A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Use of pHluorin to Assess the Dynamics of Axon Guidance Receptors in Cell Culture and in the Chick Embryo
* These authors contributed equally
In This Article
Summary
We describe here the use of a pH-sensitive green fluorescent protein variant, pHluorin, to study the spatio-temporal dynamics of axon guidance receptors trafficking at the cell surface. The pHluorin-tagged receptor is expressed both in cell culture and in vivo, using electroporation of the chick embryo.
Abstract
During development, axon guidance receptors play a crucial role in regulating axons sensitivity to both attractive and repulsive cues. Indeed, activation of the guidance receptors is the first step of the signaling mechanisms allowing axon tips, the growth cones, to respond to the ligands. As such, the modulation of their availability at the cell surface is one of the mechanisms that participate in setting the growth cone sensitivity. We describe here a method to precisely visualize the spatio-temporal cell surface dynamics of an axon guidance receptor both in vitro and in vivo in the developing chick spinal cord. We took advantage of the pH-dependent fluorescence property of a green fluorescent protein (GFP) variant to specifically detect the fraction of the axon guidance receptor that is addressed to the plasma membrane. We first describe the in vitro validation of such pH-dependent constructs and we further detail their use in vivo, in the chick spinal chord, to assess the spatio-temporal dynamics of the axon guidance receptor of interest.
Introduction
During their navigation, axons integrate multiple environmental cues that guide them towards their target. These cues activate guidance receptors at the surface of axon terminals, the growth cones, which in turn initiate an appropriate signaling pathway. Thus, the temporal and spatial regulation of the cell surface distribution of the receptors is critical to set the sensitivity of the growth cone1. In this context, midline crossing by commissural axons is an excellent model to investigate the regulation of receptor cell surface levels. In the developing spinal cord, commissural axons are initially attracted towards the ventral floor plate where they cross the midline. After crossing, they lose their responsiveness to the floor plate attractants and gain response to floor plate repellents so that they can exit the floor plate and navigate towards their final destination in the contralateral side of the nervous system2,3. Regulation of receptor availability at the growth cone surface is one of the mechanisms underlying the switch of responsiveness to midline cues4,5. Thus, selective monitoring of the receptors present at the plasma membrane of growth cones is of prime importance. We describe here a method based on the pH-dependent fluorescence property of a green fluorescent protein (GFP) variant to specifically visualize the axon guidance receptors that are addressed to the plasma membrane in vitro and in vivo, in the developing chick spinal cord.
Rothman and colleagues engineered by point mutations pH-sensitive variants of GFP including the ecliptic pHluorin6. Ecliptic pHluorin has the property of being nonfluorescent when exposed to acidic pH (<6), while being fluorescent at neutral pH. This allows distinguishing nonfluorescent receptors localized in intracellular acidic compartments (i.e. endosomes, trafficking vesicles) from fluorescent receptors incorporated to the plasma membrane and thus exposed to the extracellular neutral pH7. We took advantage of this to monitor the plasma membrane localization of plexinA1, an axon guidance receptor mediating the growth cone response to the midline repellent semaphorin 3B5 (Figure 1A). We describe here the in vitro characterization of a pHluorin-plexinA1 construct, along with in ovo electroporation8-10 of this construct in the developing chick spinal cord followed by the microscopic analysis of cryosections which enable to follow the axon guidance receptor dynamics in vivo with both spatial and temporal resolutions.
Protocol
1. Cloning Strategy to Tag PlexinA1 Receptor with pHluorin
- Choose an appropriate expression vector as a backbone (e.g. the mouse receptor plexinA1 expressing vector, a kind gift of Dr. Andreas Puschel11).
Note: This plexinA1 vector was engineered to achieve efficient HA- or VSV-tagged receptor insertion in the plasma membrane. - Amplify by PCR the ecliptic pHluorin coding sequence using the adequate plasmid as a template (e.g. pHluorin-tagged GABA A receptor, a kind gift of Dr. Jacob2). If necessary, add a restriction site to the 5’ end of the primer in order to facilitate the cloning step in the backbone.
- Insert the pHluorin sequence in frame between the signal peptide and the receptor coding sequence using restriction sites (e.g. NruI/Kpn2I restriction sites as described in Figure 1B).
Note: Because the signal peptide that ensures the correct targeting of the receptors is cleaved, the pHluorin should be placed after it. This warrants the recognition of the signal peptide and prevents cleavage of the pHluorin from the receptor of interest. - Sequence the constructs obtained to ensure that no mutation was introduced by PCR.
2. Characterization of pHluorin-tagged Receptor In vitro in COS7 Cells
The ability of the fusion protein to reach the plasma membrane and its reversible loss of fluorescence as pH is lowered can be confirmed using the following procedure.
- Day 1. Plate 1.5 x 105 COS7 cells in a glass-bottom 35 mm dish in 2 ml of complete Dulbecco’s Modified Eagle Medium (DMEM - 10% Fetal Bovine Serum - 1 mM sodium pyruvate - 25 U/ml penicillin/stretomycin - 2.5 µg/ml Amphotericin B - pH 7.4).
- Day 2. Transfect cells:
Note: Cells should be 70-80% confluent.- Prepare 200 µl NaCl 150 mM and add 3 µg DNA i.e. the vector encoding pHluorin-tagged receptor. Gently vortex and spin down briefly.
Note: A map of the pHluorin-plexinA1 vector used in these experiments is shown in Figure 1B. - Add 10 µl of transfection reagent (or the appropriate amount of the reagent used). Vortex immediately.
- Incubate 10 min at RT.
- Add 200 µl of the transfection reagent/DNA mix to the cells.
- Move the plate gently to achieve repartition of the mix and place the cells back to the 37 °C incubator.
- Prepare 200 µl NaCl 150 mM and add 3 µg DNA i.e. the vector encoding pHluorin-tagged receptor. Gently vortex and spin down briefly.
- Day 3. Remove transfection medium and replace it by 2 ml of fresh complete DMEM.
- Day 4. Perform live cell imaging of COS7 pHluorin-plexinA1 transfected cells:
- Prepare two aliquots of DMEM complete medium and adjust pH to 3.5 and to 9.5, respectively.
Note: For one 35 mm plate, 1.5 ml of each solution is needed to perform the experiment. - Remove cell medium and replace it by 1 ml of DMEM complete medium pH 7.4.
- Prepare a 5 ml syringe with an appropriate type of tubing in order to inject various components directly into the cell culture medium without opening the microscope chamber.
- Use a module that allows maintenance of a 37 °C, 5% CO2 humid working atmosphere.
Note: An alternate approach to the use of a CO2 chamber is to use HEPES-buffered medium (usually in the range of 10-25 mM according to the cell type). - Place cells in the chamber and adjust the tubing and the syringe.
Note: The microscope should be equilibrated before starting to avoid mechanical drift during the recording. - Open the imaging software and select the multidimensional acquisition program.
- Find transfected COS7 cells with the 40X objective and mark position in the software for each of them.
- Configure Z stack in order to have a 15 µm depth acquisition (the focus can change when adding media in the plate).
- Set up exposure for the GFP filter and the Phase.
- Configure acquisition timing.
Note: For the whole experiment with 5 fields of interest, acquisition every 20 sec for 10 min should be sufficient. - Start acquisition and take 5 control images in DMEM pH 7.4 medium.
- Pause acquisition, inject 1.25 ml of pH 3.5 complete DMEM to achieve a pH of 5.5 in the culture medium, mark event in the software and resume acquisition for 5 more time points.
Note: Green fluorescence should progressively disappear. - Pause acquisition, inject 1.2 ml of pH 9.5 complete DMEM to achieve a pH 7.4 in the culture medium, mark event in the software and resume acquisition for 5 more time points.
Note: Green fluorescence should reappear at the plasma membrane. - Analyze images.
Figure 2 shows representative images obtained with such a protocol with the pHluorin-plexinA1 construct.
- Prepare two aliquots of DMEM complete medium and adjust pH to 3.5 and to 9.5, respectively.
3. In ovo Electroporation of pHluorin-plexinA1 Construct
- Handling of eggs before electroporation:
- Store fertilized eggs in a fridge at 14 °C up to one week prior to incubation.
- Incubate eggs at 38.5 °C (101.3 °F) in an incubator with saturated humidity for 50-52 hr until the embryos reach stage HH1412.
Note: The eggs must be placed horizontally during incubation so that the embryo is properly positioned for electroporation, floating on the top of the yolk. HH14 stage is suitable to obtain expression of the plasmids in differentiated neurons in the spinal cord and in the dorsal root ganglion with an appropriate survival rate.
- Electroporate embryos8-10:
- Prepare electroporation:
- Prepare endotoxin-free DNA plasmids with a concentration superior to 2 µg/µl, to be able to dilute it as the desired concentration.
- Pull enough glass capillaries to inject the different DNA solutions.
- Prepare sterile PBS (-Ca2+; -Mg2+) - 100 U/ml penicillin/streptomycin and equilibrate at 38.5 °C.
- Sterilize the hood, curved scissors and fine forceps.
- Control the electrode spacing.
Note: A 4 mm space between the electrodes is generally used.
- Window the egg13 (Figure 3A):
- Use curved scissors to pierce the shell on the blunt side of the egg.
- Remove 2 ml of albumen using a 0.9 mm x 25 mm needle and a 5 ml syringe. Orient the needle vertically in order to avoid damaging the yolk sac.
- Cover the top of the egg with tape to maintain the integrity of the shell.
- Using curved scissors, pierce the shell in the middle of the tape to equalize the pressure when removing 2 ml of albumen from the egg. Then, cut a window large enough to visualize the embryo and be able to work on it.
- Add ~2 ml sterile warm PBS (-Ca2+; -Mg2+) - 100 U/ml penicillin/streptomycin to avoid dehydration of the embryo and make it more accessible to the manipulator.
- Inject DNA and electroporate the embryo
- Dilute plasmid in PBS (-Ca2+; -Mg2+) to a concentration between 0.5-2 µg/µl and add Fast green dye to reach a final 0.025% concentration. Load the DNA mix in a capillary. The use of an injector is recommended.
Note: Check that the capillary resistance is neither too large (meaning there could be difficulties when injecting embryos) nor too small (meaning that the capillary could be too big and could damage the embryo). Also, concentration of nucleic acids higher than 2 µg/µl may cause unspecific effects and needs to be controlled. - Puncture the yolk-sac and the neural tube at the caudal side with the loaded capillary. Enter the neural tube with a shallow angle and fill the lumen from the tail to the head with the DNA mix (Figure 3B).
Note: Fast green allows controlling the accuracy of the injection. - Quickly place the 4 mm platinum electrodes on either side of the neural tube at the level you wish to electroporate and apply 3 pulses at 31 V for 50 msec with 500 msec intervals (Figure 3C).
Note: Avoid placing the electrodes on the heart or on big extra embryonic vessels to avoid damaging the developing embryo. Bubbles should form on the electrodes. - With the needle, remove 2 ml of albumen to decrease the level in the shell.
- Seal the window and the blunt side hermetically with tape.
- Put the eggs back at 38.5 °C in the incubator until they reach the desired stage.
- Dilute plasmid in PBS (-Ca2+; -Mg2+) to a concentration between 0.5-2 µg/µl and add Fast green dye to reach a final 0.025% concentration. Load the DNA mix in a capillary. The use of an injector is recommended.
- Prepare electroporation:
4. Embryos Embedding and Cryosectioning
- 48 hr after electroporation, carefully harvest electroporated embryos (HH24 stage). Cut the tape and half of the chorioallantoid membrane. To prevent embryos from sinking into the yolk, position a colander under the embryo and cut the second half of the chorioallantoid membrane.
- Transfer the embryo to a dissection dish filled with ice cold PBS.
- Check electroporation efficiency by looking for fluorescence in the neural tube with a fluorescence stereo microscope.
Note : Coelectroporation of a control plasmid encoding the RFP may help to visualize the electroporated area. - Dissect embryos using a microscalpel in order to select the electroporated area of the spinal cord.
- Transfer dissected embryos to a 24-well plate and fix in pH 7.4 4% Paraformaldehyde (PFA) - Phosphate Buffer Saline (PBS), O/N at 4 °C.
Note: Fixation step is crucial to allow the stabilization of pHluorin in its “live” conformation and thus to be able to use pHluorin in fixed/permeabilized tissue. Although fixation slows drastically pH-dependent change of fluorescence, one must take into account that conformational/protonation changes can still occur after fixation. Thus, the following protocol (embedding, cryosections and observation) has to be performed within 3 days after the fixation step, with all buffers at pH 7. If fixation is not required, performing observation on live tissue sections is recommended. - Remove 4% PFA and wash embryos in pH 7.4 PBS.
- Incubate embryos in PBS-15% sucrose and keep at 4 °C until the embryos sink.
- Incubate fixed embryos in pH 7.4 7.5% gelatin- 15% sucrose for 45 min at 37 °C so embryos are completely embedded.
- Place embedding molds on ice and add 400 µl of pH 7.4 7.5% gelatin- 15% sucrose to achieve a solid 2 mm base.
- Aspirate the embedded embryo with a cut tip and place the embryo on the solid gelatin base.
- Cover with pH 7.4 7.5% gelatin- 15% sucrose and position the embryo with forceps before the gelatin solidifies.
- Once the gelatin is solid, prepare a -40 °C isopropanol bath (use dry ice or liquid nitrogen) and freeze the gelatin block for 5 min.
- Keep the frozen blocks at -80 °C.
- Place the frozen block at -20 °C for 1 hr.
- Remove the mold and fix the block on a chuck with a polyethylene glycol medium.
- Once the block is tightly fixed, place the chuck in the cryostat system.
Note: Use coated slides to avoid tissue loss during staining. - Perform serial cryosections (20 µm cryosections are usually performed).
- Let the cryosections dry for 15 min at RT.
Note: cryosections should be protected from unnecessary light exposure in order to avoid bleaching of the GFP fluorescence.
5. Microscopic Analysis of Cryosections
- Rehydrate cryosections in pH 7.4 PBS at RT for 10 min.
Note: If needed, nuclei can be stained with Hoechst. - Use a 0.5 µg/ml Hoechst solution in PBS and incubate cryosections for 15 min.
- Rinse the slides 3x with pH 7.4 PBS for 5 min.
- Proceed to slide mounting. A pH 7.4 (or more basic) polyvinyl alcohol mounting solution that hardens O/N can be used: carefully position the coverslip to avoid the formation of air bubbles between the slide and the coverslip.
- Let the mounting medium harden O/N at 4 °C in the dark.
- Use an inverted confocal microscope to precisely visualize the pHluorin-plexinA1 fusion protein: perform z-stack at the optimal pinhole and optical resolution and use a 20X (NA 0.75) or 40X (NA 1.3) lenses.
Note: pHluorin is detectable with the same parameters used to detect GFP (i.e. emission peak at 509 nm). Wavelength excitation and detection filter settings are optimally defined by the imaging software. Hoechst is detected between 425-460 nm (excitation is at 405 nm), GFP or pHluorin is detected between 485-545 nm (excitation is at 473 nm) and RFP is detected between 575-675 nm (excitation is at 559 nm).
Representative images of pHluorin-plexinA1 and eGFP expression in the chick embryo spinal cord are shown in Figure 4.
Results
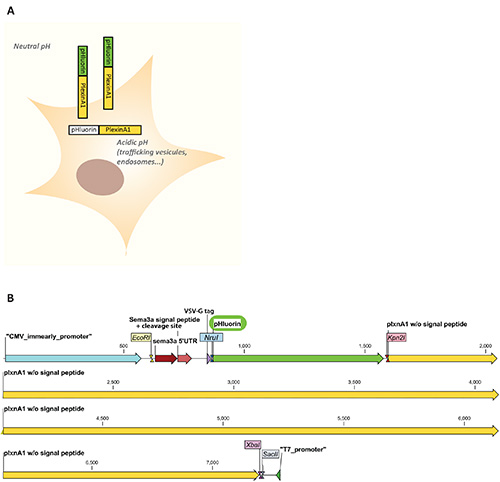
Figure 1. A. Scheme of the pHluorin-plexinA1 fluorescence properties in a cellular context. PHluorin is nonfluorescent in intracellular compartments where the pH is acidic (<6) such as in trafficking vesicules or in endosomes and is fluorescent when exposed to the extracellular medium where th...
Discussion
This protocol provides a step-by-step procedure to follow the dynamics of an axon guidance receptor both in cell culture and in the developmental context of the chick embryo spinal cord.
To design a de novo pHluorin tagged protein, two points need to be considered regarding the cloning strategy. First, the pHluorin tag should be exposed to the lumen of the acidic endosomes, and consequently, to the extracellular compartment in order to visualize the plasma membrane receptor pool. Thus...
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Homaira Nawabi, Frederic Moret and Isabelle Sanyas for their help. This work is supported by CNRS, Association Francaise contre les Myopathies (AFM), ANR YADDLE, Labex DevWeCan, Labex Cortex, ERC YODA to V.C.; C.D-B and A.J are supported by a La Ligue contre le cancer and Labex DevWeCan fellowships, respectively.
Materials
| Name | Company | Catalog Number | Comments |
| COS7 cells | ATCC | CRL-1651 | |
| DMEM GlutaMAX | GIBCO | 61965-026 | |
| Sodium pyruvate | GIBCO | 11360-039 | |
| Amphotericin B | Sigma | A2942 | |
| Fetal bovine serum | GIBCO | 10270-106 | |
| Penicillin/Streptomycin | GIBCO | 15140-122 | |
| Exgen500 reagent | Euromedex Fermentas | ET0250 | |
| PBS -Ca2+ -Mg2+ | GIBCO | 14190-094 | |
| Fast green dye | Sigma | F7252 | |
| 32% Paraformaldehyde aqueous solution | Electron Microscopy | 15714-S | Dilute extemporaneously in PBS to achieve a 4% solution |
| Gelatin from cold water fish skin | Sigma | G7041 | |
| Sucrose | Sigma | S0389 | |
| Cryomount | Histolab | 00890 | |
| Hoechst 34580 | Invitrogen | H21486 | |
| Mowiol 4-88 | Fluka | 81381 | |
| Consumables | |||
| Bottom-glass 35 mm dish | MatTek | P35G-1.5-14-C | |
| 5 ml Syringe | Terumo | SS-05S | |
| Needles 0.9 mm x 25 mm | Terumo | NN-2025R | |
| Capillaries | CML | PP230PO | capillaries are stretched manually in the flame |
| Superfrost Plus Slides | Thermo Scientific | 4951PLUS | |
| Material | |||
| Curved scissors | FST | 129-10 | |
| Microscalpel | FST | 10316-14 | |
| Forceps | FST | Dumont #5 REF#11254 | |
| Equipment/software | |||
| Time lapse microscope | Zeiss | Observer 1 | |
| Temp module S | PECON for Zeiss | ||
| CO2 module S | PECON for Zeiss | ||
| Metamorph software | Metamorph | ||
| Eggs incubator | Sanyo | MIR154 | |
| Electroporator apparatus | Nepa Gene CO., LTD | CUY21 | |
| Electrodes | Nepa Gene CO., LTD | CUY611P7-4 | 4 mm platinum electrodes |
| Fluorescence stereomicroscope | LEICA | MZ10F | |
| Cryostat | MICROM | HM550 | |
| Confocal microscope | Olympus | FV1000, X81 | |
| Fluoview software | Olympus | ||
| CLC Main Workbench software | CLC Bio |
References
- Winckler, B., Mellman, I. Trafficking guidance receptors. Cold Spring Harb. Perspect. Biol. 2, (2010).
- Jacob, T. C., et al. . J. Neurosci. 25, 10469-10478 (2005).
- Nawabi, H., Castellani, V. Axonal commissures in the central nervous system: how to cross the midline. Cell Mol. Life Sci. 68, 2539-2553 (2011).
- Keleman, K., Ribeiro, C., Dickson, B. J. Comm function in commissural axon guidance: cell-autonomous sorting of Robo in vivo. Nat. Neurosci. 8, 156-163 (2005).
- Nawabi, H., et al. A midline switch of receptor processing regulates commissural axon guidance in vertebrates. Genes Dev. 24, 396-410 (2010).
- Miesenbock, G., De Angelis, D. A., Rothman, J. E. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 394, 192-195 (1998).
- Miesenbock, G. Synapto-pHluorins: genetically encoded reporters of synaptic transmission. Cold Spring Harb. Protoc.. 2012, 213-217 (2012).
- Avraham, O., Zisman, S., Hadas, Y., Vald, L., Klar, A. Deciphering axonal pathways of genetically defined groups of neurons in the chick neural tube utilizing in ovo electroporation. J. Vis. Exp. (39), 1792-17 (2010).
- Blank, M. C., Chizhikov, V., Millen, K. J. In ovo electroporations of HH stage 10 chicken embryos. J. Vis. Exp. (9), (2007).
- Wilson, N. H., Stoeckli, E. T. In ovo electroporation of miRNA-based plasmids in the developing neural tube and assessment of phenotypes by DiI injection in open-book preparations. J. Vis. Exp. (68), (2012).
- Rohm, B., Ottemeyer, A., Lohrum, M., Puschel, A. W. Plexin/neuropilin complexes mediate repulsion by the axonal guidance signal semaphorin 3A. Mech. Dev. 93, 95-104 (2000).
- Hamburger, V., Hamilton, H. L. A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn.. 195, 231-272 (1992).
- Korn, M. J., Cramer, K. S. Windowing chicken eggs for developmental studies. J. Vis. Exp. (8), (2007).
- Alberts, P., et al. Cdc42 and actin control polarized expression of TI-VAMP vesicles to neuronal growth cones and their fusion with the plasma membrane. Mol. Biol. Cell. 17, 1194-1203 (2006).
- Perret, E., Lakkaraju, A., Deborde, S., Schreiner, R., Rodriguez-Boulan, E. Evolving endosomes: how many varieties and why. Curr. Opin. Cell Biol. 17, 423-434 (2005).
- Li, Y., et al. Imaging pHluorin-tagged receptor insertion to the plasma membrane in primary cultured mouse neurons. J. Vis. Exp. (69), (2012).
- Tojima, T., Itofusa, R., Kamiguchi, H. Asymmetric clathrin-mediated endocytosis drives repulsive growth cone guidance. Neuron. 66, 370-377 (2010).
- Matsui, A., Yoshida, A. C., Kubota, M., Ogawa, M., Shimogori, T. Mouse in utero electroporation: controlled spatiotemporal gene transfection. J. Vis. Exp. (54), 3024-30 (2011).
- Falk, J., et al. Electroporation of cDNA/Morpholinos to targeted areas of embryonic CNS in Xenopus. BMC Dev. Biol. 7 (107), (2007).
- Holzhausen, L. C., Lewis, A. A., Cheong, K. K., Brockerhoff, S. E. Differential role for synaptojanin 1 in rod and cone photoreceptors. J. Comp. Neurol. 517, 633-644 (2009).
- Shang, Y., Claridge-Chang, A., Sjulson, L., Pypaert, M., Miesenbock, G. Excitatory local circuits and their implications for olfactory processing in the fly antennal lobe. Cell. 128, 601-612 (2007).
- Dittman, J. S., Kaplan, J. M. Factors regulating the abundance and localization of synaptobrevin in the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 103, 11399-11404 (2006).
- Bozza, T., McGann, J. P., Mombaerts, P., Wachowiak, M. In vivo imaging of neuronal activity by targeted expression of a genetically encoded probe in the mouse. Neuron. 42, 9-21 (2004).
- Sankaranarayanan, S., Ryan, T. A. Real-time measurements of vesicle-SNARE recycling in synapses of the central nervous system. Nat. Cell. Biol. 2, 197-204 (2000).
- Stark, D. A., Kasemeier-Kulesa, J. C., Kulesa, P. M. Photoactivation cell labeling for cell tracing in avian development. CSH Protoc.. 2008, (2008).
- Hildick, K. L., Gonzalez-Gonzalez, I. M., Jaskolski, F., Henley, J. M. Lateral diffusion and exocytosis of membrane proteins in cultured neurons assessed using fluorescence recovery and fluorescence-loss photobleaching. J. Vis. Exp. (60), (2012).
- Hanson, G. T., et al. Green fluorescent protein variants as ratiometric dual emission pH sensors. 1. Structural characterization and preliminary application. Biochemistry. 41, 15477-15488 (2002).
- Rose, T., Schoenenberger, P., Jezek, K., Oertner, T. G. Developmental refinement of vesicle cycling at schaffer collateral synapses. Neuron. 77, 1109-1121 (2013).
- Li, Y., Tsien, R. W. pHTomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat. Neurosci. 15, 1047-1053 (2012).
- de Wit, J., Toonen, R. F., Verhage, M. Matrix-dependent local retention of secretory vesicle cargo in cortical neurons. J. Neurosci. 29, 23-37 (2009).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved