A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
How to Measure Cortical Folding from MR Images: a Step-by-Step Tutorial to Compute Local Gyrification Index
In This Article
Summary
Measuring gyrification (cortical folding) at any age represents a window into early brain development. Hence, we previously developed an algorithm to measure local gyrification at thousands of points over the hemisphere1. In this paper, we detail the computation of this local gyrification index.
Abstract
Cortical folding (gyrification) is determined during the first months of life, so that adverse events occurring during this period leave traces that will be identifiable at any age. As recently reviewed by Mangin and colleagues2, several methods exist to quantify different characteristics of gyrification. For instance, sulcal morphometry can be used to measure shape descriptors such as the depth, length or indices of inter-hemispheric asymmetry3. These geometrical properties have the advantage of being easy to interpret. However, sulcal morphometry tightly relies on the accurate identification of a given set of sulci and hence provides a fragmented description of gyrification. A more fine-grained quantification of gyrification can be achieved with curvature-based measurements, where smoothed absolute mean curvature is typically computed at thousands of points over the cortical surface4. The curvature is however not straightforward to comprehend, as it remains unclear if there is any direct relationship between the curvedness and a biologically meaningful correlate such as cortical volume or surface. To address the diverse issues raised by the measurement of cortical folding, we previously developed an algorithm to quantify local gyrification with an exquisite spatial resolution and of simple interpretation. Our method is inspired of the Gyrification Index5, a method originally used in comparative neuroanatomy to evaluate the cortical folding differences across species. In our implementation, which we name local Gyrification Index (lGI1), we measure the amount of cortex buried within the sulcal folds as compared with the amount of visible cortex in circular regions of interest. Given that the cortex grows primarily through radial expansion6, our method was specifically designed to identify early defects of cortical development.
In this article, we detail the computation of local Gyrification Index, which is now freely distributed as a part of the FreeSurfer Software (http://surfer.nmr.mgh.harvard.edu/, Martinos Center for Biomedical Imaging, Massachusetts General Hospital). FreeSurfer provides a set of automated reconstruction tools of the brain's cortical surface from structural MRI data. The cortical surface extracted in the native space of the images with sub-millimeter accuracy is then further used for the creation of an outer surface, which will serve as a basis for the lGI calculation. A circular region of interest is then delineated on the outer surface, and its corresponding region of interest on the cortical surface is identified using a matching algorithm as described in our validation study1. This process is repeatedly iterated with largely overlapping regions of interest, resulting in cortical maps of gyrification for subsequent statistical comparisons (Fig. 1). Of note, another measurement of local gyrification with a similar inspiration was proposed by Toro and colleagues7, where the folding index at each point is computed as the ratio of the cortical area contained in a sphere divided by the area of a disc with the same radius. The two implementations differ in that the one by Toro et al. is based on Euclidian distances and thus considers discontinuous patches of cortical area, whereas ours uses a strict geodesic algorithm and include only the continuous patch of cortical area opening at the brain surface in a circular region of interest.
Protocol
1. Reconstruct the 3D cortical surfaces
This first part of the protocol uses the standard FreeSurfer pipeline as described in the Wiki (http://surfer.nmr.mgh.harvard.edu/fswiki). Note that the commands detailed here describe one way of achieving the cortical surface reconstructions, but equivalent commands may also be used.
- Import the raw MRI DICOM into FreeSurfer and verify the quality of the image (e.g. that the orientation is correct, the contrast sufficient and the images not moved). This process uses the following commands (replace the text between <...> (inclusive) with values appropriate to a specific instance, and "#" denotes comments):
mksubjdirs # create the folder architecture used by FreeSurfer
cd /mri # go to the mri folder of your subject
mri_convert -cm 001.mgz # convert the raw MRI in the # FreeSurfer format
tkmedit 001.mgz # visualize the converted volume
- Create the three-dimensional cortical mesh models8,9. In order to cope with the problem of buried sulci, FreeSurfer first creates a unitary white matter volume, which is used as a starting point for the initial gray-white surface. This surface is then optimized according to the local gradient of intensity and further expanded to the gray-CSF interface.
recon-all -s # launch the cortical surface reconstruction
At the end of the reconstruction process, you will get two mesh models composed of about 150,000 points for each hemisphere: a white (gray-white interface) and a pial (gray-CSF interface) surface. It is important to note that all the surfaces and volumes remain in the native space, allowing measurement such as volume, surface area, thickness or gyrification index to be measured without deformation.
- Check for the accuracy of these reconstructed surfaces:
tkmedit T1.mgz ?h.pial # the white surface is overlaid in green and the pial surface in red
where ?h denotes the hemisphere: lh.pial for the left hemisphere and rh.pial for the right hemisphere. The Figure 2 (in 2 versions: an animated gif image to be included in the movie and a static one for the website) shows an example of correct white and pial surfaces reconstructions for the "bert" subject distributed along with the FreeSurfer package. If you have to manually correct the result of the reconstruction process, you will find a tutorial on the FreeSurfer Wiki (http://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/WhiteMatterEdits , http://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/PialEdits).
2. Compute the local Gyrification Index
When you are satisfied with your surfaces, compute the local Gyrification Index (lGI) using the command:
recon-all -lgi -s
This command usually runs for about 3 hours for the two hemispheres of one study participant, depending on the power of your workstation. The different steps of lGI process are overviewed in Fig. 1. The computing begins with the creation of an outer surface using morphological closing operation. This outer surface, denoted ?h.pial_outer_smoothed, is further illustrated in Fig. 3. Then, about 800 overlapping circular regions of interest are created on the outer surface. For each one of these regions, a corresponding region of interest is defined on the pial surface. The whole computation ends up with the creation of an individual map containing one lGI value for each point of the cortical surface (i.e. ~150,000 values per hemisphere).
3. Check the result of the lGI calculation for each hemisphere
tksurfer ?h pial -overlay /surf/?h.pial_lgi -fthresh 1
The lGI values are overlaid over the cortical surface. As correct lGI values are typically comprised between 1 and 5, setting the minimum threshold at 1 (with the option fthresh) allows a rapid check: you should not see any gray cortical area. An example of correct individual result is shown in Fig. 4.
4. Statistical group comparisons
The purpose is to quantify the effect of group at each vertex over the cortical surface while controlling for the effect of gender and age. You will need to follow the same process as if you would like to compare cortical thickness at each vertex, but giving
- The first option to compare lGI results between groups use the commands listed below; further details can be obtained at https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/GroupAnalysis.
- First you will need to create a study specific template giving all your subjects in input:
make_average_subject --subjects ...
The command above will create a subject named "average". Alternatively, you can use the subject "fsaverage" distributed as a part of the FreeSurfer distribution. - Then, create the text file containing the description of the subjects involved in your study (the "FreeSurfer Group Descriptor File"). Your FSGD.txt should look like this:
GroupDescriptorFile 1
Group Control_Male
Group Control_Female
Group Patient_Male
Group Patient_Female
Variables Age
Input Patient_Male 20
Input Control_Female 23
<…> - Resample the lGI data in the space of the average subject using the following command for each hemisphere:
mris_preproc --fsgd FSGD.txt --target average --hemi ?h --meas pial_lgi --out ?h.lgi.mgh
- Smooth the data on the cortical surface to reduce the signal to noise:
mri_surf2surf --hemi ?h --s average --sval ?h.lgi.mgh --fwhm 10 --tval ?h.10.lgi.mgh
- Compute the group comparison at the level of each vertex. For that you will need to create a contrast text file (e.g. in the case of the FSGD.txt described above, the "contrast.txt" will contain the values "1 1 -1 -1 0" to compute the difference between controls and patients while controlling for age and gender). Finally run the comparison:
mri_glmfit --y ?h.10.lgi.mgh --fsgd FSGD.txt doss --glmdir ?h.lgi.glmdir --surf average ?h --C contrast.txt
- Visualize the results on your average subject using tksurfer:
tksurfer average ?h inflated
Then load as overlay the sig.mgh file located in the folder ?h.lgi.glmdir/contrast.txt/sig.mgh. Using the option "configure overlay" you can further modify the p threshold as well as correct for multiple comparisons using false discovery rate10.
- First you will need to create a study specific template giving all your subjects in input:
- The alternative option for group comparison is to use Qdec, a graphical user interface implemented in FreeSurfer. The use of Qdec with local Gyrification Index implies to pre-smooth the lGI data:
recon-all -qcache -measure pial_lgi -average -s
With Qdec, the FreeSurfer Group Descriptor File is replaced by a slightly different version, the Data Table (qdec.table.dat) that includes the description of the different groups and other confounding variables such as age. A detailed description of the utilization of Qdec is provided at http://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/QdecGroupAnalysis.
Of note, if the lGI is not available in the list of dependent variables in Qdec, you must add the following line to the .Qdecrc file located in your home directory:
MEASURE1 = pial_lgi
5. Analysis
Alternatively, the statistical analyses could eventually be computed at the level of the cortical parcellation integrated in FreeSurfer11. For that purpose, average lGI values can be extracted for the 34 gyral regions of interest for each hemisphere, and these measurements can be further compared between the different groups. This parcel-wise analysis (as opposed to the vertex-wise analysis described above) could be attractive as it limits the amount of statistical comparisons. However, the lGI at each point quantifies the gyrification in the surrounding circular area, so that the average lGI in a gyral region of interest also reflects to some extent the gyrification in the neighboring regions of interest.
Finally, although the most important issues were described in this protocol, a solution to the other problems that may be encountered during the FreeSurfer or the lGI processing can be found in the archives of the FreeSurfer mailing list (http://surfer.nmr.mgh.harvard.edu/fswiki/FreeSurferSupport).
6. Representative Results
As described in section 1c of the protocol, you should always carefully check the accuracy of the reconstruction of the cortical surfaces prior to lGI computation. While scrolling between the frontal and the occipital lobe, pay particular attention that vessels and membrane are not included in the pial surface. Check also that the white surface accurately follows the gray-white interface. An example of correct reconstruction is provided in Figure 2 (see the animated gif figure for the whole volume).
At the end of the lGI computation, you will also have to check the result for both hemispheres of each subject. There shouldn't be any cortical area with a lGI result smaller than 1. The section 3 of the protocol and the figure 4 show how to check correct if the output of the lGI computation is correct.
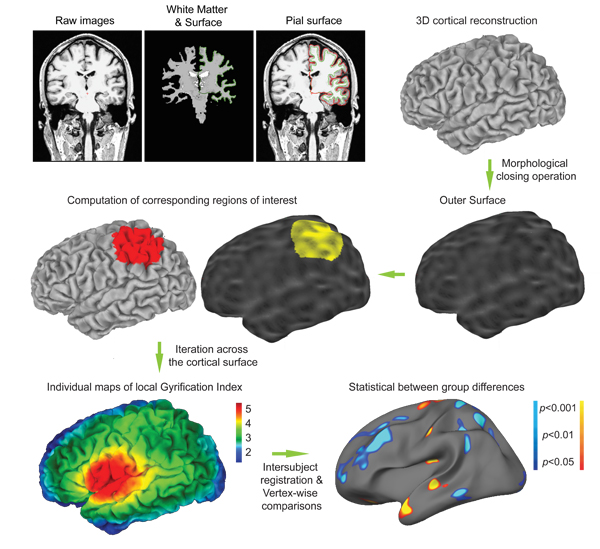
Figure 1. Overview of the lGI computation. First, three-dimensional cortical mesh models are reconstructed from the raw images using the standard FreeSurfer pipeline. These reconstruction algorithms use a binary white matter volume as starting point to overcome the issue of buried sulci. The cortical mesh models typically comprises about 150,000 vertices and are classically used to compute cortical thickness at each point. Similarly, local Gyrification Index (lGI) will be computed at each vertex. For that purpose, an outer surface is created. Then corresponding circular regions of interest are identified on the outer and cortical surface using matching algorithm. After about 800 of generating overlapping regions of interest, the process results in the creation of individual maps of lGI. These maps can be easily interpreted: an index of 5 means that there is 5 time more cortical surface invaginated within the sulci in the surrounding area that the amount of visible cortical surface; an index of 1 means that the cortex is flat in the surrounding area. Finally, statistical group comparisons are computed at the level of each vertex, similarly to cortical thickness comparisons.
Figure 1B. Individual cortical map of lGI. This small movie shows a 360-degrees rotation of an individual cortical lGI map as shown in Fig. 1. It is striking to note that the cortical regions with higher lGI values correspond to the first fold to be created during the in utero life: the sylvian fissure, the superior temporal sulcus and the intraparietal sulcus on the lateral view of the brain, and the parieto-occipital sulcus on the medial view of the brain. View movie
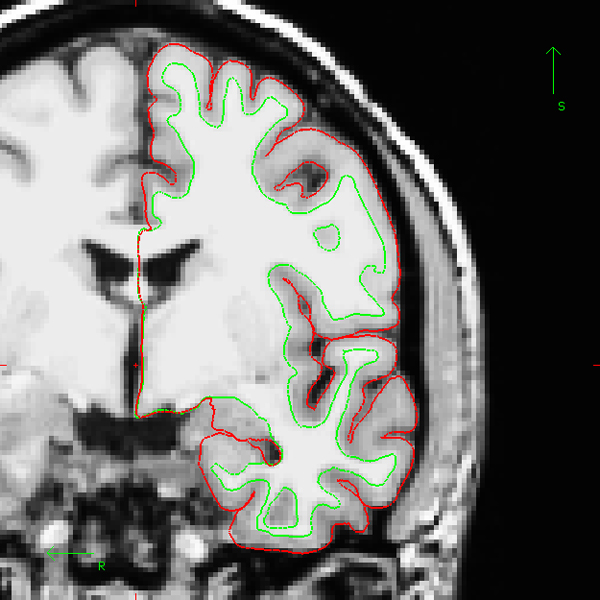
Figure 2. Example of adequate cortical surface reconstruction (one coronal section). After the end of the reconstruction process, the cortical surfaces should be accurately verified across the entire cerebral volume. The inner cortical surface (denoted white surface, in green on the image) should precisely follow the gray-white interface. The outer cortical surface (i.e. gray-CSF interface, denoted pial surface, here in red) should not include any piece of vessel or membrane. Of note, the example presented here uses the "bert" subject distributed along with the FreeSurfer package.
