A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Detection of Exosomal Biomarker by Electric Field-induced Release and Measurement (EFIRM)
In This Article
Erratum Notice
Summary
Exosomes are microvesicular structures found within biofluids that potentially carry important disease discriminatory biomarkers. Here, a novel method is used to specifically extract exosomes and rapidly test the exosomal cargo for both RNA/protein targets following the disruption of exosomes using non-uniform electric cyclic square waves.
Abstract
Exosomes are microvesicular structures that play a mediating role in intercellular communication. It is of interest to study the internal cargo of exosomes to determine if they carry disease discriminatory biomarkers. For performing exosomal analysis, it is necessary to develop a method for extracting and analyzing exosomes from target biofluids without damaging the internal content.
Electric field-induced release and measurement (EFIRM) is a method for specifically extracting exosomes from biofluids, unloading their cargo, and testing their internal RNA/protein content. Using an anti-human CD63 specific antibody magnetic microparticle, exosomes are first precipitated from biofluids. Following extraction, low-voltage electric cyclic square waves (CSW) are applied to disrupt the vesicular membrane and cause cargo unloading. The content of the exosome is hybridized to DNA primers or antibodies immobilized on an electrode surface for quantification of molecular content.
The EFIRM method is advantageous for extraction of exosomes and unloading cargo for analysis without lysis buffer. This method is capable of performing specific detection of both RNA and protein biomarker targets in the exosome. EFIRM extracts exosomes specifically based on their surface markers as opposed to size-based techniques.
Transmission electron microscopy (TEM) and assay demonstrate the functionality of the method for exosome capture and analysis. The EFIRM method was applied to exosomal analysis of 9 mice injected with human lung cancer H640 cells (a cell line transfected to express the exosome marker human CD63-GFP) in order to test their exosome profile against 11 mice receiving saline controls. Elevated levels of exosomal biomarkers (reference gene GAPDH and protein surface marker human CD63-GFP) were found for the H640 injected mice in both serum and saliva samples. Furthermore, saliva and serum samples were demonstrated to have linearity (R = 0.79). These results are suggestive for the viability of salivary exosome biomarkers for detection of distal diseases.
Introduction
Exosome research is an emerging field of investigation that examines lipid microvesicles that carry RNA1, DNA2, and protein3 cargo. Previous investigations of exosome biology have led to identification of exosomes in biofluids such as blood4, urine5, breast milk6, and saliva7. Studies have demonstrated that exosomes play a role in different cellular pathways, remotely meditating communication between different systems of the body8. Because of the role exosomes play in intercellular communication, it is hypothesized that they may package biomolecule targets (protein, RNA, and DNA) correlated with disease states. In vitro3 and animal model9 studies appear to corroborate this hypothesis. In investigating exosomal content for biomarker discovery, it is necessary to develop a methodology for selective exosome isolation from biofluids, induced expulsion of cargo from exosomes, and quantification of exosome biomolecules. In the extent of this work, exosomes will be defined as a structure having a diameter of approximately 70-100 nm and possessing surface marker CD63.
Researchers typically first purify exosomes by ultracentrifugation10 and then process exosomal content through the usage of lysis buffer kits. Usage of lysis buffer methods requires incubation times ranging from minutes to hours. This process may potentially harm exosome cargo and lead to sample degradation. For example, salivary exosome RNA released via lysis buffer into the surrounding extracellular environment possesses a half-life of under 1 min, making measurement of exosomal RNA post-lysis buffer a particularly difficult task without the addition of stabilization reagents11. The compounded effect of adding various reagents for lysis and stabilization may introduce agents that complicate and interfere with the analysis of exosomal content. An alternative approach may be helpful for rapidly unloading exosomal content and safely preserving the cargo for characterization.
In this work, we propose the usage of a non-uniform electric field for the release of exosomal content. Electric-fields have been known to carry the ability to polarize and disrupt the lipid bilayer that forms cell membranes. Our experimental work explores usage of non-uniform cyclic square waves (CSW) for disrupting the microvesicle structure of exosomes and releasing carried cargo. This method uses voltages in the several hundred millivolt range, meaning that most biomolecules will not be disrupted. We demonstrate that the usage of a cyclic-square wave is able to actuate release of salivary exosome mRNA content into the surrounding fluidic environment. This release of exosomal content is seamlessly integrated with an electrode system that can be used to quantify the biomarker expression levels12,13. This proposed method allows for rapid, sensitive, and lysis buffer free analysis of exosome content.

Figure 1. Overview of EFIRM Workflow. .The EFIRM method is broadly divided into the three major phases that are necessary for purifying and analyzing exosomes.
This CSW based exosomal content release and analysis method is used in conjunction with CD63-specific magnetic microbeads for exosome isolation. These CD63-affinity beads allow for the selective isolation of exosomes from salivary samples (and other biofluids). Following incubation and extraction of exosomes using the magnetized beads, the beads are migrated to the electrochemical sensor system for the CSW based content release and analysis portion of the experiment. Figure 1 gives an overview of the workflow of the EFIRM method.
Protocol
1. Magnetic Bead-based Exosome Extraction
- Pipette a well-mixed solution of 5 µl of Streptavidin-Coated magnetic microparticles into 495 µl of phosphate buffered saline (PBS) buffer in a microcentrifuge tube to resuspend the beads. Wash and resuspend the beads with 500 µl of PBS three times using a magnetic rack. The rack is an array of magnets on the side of a housing unit that can hold the sample microcentrifuge tubes.
- For each wash, first let the tubes sit on the rack for 1 min, and then use a pipette tip to carefully remove the supernatant buffer without disturbing the beads.
- Place the tubes on a regular rack without magnets at the side. Add 500 µl of PBS into the tubes, and use the pipette to mix the solution and beads together. Then put the tubes back on the magnetic rack to again separate the beads from the solution.
- Perform this removal of buffer via magnetization and resuspension in PBS a total of three times. This performs an initial wash of the magnetic particles.
- Resuspend the beads into 490 µl of PBS buffer, with the tube placed on the non-magnetized portion of the magnetic rack. Pipet 5 µl of biotinylated mouse anti-human CD63 antibody at 1.0 mg/ml stock concentration into the mixture of beads. Use the pipette to mix the beads and antibody in solution.
- Place the microcentrifuge tubes with bead and biotinylated antibody mixture on a sample rotator. Set the rotator parameters for the sample rotator for reciprocal rotation at 90° tilting for 5 sec and vibrating at 5° for 1 sec. Rotate the sample-bead mixture tubes at these parameters for 30 min at RT.
- Remove unbound antibody after conjugation.
- Following 30 min of rotation at RT, place the tubes back in the magnetic rack for 5 min.
- Perform three washes of beads by removing the liquid phase using a micropipette and wash with 500 µl of PBS. After the triple wash, resuspend the beads in 490 µl of casein-PBS and place on the unmagnetized portion of the rack.
- Exosome extraction using antibody-coated beads.
- Label each tube with targeted sample ID. Pipette a 10 µl sample of serum or saliva into the microcentrifuge tube. Use the pipette to mix the sample and magnetic beads by pipetting several times.
- Place the tubes with sample and anti-human CD63 antibody beads on rotator and rotate for 2 hr at RT. Use the same rotator parameters as described in step 1.2.
- Following 2 hr of sample rotating, perform a triple wash by magnetizing to separate beads from solution, removing liquid phase with micropipette, and resuspending beads in 500 µl of Tris-HCl buffer. The resultant beads are now bound to the exosomes and are ready for the electrical field release and measurement.
2. Electric Field Induced Released and Measurement of Exosomal Content
- Initial Precoating of Electrode with GADPH Primer
- Apply a plastic well to an electrode array to prevent cross-contamination of the individual electrodes. For this experiment, use a 16-sensor electrode array with each unit electrode in the array consisting of a working, counter, and reference electrode made of bare gold.
- Prepare a stock mixture of 100 nM DNA probe, 0.3 M KCl, and 10 mM pyrrole by pipetting stock reagents into a tube with ultrapure distilled water. Mix thoroughly by vortexing.
NOTE: For this study, the DNA probe selected corresponds to the GAPDH reference gene, which is known to exist within exosomes. The probe sequence used is: 5'-Biotin-AGGTCCACCACTGACACGTTG-3'. Use this mixture on all the electrodes. - Pipet 60 µl of the monomer-DNA probe mixture onto the surface of each gold electrode. Examine the electrodes to ensure that there is adequate coverage of the working, counter, and reference electrodes by the liquid mixture.
- Electropolymerize monomer-probe mixture to create a conducting polymer layer on the electrode surface by applying a cyclic square wave (CSW) electric field profile to the electrode surface. This electric field consists of applying +350 mV for 9 sec and immediately switching to +950 mV for 1 sec. Apply this cyclic square wave profile to the electrode for 10 cycles, for a total of 100 sec of applied electric field.
- Rinse sensor surface 3 times with distilled water and dry with nitrogen gas to remove liquid from the surface of the electrode. Ensure that liquid is properly removed from the electrode.
- Exosome Cargo Unloading
- Load 5 µl of 1 µM of a detector probe into 495 µl of the bead-exosome complex mixture and use a pipette to mix.
NOTE: The detector probe is a DNA primer sequence conjugated to a fluorescein molecule at the 3' end. The detector probe sequence used for this study corresponds to the GAPDH mRNA found within exosomes. The sequence of the fluorescein conjugated detector probe is: 5'-GCAGTGGGGACACGGAAGGCC-Fluorescein-3'. - Pipette 60 µl of the probe and bead-exosome complex mixture onto a gold electrode surface with a magnet array underneath. This magnet array consists of sixteen 2.54 mm diameter neodymium magnets aligned to correspond to the working electrodes of the sensor. Figure 2A illustrates the placement of the magnets and bead-exosome solution.
- Once sample is loaded on the electrode surface, apply 20 cycles of the CSW electric field with 9 sec at -300 mV and 1 sec at +200 mV (200 sec total). The exosomal cargo that is released will hybridize to the primers on the surface of the electrode. If surface markers of the exosome are the subject of investigation, skip this portion of the experiment. Figure 2B illustrates this process.
- Wash-off the unbound analytes on the electrode surface by triple rinsing the electrode surface with distilled water. Dry the electrode with nitrogen gas.
- Load 5 µl of 1 µM of a detector probe into 495 µl of the bead-exosome complex mixture and use a pipette to mix.
- Reporter Antibody and Readout
- Add 60 µl of 150 unit/ml anti-fluorescein antibody conjugated to horseradish peroxidase (HRP in 1:1,000 dilution) diluted in Casein/PBS.
- Use an electric-field driven conjugation to complex anti-fluorescein HRP to the probe sandwich. Apply -200 mV for 1 sec and +500 mV for 1 sec for 5 cycles to the electrode surface. Figure 2A shows the capture and detector probe complexes for both a protein and nucleic acid system.
- Triple wash sensor surface using distilled water and dry with nitrogen gas.
- Following the wash-off of unbound excess anti-fluorescein antibody, add 60 µl of 3,3',5,5'-Tetramethylbenzidine (TMB) substrate. Load this substrate on to each sensor surface using a multichannel pipet.
- Perform amperometric readout of the current by measuring the electrode current at -200 mV for 60 sec using an electrochemical potentiostat capable of simultaneous measurement of 16 channels. Figure 2C is an example of current profile during readout.
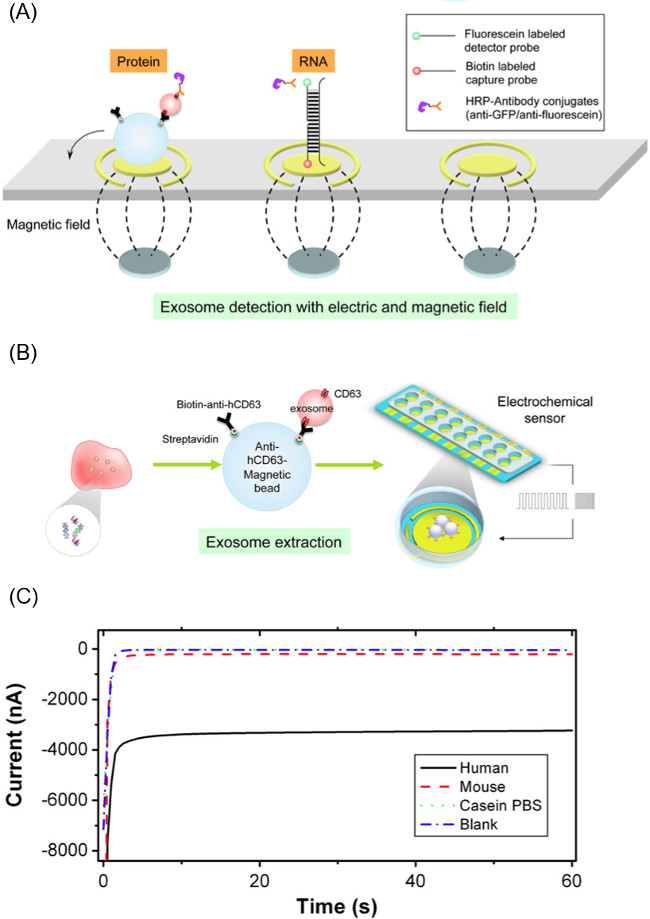
Figure 2. Components of EFIRM Method. (A) Method of extracting exosomes from biofluid using anti-human CD63 coated magnetic microparticles and then unloading exosome cargo using cyclic square waves applied to the particle-exosome complex. (B) Scheme of electrode biosensor used for detecting RNA/DNA/protein targets from the released exosome. (C) Representative example of amperometric readout from the EFIRM methodology, where larger current magnitude corresponds to higher levels of a biomolecule. This figure is from Wei et al.14 Please click here to view a larger version of this figure.
Results
Validation of Exosome Capture of Beads Using TEM
Isolation of exosomes from saliva using anti-human CD63 magnetic beads was validated following extraction protocol by using transmission electron microscopy (TEM) images. TEM shows magnetic beads with 70-100 nm granules immediately adjacent (see Figure 3A, and 3B), consistent with the known profile of exosomes. No 70-100 nm granules were observed for the magnetic beads in saliva that did not hav...
Discussion
As the results indicate, anti-human CD63 coated magnetic nanoparticles are able to specifically capture small particles that have a size ranging from 70-100 nm. This captured particle is consistent with the previously observed profile of exosomes. Furthermore, the usage of the low-voltage CSW following the capture of the particles is shown to remove them from the bead surface and cause DNA degradation profiles similar to that of a traditional lysis buffer based method for cargo release. This data indicates that the workf...
Disclosures
David Wong is co-founder of RNAmeTRIX Inc., a molecular diagnostic company. PeriRx LLC sublicensed intellectual properties pertaining to molecular diagnostics from RNAmeTRIX. David Wong is a consultant to PeriRx.
Acknowledgements
This work was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1TR000124 (to FW); the Felix & Mildred Yip Endowed Professorship and the Barnes Family Fund (to DTWW), the National Institute Of Dental & Craniofacial Research of the National Institutes of Health under Award Number T90DE022734 (to MT). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Materials
| Name | Company | Catalog Number | Comments |
| Helios 16-Channel Reader System with Chip Interface | Genefluidics, USA | RS-1000-16 | |
| 16x Sensor Chip, Bare Gold, pack of 5 chips | Genefluidics, USA | SC1000-16X-B | |
| Biotinylated anti-human CD63 Antibody | Ancell, USA | 215-030 | |
| Dynabeads MyOne Streptavidin T1 | Invitrogen, USA | 65601 | |
| Neodynium Magnetics (1/10" dia. x 1/32" thick) | K&J Magnetics, USA | DH101 | |
| Ultrapure Distilled Water | Life Technologies, USA | 10977-023 | |
| Mettler Toldeo 3 M KCl Solution | Fisher Scientific, USA | 1911512 | |
| Pyrrole | Sigma-Aldrich, USA | W338605-100g | |
| Anti-Fluorescein-POD, Fab fragments | Roche, Germany | 11426346910 | |
| 3,3′,5,5′-tetramethylbenzidine substrate (TMB/H2O2, low activity) | Neogen, Usa | 330175 | |
| Phosphate Buffered Saline Solution | Life Technologies, USA | 10010023 | |
| Casein/PBS | Fisher Scientific, USA | 37532 |
References
- Rabinowits, G., Gerçel-Taylor, C., Day, J. M., Taylor, D. D., Kloecker, G. H. Exosomal MicroRNA: A Diagnostic Marker for Lung Cancer. Clinical Lung Cancer. 10 (1), 42-46 (2009).
- Thakur, B. K., et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Research. 24 (6), 766-769 (2014).
- Lau, C. S., Wong, D. T. W. Breast Cancer Exosome-like Microvesicles and Salivary Gland Cells Interplay Alters Salivary Gland Cell-Derived Exosome-like Microvesicles In. Vitro. PLoS ONE. 7 (3), e33037 (2012).
- Bala, S., et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatolog. 56 (5), 1946-1957 (2012).
- Dear, J. W., Street, J. M., Bailey, M. A. Urinary exosomes: A reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics. 13 (10-11), 1572-1580 (2013).
- Lässer, C., et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. Journal of Translational Medicine. 9 (1), 9 (2011).
- Palanisamy, V., et al. Nanostructural and Transcriptomic Analyses of Human Saliva Derived Exosomes. PLoS ONE. 5 (1), e8577 (2010).
- Camussi, G., et al. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney International. 78 (9), 838-848 (2010).
- Lau, C., et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. The Journal of Biological Chemistry. 288 (37), 26888-26897 (2013).
- Théry, C., Amigorena, S., Raposo, G., Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Current Protocols in Cell Biology. 3 (22), (2001).
- Park, N. J., Li, Y., Yu, T., Brinkman, B. M. N., Wong, D. T. Characterization of RNA in Saliva. Clinical Chemistry. 52 (6), 988-994 (2006).
- Wei, F., et al. Bio/Abiotic Interface Constructed from Nanoscale DNA Dendrimer and Conducting Polymer for Ultrasensitive Biomolecular Diagnosis. Small. 5 (15), 1784-1790 (2009).
- Wei, F., et al. Electrochemical Sensor for Multiplex Biomarkers Detection. Clinical Cancer Research. 15 (13), 4446-4452 (2009).
- Wei, F., Yang, J., Wong, D. T. W. Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM). Biosensors and Bioelectronics. 44, 115-121 (2013).
Erratum
Formal Correction: Errata: Detection of Exosomal Biomarker by Electric Field-induced Release and Measurement (EFIRM)
Posted by JoVE Editors on 1/01/1970. Citeable Link.
An erratum was issued for Detection of Exosomal Biomarker by Electric Field-induced Release and Measurement (EFIRM). The disclosures were updated.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved