Zum Anzeigen dieser Inhalte ist ein JoVE-Abonnement erforderlich. Melden Sie sich an oder starten Sie Ihre kostenlose Testversion.
Reporter Gene Repression Assay to Study Translational Regulation of a Target Gene
In diesem Artikel
Overview
This video demonstrates an in vivo assay in bacteria to study the interaction of RNA-binding proteins (RBPs) with RNA. The bacterial cells are transformed with two plasmid constructs — a binding-site plasmid encoding an mRNA containing a fluorescent reporter gene downstream of an RBP-binding site — while an RBP plasmid expresses the RBP proteins under the control of an inducer. Upon inducer-mediated increase in RBP production, the RBPs bind to the mRNA, resulting in translational repression of the reporter via inhibition of ribosome binding.
Protokoll
1. System Preparation
- Design of binding-site plasmids
- Design the binding site cassette as depicted in Figure 1. Each minigene contains the following parts (5' to 3'): Eagl restriction site, ∼40 bases of the 5' end of the kanamycin (Kan) resistance gene, pLac-Ara promoter, ribosome binding site (RBS), AUG of the mCherry gene, a spacer (δ), an RBP binding site, 80 bases of the 5' end of the mCherry gene, and an ApaLI restriction site.
NOTE: To increase the success rate of the assay, design three binding-site cassettes for each binding site, with spacers consisting of at least one, two, and three bases. See the Representative Results section for further guidelines.
- Design the binding site cassette as depicted in Figure 1. Each minigene contains the following parts (5' to 3'): Eagl restriction site, ∼40 bases of the 5' end of the kanamycin (Kan) resistance gene, pLac-Ara promoter, ribosome binding site (RBS), AUG of the mCherry gene, a spacer (δ), an RBP binding site, 80 bases of the 5' end of the mCherry gene, and an ApaLI restriction site.
- Cloning of binding site plasmids
- Order the binding-site cassettes as double-stranded DNA (dsDNA) minigenes. Each minigene is ∼500 bp long and contains an Eagl restriction site and an ApaLI restriction site at the 5' and 3' ends, respectively (see step 1.1.1).
NOTE: In this experiment, mini-genes with half of the kanamycin gene were ordered to facilitate screening for positive colonies. However, Gibson assembly is also suitable here, in which case the binding site can be ordered as two shorter complementary single-stranded DNA oligos. - Double-digest both the mini-genes and the target vector with Eagl-HF and ApaLI by the restriction protocol, and column purify.
- Ligate the digested minigenes to the binding-site backbone containing the rest of the mCherry reporter gene, terminator, and a kanamycin resistance gene.
- Transform the ligation solution into Escherichia coli TOP10 cells.
- Identify positive transformants via Sanger sequencing.
- Design a primer 100 bases upstream to the region of interest (see Table 1 for primer sequences).
- Miniprep a few bacterial colonies.
- Prepare 5 µL of a 5 mM solution of the primer and 10 µL of the DNA at 80 ng/µL concentration.
- Send the two solutions to a convenient facility for Sanger sequencing.
- Store purified plasmids at -20 °C, and bacterial strains as glycerol stocks, both in the 96-well format. DNA will then be used for transformation into E. coli TOP10 cells containing one of four fusion-RBP plasmids (see step 1.3.5).
- Order the binding-site cassettes as double-stranded DNA (dsDNA) minigenes. Each minigene is ∼500 bp long and contains an Eagl restriction site and an ApaLI restriction site at the 5' and 3' ends, respectively (see step 1.1.1).
- Design and construction of the RBP plasmid
NOTE: Amino acid and nucleotide sequences of the coat proteins used in this study are listed in Table 2.- Order the required RBP sequence lacking a stop codon as a custom-ordered dsDNA minigene lacking a stop codon with restriction sites at the ends (Figure 1).
- Clone the tested RBP lacking a stop codon immediately downstream of an inducible promoter and upstream of a fluorescent protein lacking a start codon (Figure 1), similar to steps 1.2.2-1.2.4. Make sure that the RBP plasmid contains a different antibiotic resistance gene than the binding-site plasmid.
- Identify positive transformants via Sanger sequencing, similar to step 1.2.5 (see Table 1 for primer sequences).
- Choose one positive transformant and make it chemically competent. Store as glycerol purified plasmids at -20 °C and glycerol stocks of bacterial strains at -80 °C in 96-well plates.
- Transform the binding-site plasmids (from step 1.2.6) stored in 96-well plates into chemically-competent bacterial cells already containing an RBP-mCerulean plasmid. To save time, instead of plating the cells on Petri dishes, plate them using an 8-channel pipettor on 8-lane plates containing Luria-Bertani (LB) agar with relevant antibiotics (Kan and Amp). Colonies should appear in 16 h.
- Select a single colony for each double transformant and grow overnight in LB medium with the relevant antibiotics (Kan and Amp) and store as glycerol stocks at -80 °C in 96-well plates.
2. Experiment Setup
NOTE: The protocol presented here was performed using a liquid-handling robotic system in combination with an incubator and a plate reader. Each measurement was carried out for 24 inducer concentrations, with two duplicates for each strain + inducer combination. Using this robotic system, data for 16 strains per day with 24 inducer concentrations was collected. However, if such a device is unavailable, or if fewer experiments are necessary, these can easily be done by hand using an 8-channel multi-pipette and adapting the protocol accordingly. For example, preliminary results for four strains per day with 12 inducer concentrations and four time points were acquired in this manner.
- Prepare, in advance, 1 L of bioassay buffer (BA) by mixing 0.5 g of tryptone, 0.3 mL of glycerol, 5.8 g of NaCl, 50 mL of 1 M MgSO4, 1 mL of 10x phosphate-buffered saline (PBS) buffer pH 7.4, and 950 mL of double distilled water (DDW). Autoclave or sterile filter the BA buffer.
- Grow the double-transformant strains at 37 °C and 250 rpm shaking in 1.5 mL LB with appropriate antibiotics (kanamycin at a final concentration of 25 μg/mL and ampicillin at a final concentration of 100 μg/mL), in 48-well plates, over a period of 18 h (overnight).
- In the morning, make the following preparations.
- Inducer plate. In a clean 96-well plate, prepare wells with semi-poor medium (SPM) consisting of 95% BA and 5% LB26 in the incubator at 37 °C. The number of wells corresponds to the desired number of inducer concentrations. Add C4-HSL to the wells in the inducer plate that will contain the highest inducer concentration (218 nM).
- Program the robot to serially dilute medium from each of the highest-concentration wells into 23 lower concentrations ranging from 0 to 218 nM. The volume of each inducer dilution should be sufficient for all strains (including duplicates).
- While the inducer dilutions are being prepared, warm 180 μL of SPM in the incubator at 37 °C, in 96-well plates.
- Dilute the overnight strains from step 2.2 by a factor of 100 by serial dilutions: first, dilute by a factor of 10 by mixing 100 μL of bacteria with 900 μL of SPM in 48-well plates, and then dilute again by a factor of 10 by taking 20 μL from the diluted solution into 180 μL of pre-warmed SPM, in 96-well plates suitable for fluorescent measurements.
- Add the diluted inducer from the inducer plate to the 96-well plates with the diluted strains according to the final concentrations.
- Shake the 96-well plates at 37 °C for 6 h, while taking measurements of optical density at 595 nm (OD595), mCherry (560 nm/612 nm), and mCerulean (460 nm/510 nm) fluorescence via a plate reader every 30 min. For normalization purposes, measure the growth of SMP with no cells added.
Ergebnisse
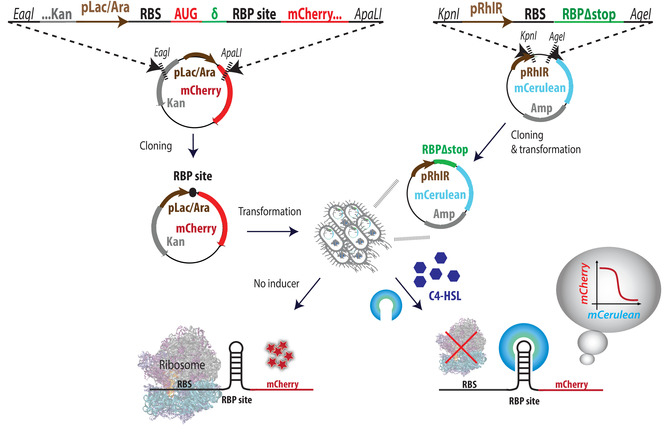
Figure 1: Overview of system design and cloning steps. Illustration of the cassette design for the binding site plasmid (left) and RBP-mCerulean plasmid (right). The next step is consecutive transformations of both plasmids into competent E. coli cells, with RBP plasmids first. Double-transformants are then tested for their mCherry expression levels in increasing inducer concentrations; ...
Offenlegungen
Materialien
| Name | Company | Catalog Number | Comments |
| Ampicillin sodium salt | SIGMA | A9518 | |
| Magnesium sulfate (MgSO4) | ALFA AESAR | 33337 | |
| 48 plates | Axygen | P-5ML-48-C-S | |
| 8-lane plates | Axygen | RESMW8I | |
| 96-well plates | Axygen | P-DW-20-C | |
| 96-well plates for plate reader | Perkin Elmer | 6005029 | |
| ApaLI | NEB | R0507 | |
| Binding site sequences | Gen9 Inc. and Twist Bioscience | see Table 1 | |
| E. coli TOP10 cells | Invitrogen | C404006 | |
| Eagl-HF | NEB | R3505 | |
| Glycerol | BIO LAB | 71205 | |
| Incubator | TECAN | Liconic incubator | |
| Kanamycin solfate | SIGMA | K4000 | |
| KpnI- HF | NEB | R0142 | |
| Ligase | NEB | B0202S | |
| Liquid-handling robotic system | TECAN | EVO 100, MCA 96-channel | |
| MATLAB analysis software | Mathworks | ||
| Multi-pipette 8 lanes | Axygen | BR703710 | |
| N-butanoyl-L-homoserine lactone (C4-HSL) | Cayman | K40982552 019 | |
| PBS buffer | Biological Industries | 020235A | |
| Plate reader | TECAN | Infinite F200 PRO | |
| Q5 HotStart Polymerase | NEB | M0493 | |
| RBP sequences | Addgene | 27121 & 40650 | see Table 2 |
| Sodium Chloride (NaCl) | BIO LAB | 190305 | |
| SV Gel and PCR Clean-Up System | Promega | A9281 | |
| Tryptone | BD | 211705 |
This article has been published
Video Coming Soon
Source: Katz, N., et al. An Assay for Quantifying Protein-RNA Binding in Bacteria. J. Vis. Exp. (2019)
Copyright © 2025 MyJoVE Corporation. Alle Rechte vorbehalten