Figure 2B. Example of adequate cortical surface reconstruction (full volume). This animated gif image shows the cortical surface of the left hemisphere of the "bert" subject on each coronal section, as seen by scrolling from the most frontal to the most occipital coronal sections with FreeSurfer. View movie

Figure 3. Example of outer surface computed as a part of the lGI process (one coronal section). The first step in the lGI computation is the creation of an outer surface enveloping the hemisphere. This surface (denoted ?h.pial_outer_smoothed in FreeSurfer) can be checked using tkmedit. Here, the "bert" subject distributed with FreeSurfer is used as an example.
Figure 3B. Example of outer surface computed as a part of the lGI process (full volume). This animated gif image shows the outer surface of the left hemisphere on each coronal section, as seen by scrolling from the most frontal to the most occipital coronal sections with tkmedit in FreeSurfer. View movie
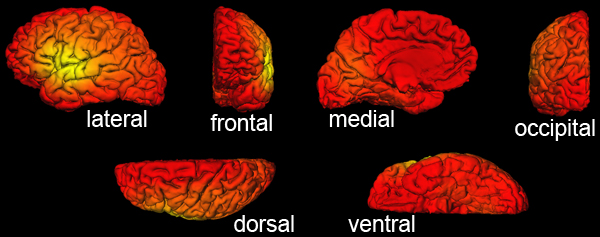
Figure 4. Example of correct lGI output as viewed with FreeSurfer. Different orientations of the cortical surface of the "bert" subject with lGI values overlaid. The color-code is the default "heat" overlay as seen with tksurfer in FreeSurfer. Using a minimum threshold of 1, all the vertices must be colored and no cortical area should appear in gray. Of note, the color overlay can be modified using the option "Configure Overlay" in tksurfer, where the minimum and maximum values, as well as the histogram of the overall distribution of the lGI can also be checked.
Discussion
The protocol above describes how to measure local Gyrification Index based on cerebral T1-weighted MRI and conduct statistical group comparisons. Our method has been specifically designed to localize early disruption in the cortical expansion process and as such is of particular interest in many neurodevelopmental or psychiatric conditions. Examples of group comparisons in clinical samples can be found in publications by our group1,12 or by others13-16. The process is fully automated and requ...
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported by the National Center of Competence in Research (NCCR) "SYNAPSY - The Synaptic Bases of Mental Diseases" financed by the Swiss National Science Foundation (n° 51AU40_125759). Development of the local Gyrification Index was supported by grants from the Swiss National Research Fund to Dr. Marie Schaer (323500-111165) and to Dr. Stephan Eliez (3200-063135.00/1, 3232-063134.00/1, PP0033-102864 and 32473B-121996) and by the Center for Biomedical Imaging (CIBM) of the Geneva-Lausanne Universities and the EPFL, as well as the foundations Leenaards and Louis-Jeantet. Support for the development of FreeSurfer software was provided in part by the National Center for Research Resources (P41-RR14075, and the NCRR BIRN Morphometric Project BIRN002, U24 RR021382), the National Institute for Biomedical Imaging and Bioengineering (R01 EB001550, R01EB006758), the National Institute for Neurological Disorders and Stroke (R01 NS052585-01) as well as the Mental Illness and Neuroscience Discovery (MIND) Institute, and is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Additional support was provided by The Autism & Dyslexia Project funded by the Ellison Medical Foundation.
Materials
| Name | Company | Catalog Number | Comments |
| Material: a Unix or Mac workstation with a processor of 2GHz or faster and a minimum of 4GB of RAM, with FreeSurfer installed (http://surfer.nmr.mgh.harvard.edu/fswiki, preferably the latest version, but no older than version 4.0.3). In order to compute the local Gyrification Index, MATLAB is also required (http://www.mathworks.com/) along with the Image Processing Toolbox. | |||
| Data: A sample of good quality (high-resolution, high contrast) cerebral MRI T1-weighted dataset. Your group of subjects must be preferably matched for age and gender. Given the normal inter-individual variability in cerebral morphology, the number of subjects in each group should be sufficient to identify an existing group difference (the more - the better). A reasonable minimum sample size would be around 20 subjects per group (although you can probably go for less if the intensity of changes is large and if your groups are tightly matched for gender and age). | |||
| FreeSurfer | Martinos Center for Biomedical Imaging, MGH | Version newer than 4.0.3 | |
| Matlab | Mathworks | Image Processing Toolbox | |
References
- Schaer, M. A surface-based approach to quantify local cortical gyrification. IEEE. Trans. Med. Imaging. 27, 161-170 (2008).
- Mangin, J. F., Jouvent, E., Cachia, A. In-vivo measurement of cortical morphology: means and meanings. Curr. Opin. Neurol. 23, 359-367 (2010).
- Mangin, J. F. A framework to study the cortical folding patterns. Neuroimage. 23, S129-S138 (2004).
- Luders, E. A curvature-based approach to estimate local gyrification on the cortical surface. Neuroimage. 29, 1224-1230 (2006).
- Zilles, K., Armstrong, E., Schleicher, A., Kretschmann, H. J. The human pattern of gyrification in the cerebral cortex. Anat. Embryol. (Berl). 179, 173-179 (1988).
- Rakic, P. Specification of cerebral cortical areas. Science. 241, 170-176 (1988).
- Toro, R. Brain size and folding of the human cerebral cortex. Cereb. Cortex. 18, 2352-2357 (2008).
- Fischl, B., Sereno, M. I., Dale, A. M. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 9, 195-207 (1999).
- Dale, A. M., Fischl, B., Sereno, M. I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 9, 179-194 (1999).
- Genovese, C. R., Lazar, N. A., Nichols, T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 15, 870-878 (2002).
- Desikan, R. S. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 31, 968-980 (2006).
- Schaer, M. Congenital heart disease affects local gyrification in 22q11.2 deletion syndrome. Dev. Med. Child. Neurol. 51, 746-753 (2009).
- Palaniyappan, L., Mallikarjun, P., Joseph, V., White, T. P., Liddle, P. F. Folding of the Prefrontal Cortex in Schizophrenia: Regional Differences in Gyrification. Biol. Psychiatry. , (2011).
- Zhang, Y. Decreased gyrification in major depressive disorder. Neuroreport. 20, 378-380 (2009).
- Juranek, J., Salman, M. S. Anomalous development of brain structure and function in spina bifida myelomeningocele. Dev. Disabil. Res. Rev. 16, 23-30 (2010).
- Zhang, Y. Reduced cortical folding in mental retardation. AJNR. Am. J. Neuroradiol. 31, 1063-1067 (2010).
- Kuperberg, G. R. Regionally localized thinning of the cerebral cortex in schizophrenia. Archives of general psychiatry. 60, 878-888 (2003).
- Milad, M. R. Thickness of ventromedial prefrontal cortex in humans is correlated with extinction memory. Proc. Natl. Acad. Sci. U. S. A. 102, 10706-10711 (2005).
- Rauch, S. L. A magnetic resonance imaging study of cortical thickness in animal phobia. Biol. Psychiatry. 55, 946-952 (2004).
- Fjell, A. M. Selective increase of cortical thickness in high-performing elderly--structural indices of optimal cognitive aging. Neuroimage. 29, 984-994 (2006).
- Walhovd, K. B. Regional cortical thickness matters in recall after months more than minutes. Neuroimage. 31, 1343-1351 (2006).
- Gold, B. T. Differing neuropsychological and neuroanatomical correlates of abnormal reading in early-stage semantic dementia and dementia of the Alzheimer type. Neuropsychologia. 43, 833-846 (2005).
- Salat, D. H. Thinning of the cerebral cortex in aging. Cereb. Cortex. 14, 721-730 (2004).
- Schaer, M., Eliez, S. Contribution of structural brain imaging to our understanding of cortical development process. European Psychiatry Reviews. 2, 13-16 (2009).
- Shaw, P. Neurodevelopmental trajectories of the human cerebral cortex. J. Neurosci. 28, 3586-3594 (2008).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved