A subscription to JoVE is required to view this content. Sign in or start your free trial.
Epithelial Cell Infection Analyses with Shigella
In This Article
Summary
The present protocol describes infection assays to interrogate Shigella adherence, invasion, and intracellular replication using in vitro epithelial cell lines.
Abstract
The human-adapted enteric bacterial pathogen Shigella causes millions of infections each year, creates long-term growth effects among pediatric patients, and is a leading cause of diarrheal deaths worldwide. Infection induces watery or bloody diarrhea as a result of the pathogen transiting the gastrointestinal tract and infecting the epithelial cells lining the colon. With staggering increases in antibiotic resistance and the current lack of approved vaccines, standardized research protocols are critical to studying this formidable pathogen. Here, methodologies are presented to examine the molecular pathogenesis of Shigella using in vitro analyses of bacterial adherence, invasion, and intracellular replication in colonic epithelial cells. Prior to infection analyses, the virulence phenotype of Shigella colonies was verified by the uptake of the Congo red dye on agar plates. Supplemented laboratory media can also be considered during bacterial culturing to mimic in vivo conditions. Bacterial cells are then used in a standardized protocol to infect colonic epithelial cells in tissue culture plates at an established multiplicity of infection with adaptations to analyze each stage of infection. For adherence assays, Shigella cells are incubated with reduced media levels to promote bacterial contact with epithelial cells. For both invasion and intracellular replication assays, gentamicin is applied for various time intervals to eliminate extracellular bacteria and enable assessment of invasion and/or the quantification of intracellular replication rates. All infection protocols enumerate adherent, invaded, and/or intracellular bacteria by serially diluting infected epithelial cell lysates and plating bacterial colony forming units relative to infecting titers on Congo red agar plates. Together, these protocols enable independent characterization and comparisons for each stage of Shigella infection of epithelial cells to study this pathogen successfully.
Introduction
Diarrheal diseases caused by enteric bacterial pathogens are a significant global health burden. In 2016, diarrheal diseases were responsible for 1.3 million deaths worldwide and were the fourth leading cause of death in children younger than five years of age1,2. The Gram-negative, enteric bacterial pathogen Shigella is the causative agent of shigellosis, a major cause of diarrheal deaths worldwide3. Shigellosis causes significant morbidity and mortality each year in children from lower- and middle-income countries4,5, while infections in high-income countries are linked to daycare center, foodborne, and waterborne outbreaks6,7,8,9. Ineffective vaccine development10 and rising rates of antimicrobial resistance (AMR)11,12 have complicated the management of large-scale Shigella outbreaks. Recent Centers for Disease Control and Prevention data show that nearly 46% of Shigella infections in the United States displayed drug resistance in 202013,14, while the World Health Organization has declared Shigella as an AMR priority pathogen for which new therapies are urgently needed15.
Shigella infections are easily transmitted via the fecal-oral route upon ingestion of contaminated food or water, or through direct human contact. Shigella has evolved to be an efficient, human-adapted pathogen, with an infectious dose of 10-100 bacteria sufficient to cause disease16. During small intestinal transit, Shigella is exposed to environmental signals, such as elevated temperature and bile17. Detection of these signals induces transcriptional changes to express virulence factors that enhance the ability of the bacteria to infect the human colon17,18,19. Shigella does not invade the colonic epithelium from the apical surface, but rather transits across the epithelial layer following uptake into specialized antigen-presenting microfold cells (M cells) within the follicle-associated epithelium20,21,22. Following transcytosis, Shigella cells are phagocytosed by resident macrophages. Shigella rapidly escapes the phagosome and triggers macrophage cell death, resulting in the release of pro-inflammatory cytokines5,23,24. Shigella then invades colonic epithelial cells from the basolateral side, lyses the macropinocytic vacuole, and establishes a replicative niche in the cytoplasm5,25. Pro-inflammatory cytokines, particularly interleukin-8 (IL-8), recruit polymorphonuclear neutrophil leukocytes (PMNs) to the site of infection, which weakens epithelial tight junctions, and enables bacterial infiltration of the epithelial lining to exacerbate basolateral infection5. The PMNs destroy the infected epithelial lining to contain the infection, which results in the characteristic symptoms of bacillary (bloody) dysentery5. Although invasion and intracellular replication mechanisms have been thoroughly characterized, new research is demonstrating important new concepts in Shigella infection, including virulence regulation during gastrointestinal (GI) transit17, adherence19, improved basolateral access through barrier permeability26, and asymptomatic carriage in malnourished children27.
The ability of Shigella spp. to cause diarrheal disease is restricted to humans and non-human primates (NHP)28. Shigella intestinal infection models have been developed for zebrafish29, mice30, guinea pigs31, rabbits21,32,33, and pigs34,35. However, none of these model systems can accurately replicate the disease characteristics observed during human infection36. Although NHP models of shigellosis have been established to study Shigella pathogenesis, these model systems are expensive to implement and require artificially high infectious doses, up to nine orders of magnitude higher than the infectious dose of humans37,38,39,40,41,42. Thus, the remarkable adaptation of Shigella for infection of human hosts necessitates the use of human-derived cell cultures to recreate physiologically relevant models for accurate interrogation of Shigella pathogenesis.
Here, detailed procedures are described to measure the rates of Shigella adherence to, invasion of, and replication within HT-29 colonic epithelial cells. Using these standardized protocols, the molecular mechanisms by which bacterial virulence genes and environmental signals impact each step of Shigella infection can be interrogated to better understand the dynamic host-pathogen interaction relationship.
Protocol
1. Preparation of reagents and materials
NOTE: All volumes are consistent with an assay using two 6-well plates.
- TSB medium: Add 0.5 L of deionized (DI) water to 15 g of Tryptic Soy Broth (TSB, see Table of Materials) medium and autoclave. Store at room temperature.
- Bile salts medium (TSB + BS): To prepare TSB containing 0.4% (w/v) bile salts, resuspend 0.06 g of bile salts (BS, see Table of Materials) in 15 mL of autoclaved TSB. Filter sterilize using a 0.22 µm PES filter.
NOTE: The bile salts consist of a 1:1 mixture of sodium cholate and sodium deoxycholate. Prepare fresh media immediately before use. - DMEM + 10% (v/v) FBS: Add 5 mL of fetal bovine serum (FBS) to 45 mL of Dulbecco's Modified Eagle Medium (DMEM). Store at 4 °C.
- DMEM + gentamicin: To a 50 mL tube, add 50 mL of DMEM and 50 µL of 50 mg/mL gentamicin (see Table of Materials).
NOTE: Make fresh aliquot and warm in a 37 °C water bath prior to each experiment. - PBS + 1% (v/v) Triton X-100: Add 150 µL of Triton X-100 to 15 mL of Phosphate-buffered saline (PBS).
NOTE: Make fresh aliquot and warm in a 37 °C water bath prior to each experiment. - TSB + Congo red indicator plates: Add 15 g of TSB, 7.5 g of select agar, and 0.125 g of Congo red dye (see Table of Materials) to a 1 L bottle. Add 0.5 L of DI water and autoclave. Pour 10-20 mL of media into individual sterile Petri dishes (100 mm x 15 mm) and let solidify.
CAUTION: Congo red is carcinogenic and a reproductive toxin. Ensure that handling of Congo red is performed using the appropriate personal protective equipment. Consult the product safety data sheet for additional information.
NOTE: Approximately 20 plates are made from 0.5 L Congo red media. Plates can be prepared 2-3 days in advance and left inverted at room temperature until use. For long-term storage, place inverted plates in plastic sleeves at 4 °C for up to 3 months. - DMEM + 10% (v/v) FBS and 5% (v/v) dimethyl sulfoxide (DMSO): Add 42.5 mL of DMEM, 5 mL of FBS, and 2.5 mL of DMSO to a 50 mL tube. Store at 4 °C.
2. Preparation of bacteria
NOTE: All Shigella laboratory cultivation and storage protocols are adapted from Payne, S. M.43.
CAUTION: Shigella spp. are Risk Group 2 pathogens44. Perform all laboratory work in a BSL-2 environment, with additional safety measures undertaken to limit accidental exposures due to the low infectious dose of Shigella spp.
- Growth of Shigella from frozen stocks
- Transfer a small amount of frozen culture from the cryogenic vial to a TSB + Congo red agar plate using a sterile applicator.
- Flame sterilize an inoculating loop and allow it to cool. Streak inoculum back and forth across one quadrant of the plate. Flame the loop, allow it to cool, then streak from the first quadrant onto the second quadrant of the plate. Repeat to streak inoculum into the third and fourth quadrants of the plate.
NOTE: Alternatively, streak inoculum using a fresh sterile applicator between each quadrant. - Invert the plate and incubate at 37 °C overnight.
NOTE: Incubation at temperatures ≥37 °C is required for expression of Shigella virulence factors necessary for observation of Congo red-positive (CR+) phenotype45. Avirulent colonies will have a white appearance and will not be invasive. - Seal the plate with paraffin film and store refrigerated at 4 °C.
NOTE: Bacterial colonies will remain viable on agar plates for 1-2 weeks.
- Overnight growth of Shigella in liquid culture
- Aliquot 3 mL of TSB media into sterile 14 mL culture tubes.
- Pick a single, well-isolated red (CR+) colony using a sterile applicator and resuspend in liquid media.
- Incubate cultures overnight (16-18 h) at 37 °C with shaking at 250 rotations per minute (rpm).
3. Preparation of HT-29 eukaryotic cells
NOTE: All volumes are consistent with an assay using two 6-well plates. HT-29 cell lines were acquired from the American Type Culture Collection (ATCC). HT-29 maintenance protocols are adapted from ATCC recommendations46. All media should be pre-warmed in a water bath at 37 °C prior to use. All HT-29 maintenance protocols should be performed in a biosafety cabinet. Refrain from producing bubbles when mixing/working with HT-29 cells in media to avoid dramatic changes in pH.
- Thawing HT-29 cells from frozen stock
- Thaw the vial of HT-29 cells in a 37 °C water bath.
NOTE: Ensure the cap stays fully above the water to avoid contamination. Thawing should take less than 2 min. - Remove the vial from the water immediately after the culture is fully thawed and decontaminate with 70% ethanol. Ensure that all steps from this point are performed using aseptic techniques.
- Add all the contents of the vial to a 15 mL centrifuge tube containing 9 mL of DMEM + 10% FBS. Centrifuge at 125 x g for 5 min at room temperature.
- Decant the supernatant into a waste container and resuspend the pellet in 10 mL of warm DMEM + 10% FBS. Transfer resuspended cells to a 75 cm2 tissue culture flask (T75) containing 10 mL of warm DMEM + 10% FBS (total volume of 20 mL).
- Incubate cells at 37 °C with 5% CO2 until the cells reach 90% confluency (approximately 6-7 days).
NOTE: Confluency is estimated through visual approximation.
- Thaw the vial of HT-29 cells in a 37 °C water bath.
- Seeding HT-29 cells
- Pre-warm 20 mL of PBS and 50 mL of DMEM + 10% FBS in a 37 °C water bath and pre-warm 3 mL of 0.25% (w/v) Trypsin-EDTA to room temperature.
- Once HT-29 cells (from step 3.1) reach 90% confluency, decant HT-29 cell culture media from the T75 flask into a waste container. Pour ~10 mL of warm PBS into the flask and swirl gently to wash. Decant the PBS into a waste container. Wash with warm PBS again and decant.
- Add 2-3 mL of 0.25% (w/v) Trypsin-EDTA and gently swirl across the entire surface area. Incubate at 37 °C with 5% CO2 for 4 min.
- Remove the flask from the incubator and gently swirl the Trypsin-EDTA, visually ensuring that all cells detach from the surface.
- Immediately add 6 mL of warm DMEM + 10% FBS to deactivate the Trypsin. Pipette up and down to thoroughly mix.
- Transfer all the contents to a 15 mL centrifuge tube and spin it at 500 x g for 5 min at room temperature.
- Gently decant supernatant into a waste container and resuspend the pellet in 6 mL of warm DMEM + 10% FBS.
- Immediately after resuspension, transfer 10 µL of suspended HT-29 cells from the middle of the culture to a 0.2 mL PCR tube. Add 10 µL of Trypan blue dye to the PCR tube and mix.
- Add 10 µL of HT-29 cell/Trypan blue mix to a disposable Countess cell counter chamber slide (see Table of Materials). Enumerate the number of live cells and calculate cell viability.
NOTE: When documenting the number of cells in the sample, read the number under the "live" cell count, not the total cell count. Alternatively, cell enumeration can be performed manually using a hemocytometer. - Seed resuspended HT-29 cells into a fresh T75 flask or 6-well plate.
- For T75 flask:
- Pipet gently to mix, then transfer 2.5 x 106 cells to a fresh T75 flask according to the equation below:
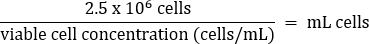
- Add warm DMEM + 10% FBS media to a final volume of 20 mL (final concentration of 1.25 x 105 cells/mL).
- Disperse cells evenly across the flask by gently rocking back and forth.
- Incubate at 37 °C with 5% CO2 until cells reach 80% confluency.
NOTE: For optimal growth, replace the DMEM + 10% FBS media in the T75 flask every ~3 days. Decant media into a waste container and add 10 mL of warm PBS to the flask. Swirl the PBS around gently and decant it into the waste container. Then add 20 mL of fresh, warm DMEM + 10% FBS to the flask and return to the 37 °C, 5% CO2 incubator.
- Pipet gently to mix, then transfer 2.5 x 106 cells to a fresh T75 flask according to the equation below:
- For 6-well plate:
- Pipet gently to mix, then transfer 5.85 x 106 cells to a fresh 50 mL conical tube according to the equation below:
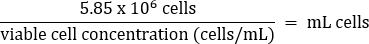
- Add warm DMEM + 10% FBS media to a final volume of 26 mL (final concentration of 2.25 x 105 cells/mL).
- Pipet gently to mix, then dispense 2 mL (4.5 x 105 cells) into individual wells of 6-well plates.
- Disperse cells evenly across the well by gently rocking up/down and left/right 2-3x.
- Incubate at 37 °C with 5% CO2 until cells reach 80%-95% confluency (approximately 3-4 days).
NOTE: 85% confluency is recommended for invasion and intracellular replication assays, while 90%-95% confluency is recommended for adherence assays. Cells should reach ~85% confluency after 48 h incubation with a final concentration of approximately 1 x 106 cells/well. Adjustments to the number of cells seeded and length of incubation may be required.
- Pipet gently to mix, then transfer 5.85 x 106 cells to a fresh 50 mL conical tube according to the equation below:
- For T75 flask:
- Making frozen HT-29 stocks
- Aliquot 1 mL of DMEM + 10% FBS + 5% DMSO media into individual cryogenic vials.
- Add 1 x 106 HT-29 cells from step 3.2.7 to each vial. Calculate the volume of cells according to the formula below:
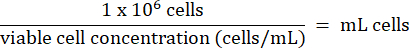
- Store HT-29 cells long-term below -130 °C in liquid nitrogen vapor storage freezer.
4. Adherence assay
NOTE: All volumes are consistent with an assay using two 6-well plates.
- Subculture overnight Shigella cultures via 1:50 dilution into fresh media.
- Vortex, then add 100 µL of each overnight culture to 5 mL of fresh TSB or TSB + BS in a suitably sized culture tube.
NOTE: Limit culture volume to <20% of culture flask or tube volume to ensure proper aeration. - Incubate at 37 °C with shaking at 250 rpm until cells reach an optical density (OD600) of 0.7 (mid-log phase of Shigella growth); about 2-2.5 h.
NOTE: During the subculture, aliquot 50 mL of DMEM and a sufficient volume of PBS for all washing steps and place in a 37 °C water bath. Allow media to reach 37 °C before use.
- Vortex, then add 100 µL of each overnight culture to 5 mL of fresh TSB or TSB + BS in a suitably sized culture tube.
- Transfer 2 x 108 colony forming units (CFUs) subcultured Shigella to individual 2 mL microcentrifuge tubes.
NOTE: 2 x 108 CFUs corresponds to approximately 1 mL of bacterial cells at an OD600 of 0.7. Use OD600 readings to approximate CFU/mL according to the calibration of each individual spectrophotometer. - Wash each Shigella sample 2x with PBS.
- Pellet cells by centrifugation at 17,000 x g for 2 min at room temperature. Aspirate the supernatant, then add 1 mL of warm PBS and resuspend the pellet well, gently pipetting the sample up and down until the mixture is fully homogeneous (8-10x).
- Repeat step 4.3.1 1x additional time.
- Pellet cells by centrifugation at 17,000 x g for 2 min at room temperature, aspirate the supernatant, and resuspend the pellets in 2 mL of warm DMEM.
NOTE: The final concentration of resuspended bacteria will be 1 x 108 CFU/mL.
- Vortex, then add 1 mL (1 x 108 CFUs) of resuspended Shigella to each well of the prepared HT-29 colonic epithelial monolayers in 6-well plates (from step 3.2.10.2).
NOTE: Infections are normally performed at a multiplicity of infection (MOI; ratio of bacterial to epithelial cells) of 100. To test different MOIs, dilute resuspended Shigella in warm DMEM to the desired concentration, then add 1 mL of diluted bacteria to HT-29 monolayers. For example, to test an MOI of 10, dilute bacteria 1:10 by adding 150 µL of 1 x 108 CFU/mL bacteria to 1.35 mL of warm DMEM, then apply 1 mL (1 x 107 CFUs) to HT-29 cells. - Incubate the 6-well plates at 37 °C with 5% CO2 for 3 h.
- During the incubation, determine the bacterial infection titer.
- Prepare 10-fold serial dilutions of resuspended Shigella cells (from step 4.3.3) into PBS.
- Plate 100 µL of the 1 x 10-5 and 1 x 10-6 dilutions onto TSB + Congo red plates and incubate overnight at 37 °C.
NOTE: Plating 100 µL from the 1 x 10-5 and 1 x 10-6 dilutions corresponds to a final dilution factor of 1 x 10-6 and 1 x 10-7, respectively.
- After incubation, wash the monolayers 4-5x with PBS.
- Aspirate media from each well.
NOTE: When aspirating media from 6-well plates, guide the tip of the aspirator along the bottom side of the wells, trying to avoid contact with the HT-29 cells. - Add 1 mL of warm PBS to each well and wash gently.
NOTE: To gently wash 6-well monolayers with PBS, move the plate up and down and side to side on the benchtop. Washing plates in a circular motion and/or removing the plate from the benchtop surface can cause the mechanical removal of cells from plastic. - Repeat steps 4.7.1 and 4.7.2 4x additional times.
- Aspirate media from each well.
- Remove PBS by aspiration and lyse HT-29 cells by adding 1 mL of PBS + 1% Triton X-100 to each well.
- Incubate 6-well plates at 37 °C for 5 min.
- Use a cell scraper or a bent pipette tip to scrape the lysed cells from the bottom of the well and transfer the full 1 mL into a fresh 1.7 mL microcentrifuge tube.
- Determine the number of cell-associated bacteria.
- Vortex each tube (from step 4.10) for at least 30 s to further displace Shigella from the lysed eukaryotic cells.
- Prepare 10-fold serial dilutions of lysates into PBS.
- Plate 100 µL of the 1 x 10-2, 1 x 10-3, and 1 x 10-4 dilutions onto TSB + Congo red plates and incubate overnight at 37 °C.
NOTE: Plating 100 µL from the 1 x 10-2, 1 x 10-3, and 1 x 10-4 dilutions corresponds to a final dilution factor of 1 x 10-3, 1 x 10-4, and 1 x 10-5, respectively.
5. Invasion assay
NOTE: All volumes are consistent with an assay using two 6-well plates.
- Subculture overnight Shigella cultures via 1:50 dilution into fresh media.
- Vortex, then add 100 µL of each overnight culture to 5 mL of fresh TSB or TSB + BS in a suitably sized culture tube.
NOTE: Limit culture volume to <20% of culture flask or tube volume to ensure proper aeration. - Incubate at 37 °C with shaking at 250 rpm until cells reach an OD600 of 0.7 (mid-log phase of Shigella growth); about 2-2.5 h.
NOTE: During the subculture, aliquot 50 mL of DMEM + 50 mg/mL gentamicin and a sufficient volume of PBS for all washing steps and place in a 37 °C water bath. Allow media to reach 37 °C before use.
- Vortex, then add 100 µL of each overnight culture to 5 mL of fresh TSB or TSB + BS in a suitably sized culture tube.
- Transfer 2 x 108 CFUs subcultured Shigella to individual 2 mL microcentrifuge tubes.
NOTE: 2 x 108 CFUs corresponds to approximately 1 mL of bacterial cells at an OD600 of 0.7. Use OD600 readings to approximate CFU/mL according to the calibration of each individual spectrophotometer. - Wash Shigella samples 1x with PBS.
- Pellet cells by centrifugation at 17,000 x g for 2 min at room temperature. Aspirate the supernatant, then add 1 mL of warm PBS and resuspend the pellet well, gently pipetting the sample up and down until the mixture is fully homogeneous (8-10x).
- Repeat step 5.3.1. 1x additional time.
- Pellet cells by centrifugation at 17,000 x g for 2 min at room temperature, aspirate the supernatant, and resuspend the pellets in 2 mL of warm DMEM.
NOTE: The final concentration of resuspended bacteria will be 1 x 108 CFU/mL.
- Vortex, then add 1 mL (1 x 108 CFUs) of resuspended Shigella plus 1 mL of DMEM to each well of the prepared HT-29 colonic epithelial monolayers in 6-well plates (from step 3.2.10.2).
NOTE: Infections are normally performed at a multiplicity of infection (MOI; ratio of bacterial to epithelial cells) of 100. To test different MOIs, dilute resuspended Shigella in DMEM to the desired concentration, then add 1 mL of diluted bacteria to HT-29 monolayers. For example, to test an MOI of 10, dilute bacteria 1:10 by adding 150 µL of 1 x 108 CFU/mL bacteria to 1.35 mL of DMEM, then add 1 mL (1 x 107 CFUs) to HT-29 cells. - To promote bacterial contact with the HT-29 cells, centrifuge the 6-well plates at 2,000 x g for 10 min at room temperature or 37 °C if the temperature setting can be adjusted.
NOTE: Centrifugation promotes bacterial contact with the HT-29 cells, which bypasses the need for adherence factors and allows the bacteria to quickly invade the cells. - Incubate 6-well plates at 37 °C with 5% CO2 for 45 min.
- During the incubation, determine the bacterial infection titer.
- Prepare 10-fold serial dilutions of resuspended Shigella cells (from step 5.3.3) into PBS.
- Plate 100 µL of the 1 x 10-5 and 1 x 10-6 dilutions onto TSB + Congo red plates and incubate overnight at 37 °C.
NOTE: Plating 100 µL from the 1 x 10-5 and 1 x 10-6 dilutions corresponds to a final dilution factor of 1 x 10-6 and 1 x 10-7, respectively.
- Thoroughly wash infected HT-29 cells 3x with 1 mL of PBS.
- Aspirate media from each well.
NOTE: When aspirating media from 6-well plates, guide the tip of the aspirator along the bottom side of the wells, trying to avoid contact with the HT-29 cells. - Add 1 mL of warm PBS to each well and wash gently.
NOTE: To gently wash 6-well monolayers with PBS, move the plate up and down and side to side on the benchtop. Washing plates in a circular motion and/or removing the plate from the benchtop surface can cause the mechanical removal of cells from plastic. - Repeat steps 5.8.1 and 5.8.2 2x additional times.
- Aspirate media from each well.
- Remove PBS by aspiration, then add 2 mL of warm DMEM supplemented with 50 µg/mL gentamicin to each well and incubate for 30 min at 37 °C with 5% CO2.
- Thoroughly wash infected HT-29 cells 3x with 1 mL of PBS.
- Repeat washing step 5.8.
- Remove PBS by aspiration, then add 2 mL of warm DMEM supplemented with 50 µg/mL gentamicin to each well and incubate for 60 min at 37 °C with 5% CO2.
- Thoroughly wash infected HT-29 cells 3x with 1 mL of PBS.
- Repeat washing step 5.8.
- Remove PBS by aspiration and lyse HT-29 cells by adding 1 mL of PBS + 1% Triton X-100 to each well.
- Incubate 6-well plates at 37 °C for 5 min.
- Use a cell scraper or a bent pipette tip to scrape the lysed cells from the bottom of the well and transfer the full 1 mL into a fresh 1.7 mL microcentrifuge tube.
- Determine the number of intracellular bacteria.
- Vortex each tube (from step 5.15) for at least 30 s to further displace Shigella from the lysed eukaryotic cells.
- Prepare 10-fold serial dilutions of lysates into PBS.
- Plate 100 µL of the 1 x 10-2 and 1 x 10-3 dilutions onto TSB + Congo red plates and incubate overnight at 37 °C.
NOTE: Plating 100 µL from the 1 x 10-2 and 1 x 10-3 dilutions corresponds to a final dilution factor of 1 x 10-3 and 1 x 10-4, respectively.
6. Intracellular replication assay
NOTE: All volumes are consistent with an assay using two 6-well plates.
- Subculture overnight Shigella cultures via 1:50 dilution into fresh media.
- Vortex, then add 100 µL of each overnight culture to 5 mL of fresh TSB or TSB + BS in a suitably sized culture tube.
NOTE: Limit culture volume to <20% of culture flask or tube volume to ensure proper aeration. - Incubate at 37 °C with shaking at 250 rpm until cells reach an OD600 of 0.7 (mid-log phase of Shigella growth); about 2-2.5 h.
NOTE: During the subculture, aliquot 50 mL of DMEM + 50 mg/mL gentamicin and a sufficient volume of PBS for all washing steps and place in a 37 °C water bath. Allow media to reach 37 °C before use.
- Vortex, then add 100 µL of each overnight culture to 5 mL of fresh TSB or TSB + BS in a suitably sized culture tube.
- Transfer 2 x 108 CFUs subcultured Shigella to individual 2 mL microcentrifuge tubes.
NOTE: 2 x 108 CFUs corresponds to approximately 1 mL of bacterial cells at an OD600 of 0.7. Use OD600 readings to approximate CFU/mL according to the calibration of each individual spectrophotometer. - Wash Shigella samples 1x with PBS.
- Pellet cells by centrifugation at 17,000 x g for 2 min at room temperature. Aspirate the supernatant, then add 1 mL of warm PBS and resuspend the pellet well, gently pipetting the sample up and down until the mixture is fully homogeneous (8-10x).
- Repeat step 6.3.1. 1x additional time.
- Pellet cells by centrifugation at 17,000 x g for 2 min at room temperature, aspirate the supernatant, and resuspend the pellets in 2 mL of warm DMEM.
NOTE: The final concentration of resuspended bacteria will be 1 x 108 CFU/mL.
- Vortex, then add 1 mL (1 x 108 CFUs) of resuspended Shigella plus 1 mL of DMEM to each well of prepared HT-29 colonic epithelial monolayers in 6-well plates (from step 3.2.10.2).
NOTE: Infections are normally performed at a multiplicity of infection (MOI; ratio of bacterial to epithelial cells) of 100. To test different MOIs, dilute resuspended Shigella in DMEM to the desired concentration, then add 1 mL of diluted bacteria to HT-29 monolayers. For example, to test an MOI of 10, dilute bacteria 1:10 by adding 150 µL of 1 x 108 CFU/mL bacteria to 1.35 mL of DMEM, then apply 1 mL (1 x 107 CFUs) to HT-29 cells. - To promote bacterial contact with the HT-29 cells, centrifuge the 6-well plates at 2,000 x g for 10 min at room temperature or 37 °C if the temperature setting can be adjusted.
NOTE: Centrifugation promotes bacterial contact with the HT-29 cells, which bypasses the need for adherence factors and allows the bacteria to quickly invade the cells. - Incubate 6-well plates at 37 °C with 5% CO2 for 45 min.
- During the incubation, determine the bacterial infection titer.
- Prepare 10-fold serial dilutions of resuspended Shigella cells (from step 6.3.3) into PBS.
- Plate 100 µL of the 1 x 10-5 and 1 x 10-6 dilutions onto TSB + Congo red plates and incubate overnight at 37 °C.
NOTE: Plating 100 µL from the 1 x 10-5 and 1 x 10-6 dilutions corresponds to a final dilution factor of 1 x 10-6 and 1 x 10-7, respectively.
- Thoroughly wash infected HT-29 cells 3x with 1 mL of PBS.
- Aspirate media from each well.
NOTE: When aspirating media from 6-well plates, guide the tip of the aspirator along the bottom side of the wells, trying to avoid contact with the HT-29 cells. - Add 1 mL of warm PBS to each well and wash gently.
NOTE: To gently wash 6-well monolayers with PBS, move the plate up and down and side to side on the benchtop. Washing plates in a circular motion and/or removing the plate from the benchtop surface can cause the mechanical removal of cells from plastic. - Repeat steps 6.8.1 and 6.8.2 2x additional times.
- Aspirate media from each well.
- Remove PBS by aspiration, then add 2 mL of warm DMEM supplemented with 50 µg/mL gentamicin to each well and incubate for 30 min at 37 °C with 5% CO2.
- Thoroughly wash infected HT-29 cells 3x with 1 mL of PBS.
- Repeat washing step 6.8.
- Remove PBS by aspiration, then add 2 mL of warm DMEM with 50 µg/mL gentamicin to each well of the 6-well plates and incubate at 37 °C with 5% CO2 for the desired length of time to allow for intracellular replication (up to 24 h).
- Thoroughly wash cells 2x with 1 mL of PBS.
- Repeat washing step 6.8.
- Remove PBS by aspiration and lyse HT-29 cells by adding 1 mL of PBS + 1% Triton X-100 to each well.
- Incubate 6-well plates at 37 °C for 5 min.
- Use a cell scraper or bent pipette tip to scrape the lysed cells from the bottom of the well and transfer the full 1 mL into a fresh 1.7 mL microcentrifuge tube.
- Determine the number of intracellular bacteria.
- Vortex each tube (from step 6.15) for at least 30 s to further displace Shigella from the lysed eukaryotic cells.
- Prepare 10-fold serial dilutions of lysates into PBS.
- Plate 100 µL of the 1 x 10-2, 1 x 10-3, and 1 x 10-4 dilutions onto TSB + Congo Red plates and incubate overnight at 37 °C.
NOTE: Plating 100 µL from the 1 x 10-2, 1 x 10-3, and 1 x 10-4 dilutions corresponds to a final dilution factor of 1 x 10-3, 1 x 10-4, and 1 x 10-5, respectively.
Results
Adherence, invasion, and intracellular replication assays were performed comparing S. flexneri 2457T wild type (WT) to S. flexneri ΔVF (ΔVF), a mutant hypothesized to negatively regulate Shigella virulence. Since Shigella uses bile salts as a signal to regulate virulence17,18,47, experiments were performed after bacterial subculture in TSB media as well as TSB supplemented w...
Discussion
This protocol describes a set of three standardized assays to study Shigella adherence, invasion, and intracellular replication of intestinal epithelial cells. Although these methods are merely modified versions of classical gentamicin assays used to study the invasion and intracellular replication of various bacterial pathogens within host cells49,50,51, special considerations must be applied when studying Shigella...
Disclosures
The authors declare no conflicts of interest.
Acknowledgements
Support for the authors includes Massachusetts General Hospital's Department of Pediatrics, the Executive Committee on Research Interim Support Funding (ISF) award 2022A009041, the National Institute of Allergy and Infectious Diseases grant R21AI146405, and the National Institute of Diabetes and Digestive and Kidney Diseases grant Nutrition Obesity Research Center at Harvard (NORCH) 2P30DK040561-26. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Materials
| Name | Company | Catalog Number | Comments |
| 0.22 μm PES filter | Millipore-Sigma | SCGP00525 | Sterile, polyethersulfone filter for sterilizing up to 50 mL media |
| 14 mL culture tubes | Corning | 352059 | 17 mm x 100 mm polypropylene test tubes with cap |
| 50 mL conical tubes | Corning | 430829 | 50 mL clear polypropylene conical bottom centrifuge tubes with leak-proof cap |
| 6-well tissue culture plates | Corning | 3516 | Plates are treated for optimal cell attachment |
| Bile salts | Sigma-Aldrich | B8756 | 1:1 ratio of cholate to deoxycholate |
| Congo red dye | Sigma-Aldrich | C6277 | A benzidine-based anionic diazo dye, >85% purity |
| Countess cell counting chamber slide | Invitrogen | C10283 | To be used with the Countess Automated Cell Counter |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D8418 | A a highly polar organic reagent |
| Dulbecco’s Modified Eagle Medium (DMEM) | Gibco | 10569-010 | DMEM is supplemented with high glucose, sodium pyruvate, GlutaMAX, and Phenol Red |
| Fetal Bovine Serum (FBS) | Sigma-Aldrich | F4135 | Heat-inactivated, sterile |
| Gentamicin | Sigma-Aldrich | G3632 | Stock concentration is 50 mg/mL |
| HT-29 cell line | ATCC | HTB-38 | Adenocarcinoma cell line; colorectal in origin |
| Paraffin film | Bemis | PM999 | Laboratory sealing film |
| Petri dishes | Thermo Fisher Scientific | FB0875713 | 100 mm x 15 mm Petri dishes for solid media |
| Phosphate-buffered saline (PBS) | Thermo Fisher Scientific | 10010049 | 1x concentration; pH 7.4 |
| Select agar | Invitrogen | 30391023 | A mixture of polysaccharides extracted from red seaweed cell walls to make bacterial plating media |
| T75 flasks | Corning | 430641U | Tissue culture flasks |
| Triton X-100 | Sigma-Aldrich | T8787 | A common non-ionic surfactant and emulsifier |
| Trypan blue stain | Invitrogen | T10282 | A dye to detect dead tissue culture cells; only live cells can exclude the dye |
| Trypsin-EDTA | Gibco | 25200-056 | Reagent for cell dissociation for cell line maintenance and passaging |
| Tryptic Soy Broth (TSB) | Sigma-Aldrich | T8907 | Bacterial growth media |
References
- Karambizi, N. U., McMahan, C. S., Blue, C. N., Temesvari, L. A. Global estimated Disability-Adjusted Life-Years (DALYs) of diarrheal diseases: A systematic analysis of data from 28 years of the global burden of disease study. PloS one. 16 (10), e0259077 (2021).
- WHO. WHO methods and data sources for country-level causes of death 2000-2016. World Health Organization. , (2018).
- Kotloff, K. L. Shigella infection in children and adults: a formidable foe. Lancet Glob Health. 5 (12), e1166-e1167 (2017).
- Kotloff, K. L., et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet. 382 (9888), 209-222 (2013).
- Schroeder, G. N., Hilbi, H. Molecular pathogenesis of Shigella spp.: Controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev. 21 (1), 134-156 (2008).
- Arvelo, W., et al. Transmission risk factors and treatment of pediatric shigellosis during a large daycare center-associated outbreak of multidrug resistant shigella sonnei: Implications for the management of shigellosis outbreaks among children. Pediatr Infect Dis J. 28 (11), 976-980 (2009).
- Kozyreva, V. K., et al. Recent outbreaks of Shigellosis in California caused by two distinct populations of Shigella sonnei with either increased virulence or fluoroquinolone resistance. mSphere. 1 (6), 1-18 (2016).
- Bowen, A., et al. Importation and domestic transmission of Shigella sonnei resistant to ciprofloxacin - United States, May 2014-February 2015. MMWR Morb Mortal Wkly Rep. 64 (12), 318-320 (2015).
- Tansarli, G. S., et al. Genomic reconstruction and directed interventions in a multidrug-resistant Shigellosis outbreak in Seattle, WA, USA: a genomic surveillance study. Lancet. 3099 (22), 1-11 (2023).
- Barry, E. M., et al. Progress and pitfalls in Shigella vaccine research. Nat Rev Gastroenterol Hepatol. 10 (4), 245-255 (2013).
- Increase in Extensively Drug-Resistant Shigellosis in the United States. CDC Health Alert Network. Centers for Disease Control and Prevention Available from: https://emergency.cdc.gov/han/2023/han00486.asp?ACSTrackingID=USCDC_511-DM100260&ACSTrackingLabel=HAN%20486%20-%20General%20Public&deliveryName=USCDC_511-DM100260 (2023)
- Shiferaw, B., et al. Antimicrobial susceptibility patterns of Shigella isolates in Foodborne Diseases Active Surveillance Network (FoodNet) sites, 2000-2010. Clin Infect Dis. 54, S458-S463 (2012).
- Centers for Disease Control and Prevention. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022. Atlanta, GA: U.S. Department of Health and Human Services. CDC. , (2022).
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. CDC. 10 (1), (2019).
- WHO. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. WHO. , (2017).
- DuPont, H. L., Levine, M. M., Hornick, R. B., Formal, S. B. Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis. 159 (6), 1126-1128 (1989).
- Nickerson, K. P., et al. Analysis of Shigella flexneri resistance, biofilm formation, and transcriptional profile in response to bile salts. Infect Immun. 85 (6), 1-18 (2017).
- Faherty, C. S., Redman, J. C., Rasko, D. A. Shigella flexneri effectors OspE1 and OspE2 mediate induced adherence to the colonic epithelium following bile salts exposure. Mol Microbiol. 85 (1), 107-121 (2012).
- Chanin, R. B., et al. Shigella flexneri adherence factor expression in in vivo-like conditions. mSphere. 4 (6), e00751 (2019).
- Baranov, V., Hammarström, S. Carcinoembryonic antigen (CEA) and CEA-related cell adhesion molecule 1 (CEACAM1), apically expressed on human colonic M cells, are potential receptors for microbial adhesion. Histochem Cell Biol. 121 (2), 83-89 (2004).
- Wassef, J. S., Keren, D. F., Mailloux, J. L. Role of M cells in initial antigen uptake and in ulcer formation in the rabbit intestinal loop model of shigellosis. Infect Immun. 57 (3), 858-863 (1989).
- Sansonetti, P. J., Arondel, J., Cantey, J. R., Prévost, M. C., Huerre, M. Infection of rabbit Peyer's patches by Shigella flexneri: Effect of adhesive or invasive bacterial phenotypes on follicle-associated epithelium. Infect Immun. 64 (7), 2752-2764 (1996).
- Sansonetti, P. J., et al. Caspase-1 activation of IL-1beta and IL-18 are essential for Shigella flexneri-induced inflammation. Immunity. 12 (5), 581-590 (2000).
- Zychlinsky, A., Fitting, C., Cavaillon, J. M., Sansonetti, P. J. Interleukin 1 is released by murine macrophages during apoptosis induced by Shigella flexneri. J Clin Invest. 94 (3), 1328-1332 (1994).
- Sansonetti, P. J., Ryter, A., Clerc, P., Maurelli, A. T., Mounier, J. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect Immun. 51 (2), 461-469 (1986).
- Maldonado-Contreras, A., et al. Shigella depends on SepA to destabilize the intestinal epithelial integrity via cofilin activation. Gut Microbes. 8 (6), 544-560 (2017).
- Collard, J. -. M., et al. High prevalence of small intestine bacteria overgrowth and asymptomatic carriage of enteric pathogens in stunted children in Antananarivo, Madagascar. PLoS Negl Trop Dis. 16 (5), e0009849 (2022).
- Mattock, E., Blocker, A. J. How do the virulence factors of shigella work together to cause disease. Front Cell Infect Microbiol. 7, 1-24 (2017).
- Mostowy, S., et al. The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog. 9 (9), e1003588 (2013).
- Martinez-Becerra, F. J., et al. Parenteral immunization with IpaB/IpaD protects mice against lethal pulmonary infection by Shigella. Vaccine. 31 (24), 2667-2672 (2013).
- Shim, D. -. H., et al. New animal model of shigellosis in the Guinea pig: its usefulness for protective efficacy studies. J Immunol. 178 (4), 2476-2482 (2007).
- Marteyn, B., et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 465 (7296), 355-358 (2010).
- West, N. P., et al. Optimization of virulence functions through glucosylation of Shigella LPS. Science. 307 (5713), 1313-1317 (2005).
- Maurelli, A. T., et al. Shigella infection as observed in the experimentally inoculated domestic pig, Sus scrofa domestica. Microbial Pathog. 25 (4), 189-196 (1998).
- Jeong, K. -. I., Zhang, Q., Nunnari, J., Tzipori, S. A piglet model of acute gastroenteritis induced by Shigella dysenteriae Type 1. J Infect Dis. 201 (6), 903-911 (2010).
- Kim, Y. -. J., Yeo, S. -. G., Park, J. -. H., Ko, H. -. J. Shigella vaccine development: prospective animal models and current status. Curr Pharm Biotechnol. 14 (10), 903-912 (2013).
- Kent, T. H., Formal, S. B., LaBrec, E. H., Sprinz, H., Maenza, R. M. Gastric shigellosis in rhesus monkeys. Am J Pathol. 51 (2), 259-267 (1967).
- Shipley, S. T., et al. A challenge model for Shigella dysenteriae 1 in cynomolgus monkeys (Macaca fascicularis). Comp Med. 60 (1), 54-61 (2010).
- Higgins, R., Sauvageau, R., Bonin, P. Shigella flexneri Type 2 Infection in captive nonhuman primates. Can Vet J. 26 (12), 402-403 (1985).
- Oaks, E. V., Hale, T. L., Formal, S. B. Serum immune response to Shigella protein antigens in rhesus monkeys and humans infected with Shigella spp. Infect Immun. 53 (1), 57-63 (1986).
- Formal, S. B., et al. Protection of monkeys against experimental shigellosis with a living attenuated oral polyvalent dysentery vaccine. J Bacteriol. 92 (1), 17-22 (1966).
- Levine, M. M., Kotloff, K. L., Barry, E. M., Pasetti, M. F., Sztein, M. B. Clinical trials of Shigella vaccines: two steps forward and one step back on a long, hard road. Nat Rev Microbiol. 5 (7), 540-553 (2007).
- Payne, S. M. Laboratory cultivation and storage of Shigella. Curr Protoc Microbiol. 55 (1), 93 (2019).
- NIH Guidelines. NIH guidelines for research involving recombinant or synthetic nucleic acid molecules. NIH Guidelines. 2, 142 (2019).
- Maurelli, A. T., Blackmon, B., Curtiss, R. Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect Immun. 43 (1), 397-401 (1984).
- HT-29 cell line product sheet. ATCC Available from: https://www.atcc.org/products/htb-38 (2023)
- Sistrunk, J. R., Nickerson, K. P., Chanin, R. B., Rasko, D. A., Faherty, C. S. Survival of the fittest: How bacterial pathogens utilize bile to enhance infection. Clin Microbiol Rev. 29 (4), 819-836 (2016).
- Stensrud, K. F., et al. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J Biol Chem. 283 (27), 18646-18654 (2008).
- Mandell, G. L. Interaction of intraleukocytic bacteria and antibiotics. J Clin Invest. 52 (7), 1673-1679 (1973).
- Elsinghorst, E. A. Measurement of invasion by gentamicin resistance. Methods Enzymo. 236 (1979), 405-420 (1994).
- Elsinghorst, E. A., Weitz, J. A. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect Immun. 62 (8), 3463-3471 (1994).
- Dorman, C. J., McKenna, S., Beloin, C. Regulation of virulence gene expression in Shigella flexneri, a facultative intracellular pathogen. Int J Med Microbiol. 291 (2), 89-96 (2001).
- Porter, M. E., Dorman, C. J. Positive regulation of Shigella flexneri virulence genes by integration host factor. J Bacteriol. 179 (21), 6537-6550 (1997).
- Maurelli, A. T., Blackmon, B., Curtiss, R. Temperature-dependent expression of virulence genes in Shigella species. Infect Immun. 43 (1), 195-201 (1984).
- Schuch, R., Maurelli, A. T. Virulence plasmid instability in Shigella flexneri 2a is induced by virulence gene expression. Infect Immun. 65 (9), 3686-3692 (1997).
- Formal, S. B., Hale, T. L., Sansonetti, P. J. Invasive enteric pathogens. Rev Infect Dis. 5, S702-S707 (1983).
- Pál, T., Hale, T. L. Plasmid-associated adherence of Shigella flexneri in a HeLa cell model. Infect Immun. 57 (8), 2580-2582 (1989).
- Noben, M., et al. Human intestinal epithelium in a dish: Current models for research into gastrointestinal pathophysiology. United European Gastroenterol J. 5 (8), 1073-1081 (2017).
- Liévin-Le Moal, V., Servin, A. L. Pathogenesis of human enterovirulent bacteria: lessons from cultured, fully differentiated human colon cancer cell lines. Microbiol Mol Biol Rev R. 77 (3), 380-439 (2013).
- Mitchell, D. M., Ball, J. M. Characterization of a spontaneously polarizing HT-29 cell line, HT-29/cl.f8. In Vitro Cell Dev Biol - Anim. 40 (10), 297-302 (2004).
- Gagnon, M., Zihler Berner, A., Chervet, N., Chassard, C., Lacroix, C. Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J Microbiol Methods. 94 (3), 274-279 (2013).
- Koestler, B. J., et al. Human intestinal enteroids as a model system of Shigella pathogenesis. Infect Immun. 87 (4), 00733 (2019).
- Ranganathan, S., et al. Evaluating Shigella flexneri pathogenesis in the human enteroid model. Infect Immun. 87 (4), (2019).
- Nickerson, K. P., et al. A versatile human intestinal organoid-derived epithelial monolayer model for the study of enteric pathogens. Microbiol Spectr. 9 (1), 1-17 (2021).
- Perlman, M., Senger, S., Verma, S., Carey, J., Faherty, C. S. A foundational approach to culture and analyze malnourished organoids. Gut Microbes. 15 (2), 2248713 (2023).
- Pope, L. M., Reed, K. E., Payne, S. M. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect Immun. 63 (9), 3642-3648 (1995).
- Faherty, C. S., et al. The synthesis of OspD3 (ShET2) in Shigella flexneri is independent of OspC1. Gut Microbes. 7 (6), 486-502 (2016).
- Ridlon, J. M., Kang, D. -. J., Hylemon, P. B. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 47 (2), 241-259 (2006).
- Köseoğlu, V. K., Hall, C. P., Rodríguez-López, E. M., Agaisse, H. The Autotransporter IcsA promotes Shigella flexneri biofilm formation in the presence of bile salts. Infect Immun. 87 (7), 1-14 (2019).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved