A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Isolation of Cardiomyocyte Nuclei from Post-mortem Tissue
In This Article
Summary
Cardiac nuclei are isolated via density sedimentation and immunolabeled with antibodies against pericentriolar material 1 (PCM-1) to identify and sort cardiomyocyte nuclei by flow cytometry.
Abstract
Identification of cardiomyocyte nuclei has been challenging in tissue sections as most strategies rely only on cytoplasmic marker proteins1. Rare events in cardiac myocytes such as proliferation and apoptosis require an accurate identification of cardiac myocyte nuclei to analyze cellular renewal in homeostasis and in pathological conditions2. Here, we provide a method to isolate cardiomyocyte nuclei from post mortem tissue by density sedimentation and immunolabeling with antibodies against pericentriolar material 1 (PCM-1) and subsequent flow cytometry sorting. This strategy allows a high throughput analysis and isolation with the advantage of working equally well on fresh tissue and frozen archival material. This makes it possible to study material already collected in biobanks. This technique is applicable and tested in a wide range of species and suitable for multiple downstream applications such as carbon-14 dating3, cell-cycle analysis4, visualization of thymidine analogues (e.g. BrdU and IdU)4, transcriptome and epigenetic analysis.
Protocol
1. Isolation of the Cardiac Nuclei
- Coat ultracentrifuge tubes (Beckman Centrifuge Tubes #363664) with 10 ml of 1%BSA/PBS coating solution. Cap the tubes and let them rotate for 30 min in a tube rotator. Remove the coating solution and let the centrifuge tubes air dry (one tube per mouse heart is required for the analysis of single mouse hearts, alternately up to 5 mouse hearts or 1 g of heart tissue from a different species (e.g. human) can be processed in one tube).
- All following steps should be performed on ice. Dissect the left ventricle from fresh or snap-frozen mouse heart with a scalpel. Note, this protocol is optimized for mouse heart but can also be adapted to rat or human heart. Alternatively, use up to 1 g of heart tissue from a different species.
- Trim the specimen into small cubicles with a scalpel.
- Transfer the tissue pieces into a 50 ml Falcon tube filled with 15 ml of lysis buffer.
- Homogenize the heart tissue with a T-25 Ultra-Turrax probe homogenizer (IKA) at 24,000 rpm for 10 sec.
- Dilute the homogenate with equal volume of lysis buffer to 30 ml.
- Use a glass douncer (40 ml) to further homogenize the tissue and free the nuclei. Perform eight strokes with a large clearance pestle.
- Pass the crude nuclei isolate through a 100 μm and 70 μm nylon mesh cell strainer (BD Biosciences), consecutively.
- Spin down the crude nuclei isolate in a refrigerated centrifuge (4 °C) at 700 x g for 10 min.
- Remove the supernatant carefully by inverting the tubes and wipe the inside of the tube with paper towel. Be careful not to disturb the nuclei pellet.
- Dissolve the crude nuclei isolate in 5 ml of sucrose buffer by pipetting the solution several times up and down. Add a further 25 ml of sucrose buffer to the dissolved pellet.
- Add 10 ml of freshly prepared sucrose buffer to the coated ultracentrifuge tube (see step 1.1).
- Carefully overlay the added 10 ml of sucrose buffer with the resuspended nuclei pellet dissolved in sucrose buffer from step 1.9.
- Balance the centrifuge tubes before placing them into a JS13.1 free swinging rotor and place the rotor into a high-speed centrifuge (Beckman Avanti S-25).
- Spin the nuclei sample at 13,000 x g at 4 °C for 60 min.
- When the spin has completed, remove the tubes carefully from the rotor and discard the supernatant by inverting the tubes and wiping the remaining debris from the inside of the tubes with paper towel.
- Dissolve the nuclei pellet in 1 ml of nuclei storage buffer (NSB plus buffer). Note: NSB plus contains 1.5 mM spermine as a DNA stabilizer.
- Proceed with step 2.1, immunostaining of cardiomyocyte nuclei.
2. Immunostaining for Flow Cytometry
- Prepare the negative control for immunostaining. Take an aliquot of 20 μl out of the nuclei sample and add 980 μl of NSB plus buffer.
- Add anti-pericentriolar material 1 antibody (rabbit anti-PCM-1, Atlas Antibodies) to the nuclei sample in a dilution of 1:500 to immunolabel cardiomyocyte nuclei. Add the isotype antibody in the same dilution as the anti-PCM-1 antibody to the negative control, prepared in step 2.1.
- Incubate negative control and sample tube at 4 °C overnight.
- Wash negative control and sample at least once with NSB plus buffer (spin down tubes in a refrigerated centrifuge (4 °C) at 700 x g for 10 min. Discard supernatant and dissolve the nuclei pellet in 1 ml of NSB plus buffer).
- Add anti-rabbit fluorescent secondary antibody (FITC or APC) to negative control and sample tube in a dilution of 1:1000.
- Incubate negative control and sample tube at 4 °C for 1 h.
- Wash negative control and sample at least once with NSB plus buffer (spin down tubes in a refrigerated centrifuge (4 °C) at 700 x g for 10 min. Discard supernatant and dissolve the nuclei pellet in 1 ml of NSB plus buffer).
- Proceed with the flow cytometric analysis and sorting.
3. Flow Cytometry
- Coat nuclei collection tubes (Falcon 15 ml) with 1%BSA/PBS solution before you start flow cytometry sorting as described in step 1.1.
- Filter the sample and the negative control through a 30 μm cell strainer and load first the negative control to the flow cytometer (BD Influx). Define the first and second gate to define nuclei and singlets (single nuclei), based on forward scatter (FSC), forward scatter pulse width (FS pulse width) and side scatter (SSC) (Fig. 2a and b). Adding a DNA stain (DRAQ5 (1:500)) to the sample can help to identify the nuclei population initially.
- Load the immunolabeled sample and define the third gate to isolate cardiomyocyte nuclei (PCM-1-positve) from non-cardiomyocyte nuclei (PCM-1-negative). Start the sorting (Fig. 2c and d).
Optional: In order to analyze the nuclear DNA content (ploidy) and to perform cell cycle analysis add an appropriate DNA stain to the nuclei (e.g. Hoechst 33342 or DRAQ5) (Fig. 2e).
- After flow cytometry sorting, place the nuclei on ice and re-analyze to determine the sorting purity (Fig. 3a and b).
- Spin down the sorted nuclei in the collection tubes at 1500 x g in a refrigerated centrifuge for 15 min.
- Dissolve the nuclei pellet in a buffer compatible with the downstream application.
4. Representative Results
Nuclei morphology and integrity can be assessed by DNA stains and visualized by microscopy (Fig. 1). Successful PCM-1 labeling can be assessed by epifluorescence microscopy and by flow cytometry (Fig. 1 and Fig. 2c and d). PCM-1-positive and negative populations should be well separated from each other (Fig. 2c and d). In murine left ventricle about 30% of all nuclei should be cardiomyocyte nuclei (Fig. 2d). Sorting purity can be assessed by re-analyzing the sorted nuclei (Fig. 3a and b). Both nuclei populations should have a sorting purity exceeding 95%.
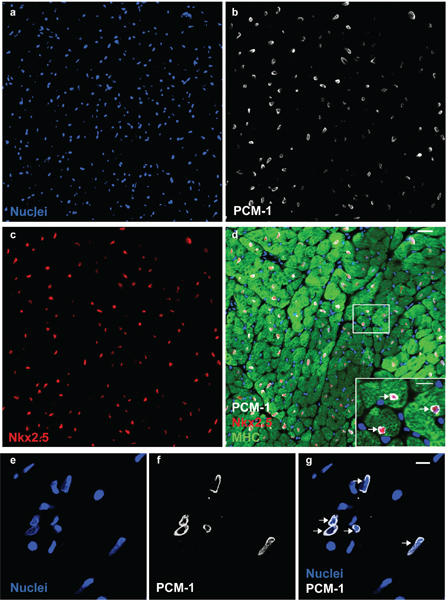
Figure 1. PCM-1 identifies cardiomyocyte nuclei. Cardiac nuclei (a) are stained with antibodies to PCM-1 (b) and to Nkx2.5 (c) in an adult mouse heart. (d) PCM-1-labeled nuclei are surrounded by cardiomyocyte cytoplasm (myosin heavy chain (MHC)) and express the transcription factor Nkx2.5, documenting the accurate identification of cardiomyocyte nuclei by PCM-1 staining (scale bars 20 μm and 10 μm (d, inset)). (e) Cardiac nuclei isolates visualized with the DNA stain DRAQ5. (f and g) Cardiomyocyte nuclei are labeled with antibodies against PCM-1 (scale bar 10 μm). Note, the epinuclear staining pattern of PCM-1 in myocyte nuclei in tissue section and in isolated nuclei (arrows).
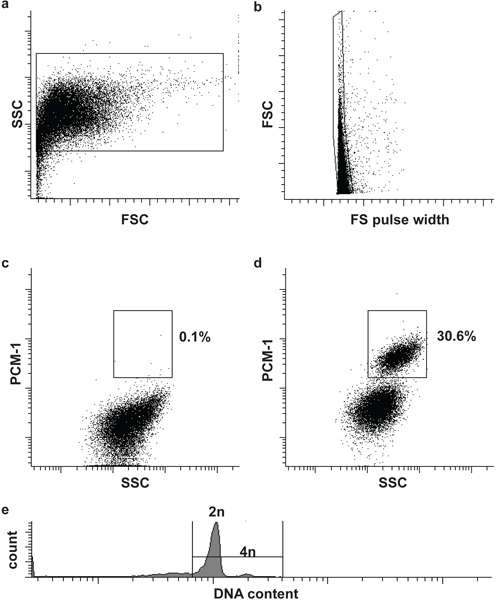
Figure 2. Flow cytometric sorting of cardiomyocyte nuclei. (a) Cardiac nuclei are identified by forward scatter (FSC) and side scatter (SSC). (b) A second gate identifies single nuclei by FSC and FS pulse width5. (c, d) Fluorescent gating allows the separation of cardiomyocyte nuclei (PCM-1-positive) and non-cardiomyocyte (PCM-1-negative) nuclei from heart tissue. (e) Mouse cardiomyocyte are mostly (>80%) diploid (2n), only a small subset is tetraploid (4n)6. Note, human cardiomyocytes contain a higher frequency of polyploidy nuclei (>2n)7,8.

Figure 3. Purity analysis of sorted cardiomyocyte and non-cardiomyocyte nuclei. Re-analysis of sorted non-cardiomyocyte (a) and cardiomyocyte nuclei (b). Both populations show a sorting purity exceeding 99%.
Discussion
Accurate identification of cardiomyocyte nuclei is crucial for the analysis of regenerative processes in the myocardium2,3. Conventional techniques to isolate cardiomyocytes from fresh tissue are mainly based on enzymatic digestion of extracellular matrix proteins and the subsequent purification from interstitial cells by low speed centrifugation. Further purification of living cardiomyocytes from embryonic stem cells (ESC) can be performed by immunolabeling with surface markers such as SIRPA9 or mi...
Disclosures
No conflicts of interest declared.
Acknowledgements
We like to acknowledge Marcelo Toro for the assistance with flow cytometry. This study was supported by Swedish Heart- and Lung Foundation, EU Commission FP7 "CardioCell", Swedish Research Council, AFA insurances and ALF. O.B. was supported by Deutsche Forschungsgemeinschaft.
Materials
| Name | Company | Catalog Number | Comments |
| 1. Lysis Buffer | |||
| Name of the reagent | |||
| 0.32 M sucrose | |||
| 10 mM Tris-HCl (pH = 8) | |||
| 5 mM CaCl2 | |||
| 5 mM magnesium acetate | |||
| 2.0 mM EDTA | |||
| 0.5 mM EGTA | |||
| 1 mM DTT | |||
| 2. Sucrose buffer | |||
| Name of the reagent | |||
| 2.1 M sucrose | |||
| 10 mM Tris-HCl (pH = 8) | |||
| 5 mM magnesium acetate | |||
| 1 mM DTT | |||
| 3. Nuclei storage buffer (NSB plus) | |||
| Name of the reagent | |||
| 0.44 M sucrose | |||
| 10 mM Tris-HCl (pH = 7.2) | |||
| 70 mM KCl | |||
| 10 mM MgCl2 | |||
| 1.5 mM spermine | |||
| Isotype rabbit IgG- ChIP Grade, #ab37415 | Abcam | ||
| Rabbit anti-PCM-1 antibody, #HPA023374 | Atlas Antibodies | ||
| Donkey sec. antibody, anti-rabbit Alexa 488 Fluor, #A-21206 or equivalent sec. fluorescent antibody | Life Technologies | ||
| DRAQ5 | Biostatus | ||
| cell strainers 30 μm, 70 μm and 100 μm | BD Biosciences | ||
| Glass douncer (40 ml) and pestle "L" | VWR (Wheaton Industries Inc.) | ||
| T-25 Ultra-Turrax Homogenizer | IKA Germany | ||
| Dispersing tool S25 N-18 G | IKA Germany | ||
| Beckman Avanti Centrifuge | Beckman Coulter | ||
| Falcon Tubes 15 ml and 50 ml | VWR | ||
| Beckman Centrifuge Tubes #363664 | Beckman Coulter | ||
| JS13.1 free swinging rotor | Beckman Coulter | ||
| Influx cytometer | Beckman Coulter | ||
| Tube Rotator | VWR |
References
- Ang, K. L. Limitations of conventional approaches to identify myocyte nuclei in histologic sections of the heart. American journal of physiology. Cell physiology. , 298-1603 (2010).
- Bergmann, O. Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Experimental cell research. 327, 188-194 (2011).
- Bergmann, O. Evidence for Cardiomyocyte Renewal in Humans. Science. 324, 98-102 (1126).
- Walsh, S. Cardiomyocyte cell cycle control and growth estimation. Cardiovascular Research. , 1-31 (2010).
- Spalding, K., Bhardwaj, R. D., Buchholz, B., Druid, H., Frisén, J. Retrospective birth dating of cells in humans. Cell. 122, 133-143 (2005).
- Adler, C. P., Friedburg, H., Herget, G. W., Neuburger, M., Schwalb, H. Variability of cardiomyocyte DNA content, ploidy level and nuclear number in mammalian hearts. Virchows Arch. 429, 159-164 (1996).
- Herget, G. W., Neuburger, M., Plagwitz, R., Adler, C. P. DNA content, ploidy level and number of nuclei in the human heart after myocardial infarction. Cardiovascular Research. 36, 45-51 (1997).
- Adler, C. P., Friedburg, H. Myocardial DNA content. ploidy level and cell number in geriatric hearts: postmortem examinations of human myocardium in old age. Mol. Cell Cardiol. 18, 3953-39 (1986).
- Dubois, N. C. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nature. 29, 1011-1018 (2011).
- Hattori, F. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nature Methods. 7, 61-66 (2010).
- Fransioli, J. Evolution of the c-kit-Positive Cell Response to Pathological Challenge in the Myocardium. Stem Cells. 26, 1315-1324 (2008).
- Elliott, D. A. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nature Methods. 8, 1037-1040 (2011).
- Laflamme, M. A. Evidence for Cardiomyocyte Repopulation by Extracardiac Progenitors in Transplanted Human Hearts. Circulation Research. 90, 634-640 (2002).
- Srsen, V., Fant, X., Heald, R., Rabouille, C., Merdes, A. Centrosome proteins form an insoluble perinuclear matrix during muscle cell differentiation. BMC cell biology. 10, 28 (2009).
- Spoelgen, R. A novel flow cytometry-based technique to measure adult neurogenesis in the brain. Journal of neurochemistry. 119, 165-175 (2011).
- Soonpaa, M. H., Kim, K. K., Pajak, L., Franklin, M., Field, L. J. Cardiomyocyte DNA synthesis and binucleation during murine development. The American journal of physiology. 271, H2183-H2189 (1996).
- Olivetti, G. Aging, cardiac hypertrophy and ischemic cardiomyopathy do not affect the proportion of mononucleated and multinucleated myocytes in the human heart. J Mol Cell Cardiol. 28, 1463-1477 (1996).
- Okada, S. Flow cytometric sorting of neuronal and glial nuclei from central nervous system tissue. Journal of cellular physiology. 226, 552-558 (2011).
- Matevossian, A., Akbarian, S. Neuronal Nuclei Isolation from Human Postmortem Brain Tissue. J. Vis. Exp. (20), e914 (2008).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved