
| ExAC_OTH | 0.304 | 0.3062 | . | . | 0.2108 | 0.0973 | . |
| ExAC_SAS | 0.4153 | 0.1409 | . | . | 0.316 | 0.165 | . |
Table 14: Filter- based annotation of SNPs in the Exome Aggregation Consortium.
Supplementary Table 1: Sequence of adding reagents for DNA amplification. Add reagents into the tube in the order provided here. Please click here to download this File.
Supplementary Table 2: Sequence of adding reagents for cDNA amplification. Add reagents into the tube in the order provided here. Please click here to download this File.
Supplementary Table 3: Example of clinical specimen sequencing results. Only mutations present in the specimens will be recorded in the documents. Please click here to download this File.
Discussion
Clinical experts unanimously concur that patients, even with the same type and stage of gastric cancer, can have markedly different responses to an identical treatment approach. Years of research have revealed to scientists that the individual variations are chiefly attributed to gastric cancer's nature as a heterogeneous, polymorphic, and diversely differentiated cellular population, leading to significant individual disparities in treatment responses28. Consequently, the acquisition of gastric cancer samples through upper gastrointestinal endoscopy or surgery and blood samples, coupled with high-throughput sequencing for genetic analysis, enables personalized gastric cancer treatment. This strategy is designed to enhance clinical treatment efficacy and reduce the risk of severe toxic side effects. The progress in ion semiconductor sequencing technology has turned personalized treatment into a practical reality29.
Here are some limitations of this test method. The kit used here is primarily for in vitro diagnosis, so it is limited to detecting the mutation of rs1801131 and rs1801133 of MTHFR gene, rs1650697 and rs442767 of DHFR gene, rs1805087 of MTR gene, rs11545078 of GGH gene and rs1051298 of SLC19A1. Mutation in other sections cannot be detected. Due to significant heterogeneity in tumor tissue, different sampling locations may affect the detection results. For paraffin embedded tissue samples stored for longer duration, DNA and RNA may be degraded to a certain extent, affecting the test results. Unreasonable sample collection, transportation, and processing, as well as improper experimental operation and experimental environment may lead to false negative or false positive results. The detection result cannot be guaranteed if the nucleic acid concentration is lower than 2 ng/µL. The test results of the kit are only for clinical reference. The selection of personalized treatment for patients should be considered in combination with their symptoms/signs, medical history, other laboratory tests and treatment reactions. Negative results cannot completely exclude the existence of target gene mutation. Negative results can also be caused by too few tumor cells in the sample, excessive degradation of nucleic acid or the concentration of target gene in the amplification reaction system below the detection limit.
Some performance indexes of the kit used are as described. Analytical sensitivity: For DNA samples, the minimum detectable amount of the total nucleic acid in this kit is 10 ng, and 5% mutation rate can be detected. For RNA samples, the minimum detectable amount of the total nucleic acid in this kit is 10 ng. Positive and negative coincidence rate: The positive and negative coincidence rate reaches 100%. Limit of detection (LOD): A total of 14 LOD references, numbered L1-L14 can be used. L1-L11 is LOD references of MTHFR, DHFR, MTR, GGH and SLC19A1 genes, and their detection results should be that the mutation types of the corresponding genes sites are positive, and the coincidence rates are 100%. Repeatability: A total of five repetitive reference materials, numbered R1-R5 can be used. R1 is a strong positive repetitive reference material (rs67376798 mutation of DPYD gene). R2, which contains this mutation in lesser number, is a weak positive repetitive reference material (rs67376798 mutation of DPYD gene), R3 is a negative repetitive reference material (6 sites of the DPYD, MTHFR and ABCB1 genes are detected as wild type). Each reference sample must be tested 10 times, ensuring that the outcomes of these repeated assessments correspond with their predefined classifications. Data volume: The effective data volume of DNA and RNA samples should be controlled above 0.05 M. The sequencing depth of DNA samples shall be controlled above 500, and the Mapped Reads of RNA samples shall be controlled above 20000. Interference test: This kit is not affected by endogenous interference substances (triglycerides and albumin) and exogenous interference substances (formalin and dehydrated alcohol).
Some precautions must be observed during the experiment. The kit used here can only be used for in vitro testing. Please read this manual carefully before the experiment and use it within the validity period. Components of the kit in different batches cannot be used interchangeably. It is recommended to use disposable consumables for this kit to prevent contamination. During the use of this kit, it is recommended to use a suction head with a filter element. In order to avoid any potential biological hazards in the sample, the test sample should be considered as infectious substances to avoid contact with skin and mucous membrane. It is recommended to handle the samples in a biosafety cabinet that can prevent the outflow of aerosols. Test tubes and suckers used in the operation should be sterilized before being discarded. The operation and disposal of samples shall meet the requirements of relevant laws and regulations: General Guidelines for Biosafety of Microbial Biomedical Laboratories and Medical Waste Management Regulations of the Ministry of Health30,31. The experimental personnel must receive professional training, operate in strict accordance with the instructions, and strictly separate the areas according to the experimental process. Special instruments and equipment shall be used at each stage of the experimental operation, and the articles at each stage of each area shall not be used interchangeably. The experimental personnel must strictly separate the areas according to the experimental process. Special instruments and equipment shall be used at each stage of the experimental operation. Take protective measures as required, such as gloves, work clothes, etc. Waste disposal shall comply with relevant national regulations.
Although the focus of this article is on the seven SNPs within five genes related to gastric cancer, the sequencing in practical applications is not confined to these five genes alone. This paper definitively establishes a significant correlation between the seven SNPs and the sensitivity to 5-FU chemotherapy in gastric cancer.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This study is supported by Exploring the Mechanism and Role of the ERK2/Snai1/AGPS/PUFA-PL Pathway in Gastric Cancer Cell's Resistance to Apatinib-Induced Ferroptosis (Item number: 82172814), supported by National Nature Science Foundation of China; Investigating the Role and Mechanism of Zinc Finger Transcription Factor 1 in Modulating Gastric Cancer Cell Resistance to Ferroptosis Induced by Apatinib via the Polyunsaturated Ether Phospholipid Pathway(Item number: 2022A1515010267), supported by Guangdong Provincial Committee for the Funding of Basic and Applied Research; and the Research and Application of Reagent for the Diagnosis of 5-FU Chemosensitivity for Gastric Cancer (Item number: 201903010072), Science and Technology Projects in Guangzhou.
Materials
| Name | Company | Catalog Number | Comments |
| 1.5 mL DNA LoBind Tubes | Eppendorf | 30108051 | |
| 50 mL tubes | Greiner Bio-One | 227261 | |
| Amplification primer of gastric cancer | Thermo Fisher | The primers are sythesized by Thermo Fshier according to the sequence in Table 1. | |
| Deparaffinization | Qiagen | 19093 | |
| DNA purification magnetic beads | Bechkman | A63881 | |
| Ethyl alcohol | Guangzhou Chemical Reagent Factory Thermo Fisher Scientific | http://www.chemicalreagent.com/ | |
| Ion AmpliSeq Library Kit 2.0 | Thermo Fisher | 4480441 | |
| Nuclease-Free Water | Life Technologies | AM9932 | |
| PCR tubes | Axygen | PCR-02D-C | |
| PCR tubes | Axygen | PCR-02D-C | |
| Pipette tips | Quality Scientific Products | https://www.qsptips.com/products/standard_pipette_tips.aspx | |
| PureLink RNA Mini Columns | Thermo Fisher | A29839 | |
| RecoverAll Total Nucleic Acid Isolation Kit | Thermo Fisher | AM1975 | |
| Tabletop mini centrifuge | SCILOGES | S1010E | |
| Thermal Cycler | Life Technologies | 4375786 | |
| Ultramicro nucleic acid analyzer | BEIJING ORIENTAL SCIENCE & TECHNOLOGY DEVELOPMENT LTD. | BD-1000 |
References
- Sung, H., et al. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71 (3), 209-249 (2021).
- Wagner, A. D., et al. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 8, (2017).
- Shah, M. A., et al. Immunotherapy and targeted therapy for advanced gastroesophageal cancer: Asco guideline. J Clin Oncol. 41 (7), 1470-1491 (2023).
- Loganayagam, A., et al. Pharmacogenetic variants in the dpyd, tyms, cda and mthfr genes are clinically significant predictors of fluoropyrimidine toxicity. Br J Cancer. 108 (12), 2505-2515 (2013).
- Giacchetti, S., et al. Phase iii multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J Clin Oncol. 18 (1), 136-147 (2000).
- Hoff, P. M., et al. Comparison of oral capecitabine versus intravenous fluorouracil plus leucovorin as first-line treatment in 605 patients with metastatic colorectal cancer: Results of a randomized phase iii study. J Clin Oncol. 19 (8), 2282-2292 (2001).
- Meulendijks, D., et al. Clinical relevance of dpyd variants c.1679t>g, c.1236g>a/hapb3, and c.1601g>a as predictors of severe fluoropyrimidine-associated toxicity: A systematic review and meta-analysis of individual patient data. Lancet Oncol. 16 (16), 1639-1650 (2015).
- Sadighi, S., Mohagheghi, M. A., Montazeri, A., Sadighi, Z. Quality of life in patients with advanced gastric cancer: A randomized trial comparing docetaxel, cisplatin, 5-fu (tcf) with epirubicin, cisplatin, 5-fu (ecf). BMC Cancer. 6, 274 (2006).
- Collins, F. S., Brooks, L. D., Chakravarti, A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 8 (12), 1229-1231 (1998).
- Ladner, R. D. The role of dutpase and uracil-DNA repair in cancer chemotherapy. Curr Protein Pept Sci. 2 (4), 361-370 (2001).
- Capitain, O., et al. The influence of fluorouracil outcome parameters on tolerance and efficacy in patients with advanced colorectal cancer. Pharmacogenomics J. 8 (4), 256-267 (2008).
- Askari, B. S., Krajinovic, M. Dihydrofolate reductase gene variations in susceptibility to disease and treatment outcomes. Curr Genomics. 11 (8), 578-583 (2010).
- Ceppi, F., et al. DNA variants in dhfr gene and response to treatment in children with childhood b all: Revisited in aieop-bfm protocol. Pharmacogenomics. 19 (2), 105-112 (2018).
- Sharp, L., Little, J. Polymorphisms in genes involved in folate metabolism and colorectal neoplasia: A huge review. Am J Epidemiol. 159 (5), 423-443 (2004).
- Yousef, A. M., et al. The association of polymorphisms in folate-metabolizing genes with response to adjuvant chemotherapy of colorectal cancer. Cancer Chemother Pharmacol. 82 (2), 237-243 (2018).
- Schneider, E., Ryan, T. J. Gamma-glutamyl hydrolase and drug resistance. Clin Chim Acta. 374 (1-2), 25-32 (2006).
- Moran, R. G. Roles of folylpoly-gamma-glutamate synthetase in therapeutics with tetrahydrofolate antimetabolites: An overview. Semin Oncol. 26 (2), 24-32 (1999).
- Maezawa, Y., et al. High gamma-glutamyl hydrolase and low folylpolyglutamate synthetase expression as prognostic biomarkers in patients with locally advanced gastric cancer who were administrated postoperative adjuvant chemotherapy with s-1. J Cancer Res Clin Oncol. 146 (1), 75-86 (2020).
- Cheng, Q., et al. A substrate specific functional polymorphism of human gamma-glutamyl hydrolase alters catalytic activity and methotrexate polyglutamate accumulation in acute lymphoblastic leukaemia cells. Pharmacogenetics. 14 (8), 557-567 (2004).
- Zhang, Q., et al. Recognition of cyclic dinucleotides and folates by human slc19a1. Nature. 612 (7938), 170-176 (2022).
- Ulrich, C. M., et al. Polymorphisms in folate-metabolizing enzymes and response to 5-fluorouracil among patients with stage ii or iii rectal cancer (int-0144; swog 9304). Cancer. 120 (21), 3329-3337 (2014).
- Zhang, X., et al. Discovery of novel biomarkers of therapeutic responses in han chinese pemetrexed-based treated advanced nsclc patients. Front Pharmacol. 10, 944 (2019).
- Corrigan, A., et al. Pharmacogenetics of pemetrexed combination therapy in lung cancer: Pathway analysis reveals novel toxicity associations. Pharmacogenomics J. 14 (5), 411-417 (2014).
- Ion 520 & ion 530 ext kit - chef user guide. Thermofisherscientific Available from: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2FMAN0015805_Ion520_530ExTKit_UG.pdf (2023)
- Ion genestudio s5 instrument user guide. Thermofisherscientific Available from: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2FMAN0017528_Ion_GeneStudio_S5_Instrument_UG.pdf (2023)
- Parkin, N. T., et al. Multi-laboratory comparison of next-generation to sanger-based sequencing for hiv-1 drug resistance genotyping. Viruses. 12 (7), 694 (2020).
- Ziller, M. J., Hansen, K. D., Meissner, A., Aryee, M. J. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat Methods. 12 (3), 230-232 (2015).
- Lordick, F., et al. Unmet needs and challenges in gastric cancer: The way forward. Cancer Treat Rev. 40 (6), 692-700 (2014).
- Kumar, K. R., Cowley, M. J., Davis, R. L. Next-generation sequencing and emerging technologies. Semin Thromb Hemost. 45 (7), 661-673 (2019).
- Chinese Centres for Disease Control and Prevention. Ministry of Health of the People's Republic of China. WS 233-2002. Chinese Centres for Disease Control and Prevention. , 1-64 (2002).
- China Environmental Protection Industry. Medical waste management regulations. China Environmental Protection Industry. (1), 6-10 (2004).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved
We use cookies to enhance your experience on our website.
By continuing to use our website or clicking “Continue”, you are agreeing to accept our cookies.