A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Genome-wide Mapping of Drug-DNA Interactions in Cells with COSMIC (Crosslinking of Small Molecules to Isolate Chromatin)
In This Article
Summary
Identifying the direct targets of genome-targeting molecules remains a major challenge. To understand how DNA-binding molecules engage the genome, we developed a method that relies on crosslinking of small molecules to isolate chromatin (COSMIC).
Abstract
The genome is the target of some of the most effective chemotherapeutics, but most of these drugs lack DNA sequence specificity, which leads to dose-limiting toxicity and many adverse side effects. Targeting the genome with sequence-specific small molecules may enable molecules with increased therapeutic index and fewer off-target effects. N-methylpyrrole/N-methylimidazole polyamides are molecules that can be rationally designed to target specific DNA sequences with exquisite precision. And unlike most natural transcription factors, polyamides can bind to methylated and chromatinized DNA without a loss in affinity. The sequence specificity of polyamides has been extensively studied in vitro with cognate site identification (CSI) and with traditional biochemical and biophysical approaches, but the study of polyamide binding to genomic targets in cells remains elusive. Here we report a method, the crosslinking of small molecules to isolate chromatin (COSMIC), that identifies polyamide binding sites across the genome. COSMIC is similar to chromatin immunoprecipitation (ChIP), but differs in two important ways: (1) a photocrosslinker is employed to enable selective, temporally-controlled capture of polyamide binding events, and (2) the biotin affinity handle is used to purify polyamide–DNA conjugates under semi-denaturing conditions to decrease DNA that is non-covalently bound. COSMIC is a general strategy that can be used to reveal the genome-wide binding events of polyamides and other genome-targeting chemotherapeutic agents.
Introduction
The information to make each cell in the human body is encoded in DNA. The selective use of that information governs the fate of a cell. Transcription factors (TFs) are proteins that bind to specific DNA sequences to express a particular subset of the genes in the genome, and the malfunction of TFs is linked to the onset of a wide array of diseases, including developmental defects, cancer, and diabetes.1,2 We have been interested in developing molecules that can selectively bind to the genome and modulate gene regulatory networks.
Polyamides composed of N-methylpyrrole and N-methylimidazole are rationally-designed molecules that can target DNA with specificities and affinities that rival natural transcription factors.3-6 These molecules bind to specific sequences in the minor groove of DNA.4,5,7-11 Polyamides have been employed to both repress and activate the expression of specific genes.4,12-19 They also have interesting antiviral20-24 and anticancer12,13,25-30 properties. One attractive feature of polyamides is their ability to access DNA sequences that are methylated31,32 and wrapped around histone proteins9,10,33.
To measure the comprehensive binding specificities of DNA-binding molecules, our lab created the cognate site identifier (CSI) method.34-39 The predicted occurrence of binding sites based on in vitro specificities (genomescapes) can be displayed on the genome, because the in vitro binding intensities are directly proportional to association constants (Ka).34,35,37 These genomescapes provide insight into polyamide occupancy across the genome, but measuring polyamide binding in live cells has been a challenge. DNA is tightly packaged in the nucleus, which could influence the accessibility of binding sites. The accessibility of these chromatinized DNA sequences to polyamides remains a mystery.
Recently, many methods to study interactions between small molecules and nucleic acids have emerged.40-48 The chemical affinity capture and massively-parallel DNA sequencing (chem-seq) is one such technique. Chem-seq uses formaldehyde to crosslink small molecules to a genomic target of interest and a biotinylated derivative of a small molecule of interest to capture the ligand–target interaction.48,49
Formaldehyde crosslinking leads to indirect interactions that can produce false positives.50 We developed a new method, the crosslinking of small molecules to isolate chromatin (COSMIC),51 with a photocrosslinker to eliminate these so-called “phantom” peaks.50 To begin, we designed and synthesized trifunctional derivatives of polyamides. These molecules contained a DNA-binding polyamide, a photocrosslinker (psoralen), and an affinity handle (biotin, Figure 1). With trifunctional polyamides, we can covalently capture polyamide–DNA interactions with 365 nm UV irradiation, a wavelength that does not damage DNA or induce non-psoralen-based crosslinking.51 Next, we fragment the genome and purify the captured DNA under stringent, semi-denaturing conditions to decrease DNA that is non-covalently bound. Thus, we view COSMIC as a method related to chem-seq, but with a more direct readout of DNA targeting. Importantly, the weak (Ka 103-104 M-1) affinity of psoralen for DNA does not detectably impact polyamide specificity.51,52 The enriched DNA fragments can be analyzed by either quantitative polymerase chain reaction51 (COSMIC-qPCR) or by next-generation sequencing53 (COSMIC-seq). These data enable an unbiased, genome-guided design of ligands that interact with their desired genomic loci and minimize off-target effects.
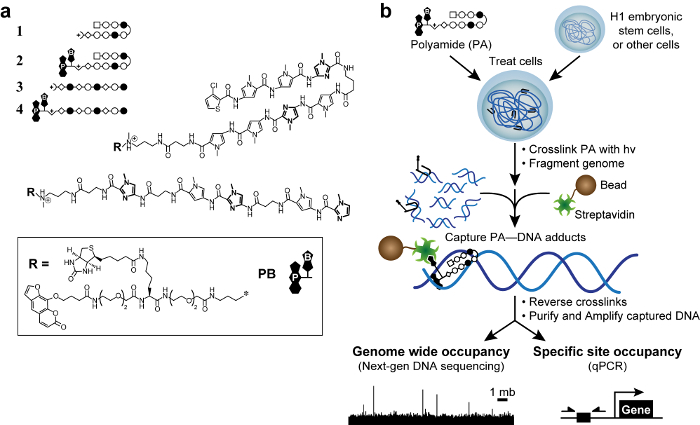
Figure 1. Bioactive polyamides and COSMIC scheme. (A) Hairpin polyamides 1–2 target the DNA sequence 5′-WACGTW-3′. Linear polyamides 3–4 target 5′-AAGAAGAAG-3′. Rings of N-methylimidazole are bolded for clarity. Open and filled circles represent N-methylpyrrole and N-methylimidazole, respectively. Square represents 3-chlorothiophene, and diamonds represent β-alanine. Psoralen and biotin are denoted by P and B, respectively. (B) COSMIC scheme. Cells are treated with trifunctional derivatives of polyamides. After crosslinking with 365 nm UV irradiation, cells are lysed and genomic DNA is sheared. Streptavidin-coated magnetic beads are added to capture polyamide–DNA adducts. The DNA is released and can be analyzed by quantitative PCR (qPCR) or by next-generation sequencing (NGS). Please click here to view a larger version of this figure.
Protocol
1. Crosslinking in Live Cells
- Begin with ~2.5x107 H1 cells, or other cultured cells.
NOTE: This number of H1 cells corresponds to five 10-cm dishes (approximately 40% confluency). - Grow cells in E8 media on dishes coated on a surface that supports pluripotent stem cells (see Materials List), and incubate them at 37 °C in humidified atmosphere of 5% CO2. Harvest cells enzymatically (see Materials List). Note: Do not allow H1 cells to exceed 90% confluency; confluency induces spontaneous differentiation of H1 cells. To count cells, grow an extra 10-cm dish of cells, lift the cells enzymatically as described in sections 1.8-1.10, and count them with a hemocytometer.
- Prepare E8 culture media as described in Chen et al.54
- Prior to adding polyamide, remove the spent culture media with a Pasteur pipette attached to a vacuum trap. Add fresh media with a serological pipette and pipette dispenser (8 ml per 10-cm dish).
NOTE: Add the media to the side of the dish in order to avoid disrupting the cells. From this point onward protect the cells from light to avoid premature photo-crosslinking. - Add polyamide with a pipette (8 µl of 400 µM polyamide in DMSO, 400 nM final concentration) directly to the culture media of each dish. Swirl the dish to disperse the polyamide evenly into the media.
NOTE: Concentration can be varied, but ensure that no cellular toxicity is observed for the treatment selected. - Incubate the cells 24 hr 37 °C in a humidified atmosphere of 5% CO2, and ensure they are protected from light.
NOTE: The incubation time can be varied to measure polyamide binding across time. - Wash each 10-cm dish with 4 ml PBS (1.05 mM potassium phosphate monobasic, 155.17 mM sodium chloride, 2.97 mM sodium phosphate dibasic) using a serological pipette and pipette dispenser. Aspirate PBS and add 3 ml E8 culture media.
- With the lights dimmed, remove the lid of 5 culture dishes and place the cells on a flat surface outside of the hood. Place a glass filter over the 5 culture dishes to filter out light with λ < 300 nm. Place the UV source on top of filter. Crosslink samples 30 min with 365 nm UV irradiation (2.4 mW/cm2).
NOTE: Crosslinking time should be determined empirically. - Transfer culture dishes back to the hood. Aspirate the media with a Pasteur pipette attached to a vacuum trap. Wash each 10-cm dish with 4 ml PBS. Aspirate the PBS. Add 3 ml enzyme for cell dissociation (see Materials List) per 10-cm dish to dissociate the cells. Incubate 5 min at 37 °C.
NOTE: Do not pre-warm the enzyme. - Quench the enzyme with 3 ml E8 media per dish. Transfer dissociated cells into one 15-ml conical tube.
NOTE: Place cells on ice from this point onward. - Centrifuge dissociated cells 5 min, 500 x g at 4 °C. Aspirate supernatant to remove enzyme and media.
NOTE: Pause point. Cells can be flash-frozen in liquid nitrogen, stored at -80 °C for subsequent processing at a later time.
2. Isolation of Chromatin
- Add 1.2 ml COSMIC buffer (20 mM Tris-HCl [pH 8.1], 2 mM EDTA, 150 mM NaCl, 1% Triton-X100, 0.1% SDS) and pipette up and down several times to resuspend the cell pellet for each sample in 1.2 ml COSMIC buffer.
- Add 133 µl each of 100 mM phenylmethylsulfonyl fluoride (PMSF), 100 mM benzamidine, and 150 µM pepstatin protease inhibitors fresh to a final concentration of 1 mM for PMSF and benzamidine and 1.5 µM for pepstatin. Split solution of cell lysate into two amber 1.7 ml microcentrifuge tubes.
- Sonicate with sonicator 35 min (10 sec on, 10 sec off, 60% power) to fragment the genome to between 100 and 500 bp.
- Keep the level of chromatin solution in the microcentrifuge tube parallel to the level of water in the reservoir. Confirm this level by visual inspection.
NOTE: Confirm that DNA was sheared with a 1.5% agarose gel.55 - Optimize the sonication time empirically. Use a minimal amount of ice in the reservoir to chill the samples, and ensure the ice is not interfering between the samples and the cup horn.
- Keep the level of chromatin solution in the microcentrifuge tube parallel to the level of water in the reservoir. Confirm this level by visual inspection.
- Centrifuge the sample 12,000 x g 10 min. Save the aqueous solution which contains soluble chromatin by transferring it to a new amber microcentrifuge tube with a pipette. Discard the pellet.
- Transfer 110 µl (10%) of sample to a new microcentrifuge tube and label it Input DNA. Store at -80 °C. Save the rest of the chromatin sample on ice for use in Step 3.3.
3. Capture of Ligand–DNA Crosslinks
- Use a pipette to dispense 60 µl streptavidin-coated magnetic beads in a microcentrifuge tube. Add 1 ml COSMIC buffer and mix on a nutator 5 min at RT. Place magnetic beads on magnetic separation rack 2 min to capture beads. Remove COSMIC buffer with a pipette.
NOTE: Do not allow the beads to dry out. Proceed immediately to the next step. - Add chromatin sample (~1 ml) to beads (60 µl) and resuspend the beads. Incubate chromatin with streptavidin-coated magnetic beads at least 4 hr on a rotating, rocking mixer at 4 °C. Incubate the samples O/N if desired.
4. Isolation of Affinity-purified DNA
- Wash beads with 7 min interval at RT with the following wash buffers to remove non-specific interactions. Use a magnetic separation rack to capture beads after each wash. Resuspend the beads after each change in washing buffer.
- Prepare wash buffers in distilled deionized water and filter (0.2 µm) before use. Store wash Buffers 1 and 2 at 4 °C for several months. Prepare Wash Buffer 3 fresh each day. Add and remove wash buffers from the sample with a pipette. For 2 and 4, add 1 and 3 (5 mM), respectively, in the washes. Samples can additionally be washed twice with COSMIC buffer (once 12 hr, once 4 hr) prior to the washes listed below.
- Wash once with Wash Buffer 1 (10 mM Tris-Cl [pH 8.0], 1 mM EDTA, 3% SDS). Wash once with Wash Buffer 2 (10 mM Tris-Cl [pH 8.0], 250 mM LiCl, 1 mM EDTA, 0.5% NP40, 1% sodium deoxycholate). Wash twice with Wash Buffer 3 (4 M urea, 10 mM Tris-Cl [pH 7.5], 1 mM EDTA, 0.1% NP-40). Wash twice with Tris EDTA (TE) buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA).
- Resuspend the beads in 200 μl TE buffer with a pipette. This sample of captured DNA is referred to as affinity-purified (AP) DNA.
- Supplement the Input and AP DNA with 10x Crosslink Reversal Buffer (100 mM Tris-Cl [pH 8.0], 1 M KOH, 4 mM EDTA) to 1x final concentration.56,57 Incubate 30 min at 90 °C.
NOTE: 254 nm UV irradiation can also be used to reverse the psoralen crosslink,58 but cyclobutyl pyrimidine dimers can be formed from irradiation at this wavelength and interfere with downstream processing of DNA. - For AP DNA, place the microcentrifuge tube containing the sample on magnetic separation rack 2 min at RT and isolating liquid (DNA) with a pipette. Transfer the AP DNA (which has been released from the beads) to a new amber microcentrifuge tube.
NOTE: The desired AP DNA is in the liquid, no longer attached to the beads. - Neutralize Input and AP DNA with concentrated HCl to pH 7 by adding approximately 1 µl 6 N HCl per 100 μl sample with a pipette. Confirm the samples are neutralized by adding ~0.5 µl onto pH paper with a pipette. WARNING: Concentrated HCl is a strong corrosive acid. Handle according to your institution’s guidelines for the appropriate personal protective equipment.
- Add RNase A (100 mg/ml) to 0.2 µg/µl final concentration to both Input and AP DNA. Incubate 1 hr at 37 °C. Add Proteinase K (20 mg/ml) to 0.2 µg/µl final concentration to both Input and AP DNA, add. Incubate 1 hr at 55 °C.
- Purify the DNA with a DNA column cleanup kit (see Materials List).55 Elute DNA in 58 μl DNA-grade H2O. Store Input and AP samples at -20 or -80 °C
- Analyze AP DNA by quantitative polymerase chain reaction (qPCR) with locus-specific primers, and/or by next-generation sequencing. For qPCR, use 2 µl AP DNA per locus with the following parameters: 1 cycle of 95 °C 10 min and 40 cycles of 95 °C 20 sec, 54 °C or 56 °C 20 sec, 72 °C 40 sec.
NOTE: The annealing temperature should be modified according to the melting temperature of the primer pairs used. Select an annealing temperature that minimizes the quantitation cycle with no nonspecific amplification.
NOTE: For analysis by next-generation sequencing, aim for at least 10 million mapped reads. The number of mapped reads can be increased by increasing the amount of AP DNA and by combining fewer samples into a run for sequencing. More reads improve the sensitivity. Standard packages for ChIP-seq analysis (e.g., HOMER,59 MACS,60 and SPP61) work with COSMIC-seq data. As with other methods that rely upon next-generation sequencing, including genome sequencing and ChIP-seq, repetitive regions create ambiguities in alignment and assembly and thus remain a technical challenge.
Results
To account for non-uniform genome fragmentation and other variables, the purified DNA should always be normalized against a reference of Input DNA. Primers specific to a locus of interest can be used. It is helpful to also analyze a locus where the molecule is not expected to bind, as a negative control. We see a >100-fold increase in polyamide occupancy upon irradiation with 365-nm light (Figure 2).
Enriched DNA can also be analyzed by next-generation sequencing. DNA is p...
Discussion
One of the primary challenges with conventional ChIP is the identification of suitable antibodies. ChIP depends heavily upon the quality of the antibody, and most commercial antibodies are unacceptable for ChIP. In fact, the Encyclopedia of DNA Elements (ENCODE) consortium found only 20% of commercial antibodies to be suitable for ChIP assays.50 With COSMIC, antibodies are replaced by streptavidin. Because polyamides are functionalized with biotin, streptavidin is used in place of an antibody to capture polyam...
Disclosures
A.Z.A. is the sole proprietor of Vista Motif, LLC and WINStep Forward.
Acknowledgements
We thank members of the Ansari lab and Prof. Parameswaran Ramanathan for helpful discussions. This work was supported by NIH grants CA133508 and HL099773, the H. I. Romnes faculty fellowship, and the W. M. Keck Medical Research Award to A.Z.A. G.S.E. was supported by a Peterson Fellowship from the Department of Biochemistry and Molecular Biosciences Training Grant NIH T32 GM07215. A.E. was supported by the Morgridge Graduate Fellowship and the Stem Cell and Regenerative Medicine Center Fellowship, and D.B. was supported by the NSEC grant from NSF.
Materials
| Name | Company | Catalog Number | Comments |
| Phenylmethylsulfonyl fluoride (PMSF) | any source | ||
| Benzamidine | any source | ||
| Pepstatin | any source | ||
| Proteinase K | any source | ||
| Dynabeads MyOne Streptavidin C1 | Life Technologies | 65001 | |
| PBS, pH 7.4 | Life Technologies | 10010-023 | Other sources can be used |
| StemPro Accutase Cell Dissociation Reagent | Life Technologies | A1110501 | |
| QIAquick PCR Purification Kit | Qiagen | 28104 | We have tried other manufacturers of DNA columns with success. |
| TruSeq ChIP Sample Prep Kit | Illumina | IP-202-1012 | This Kit can be used to prepare COSMIC DNA for next-generation sequencing |
| Matrigel Basement Membrane Matrix | BD Biosciences | 356231 | Used to coat plates in order to grow H1 ESCs |
| pH paper | any source | ||
| amber microcentrifuge tubes | any source | ||
| microcentrifuge tubes | any source | ||
| pyrex filter | any source | Pyrex baking dishes are suitable | |
| qPCR master mix | any source | ||
| RNase | any source | ||
| HCl (6 N) | any source | ||
| 10-cm tissue culture dishes | any source | ||
| Serological pipettes | any source | ||
| Pasteur pipettes | any source | ||
| Pipette tips | any source | ||
| 15-ml conical tubes | any source | ||
| centrifuge | any source | ||
| microcentrifuge | any source | ||
| nutator | any source | ||
| Magnetic separation rack | any source | ||
| UV source | CalSun | B001BH0A1A | Other UV sources can be used, but crosslinking time must be optimized empirically |
| Misonix Sonicator | Qsonica | S4000 with 431C1 cup horn | Other sonicators can be used, but sonication conditions must be optimized empirically |
| Humidified CO2 incubator | any source | ||
| Biological safety cabinet with vacuum outlet | any source |
References
- Lee, T. I., Young, R. A. Transcriptional Regulation and Its Misregulation in Disease. Cell. 152, 1237-1251 (2013).
- Vaquerizas, J. M., Kummerfeld, S. K., Teichmann, S. A., Luscombe, N. M. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 10, 252-263 (2009).
- Meier, J. L., Yu, A. S., Korf, I., Segal, D. J., Dervan, P. B. Guiding the Design of Synthetic DNA-Binding Molecules with Massively Parallel Sequencing. J. Am. Chem. Soc. 134, 17814-17822 (2012).
- Dervan, P. B. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 9, 2215-2235 (2001).
- Wemmer, D. E., Dervan, P. B. Targeting the minor groove of DNA. Curr. Opin. Struct. Biol. 7, 355-361 (1997).
- Eguchi, A., Lee, G. O., Wan, F., Erwin, G. S., Ansari, A. Z. Controlling gene networks and cell fate with precision-targeted DNA-binding proteins and small-molecule-based genome readers. Biochem. J. 462, 397-413 (2014).
- Mrksich, M., et al. Antiparallel side-by-side dimeric motif for sequence-specific recognition in the minor groove of DNA by the designed peptide 1-methylimidazole-2-carboxamide netropsin. Proc Natl Acad Sci U S A. 89, 7586-7590 (1992).
- Edayathumangalam, R. S., Weyermann, P., Gottesfeld, J. M., Dervan, P. B., Luger, K. Molecular recognition of the nucleosomal "supergroove". Proc Natl Acad Sci U S A. 101, 6864-6869 (2004).
- Suto, R. K., et al. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J. Mol. Biol. 326, 371-380 (2003).
- Gottesfeld, J. M., et al. Sequence-specific Recognition of DNA in the Nucleosome by Pyrrole-Imidazole Polyamides. J. Mol. Biol. 309, 615-629 (2001).
- Chenoweth, D. M., Dervan, P. B. Structural Basis for Cyclic Py-Im Polyamide Allosteric Inhibition of Nuclear Receptor Binding. J. Am. Chem. Soc. 132, 14521-14529 (2010).
- Raskatov, J. A., et al. Modulation of NF-κB-dependent gene transcription using programmable DNA minor groove binders. Proc. Natl. Acad. Sci. U.S.A. 109, 1023-1028 (2012).
- Yang, F., et al. Antitumor activity of a pyrrole-imidazole polyamide. Proc. Natl. Acad. Sci. U.S.A. 110, 1863-1868 (2013).
- Mapp, A. K., Ansari, A. Z., Ptashne, M., Dervan, P. B. Activation of gene expression by small molecule transcription factors. Proc. Natl. Acad. Sci. U.S.A. 97, 3930-3935 (2000).
- Ansari, A. Z., Mapp, A. K., Nguyen, D. H., Dervan, P. B., Ptashne, M. Towards a minimal motif for artificial transcriptional activators. Chem. Biol. 8, 583-592 (2001).
- Arora, P. S., Ansari, A. Z., Best, T. P., Ptashne, M., Dervan, P. B. Design of artificial transcriptional activators with rigid poly-L-proline linkers. J. Am. Chem. Soc. 124, 13067-13071 (2002).
- Nickols, N. G., Jacobs, C. S., Farkas, M. E., Dervan, P. B. Modulating Hypoxia-Inducible Transcription by Disrupting the HIF-1–DNA Interface. ACS Chemical Biology. 2, 561-571 (2007).
- Pandian, G. N., et al. A synthetic small molecule for rapid induction of multiple pluripotency genes in mouse embryonic fibroblasts. Sci. Rep. 2, 544 (2012).
- Pandian, G. N., et al. Synthetic Small Molecules for Epigenetic Activation of Pluripotency Genes in Mouse Embryonic Fibroblasts. Chem Bio Chem. 12, 2822-2828 (2011).
- He, G., et al. Binding studies of a large antiviral polyamide to a natural HPV sequence. Biochimie. 102, 83-91 (2014).
- Edwards, T. G., Vidmar, T. J., Koeller, K., Bashkin, J. K., Fisher, C. DNA Damage Repair Genes Controlling Human Papillomavirus (HPV) Episome Levels under Conditions of Stability and Extreme Instability. PLoS ONE. 8, e75406 (2013).
- Edwards, T. G., Helmus, M. J., Koeller, K., Bashkin, J. K., Fisher, C. HPV Episome Stability is Reduced by Aphidicolin and Controlled by DNA Damage Response Pathways. Journal of Virology. , (2013).
- Edwards, T. G., et al. HPV episome levels are potently decreased by pyrrole-imidazole polyamides. Antiviral Res. 91, 177-186 (2011).
- Dickinson, L. A., et al. Inhibition of RNA polymerase II transcription in human cells by synthetic DNA-binding ligands. Proc. Natl. Acad. Sci. U.S.A. 95, 12890-12895 (1998).
- Dickinson, L. A., et al. Arresting Cancer Proliferation by Small-Molecule Gene Regulation. Chem. Biol. 11, 1583-1594 (2004).
- Nickols, N. G., et al. Activity of a Py–Im Polyamide Targeted to the Estrogen Response Element. Molecular Cancer Therapeutics. 12, 675-684 (2013).
- Raskatov, J. A., Puckett, J. W., Dervan, P. B. A C-14 labeled Py–Im polyamide localizes to a subcutaneous prostate cancer tumor. Bioorg. Med. Chem. 22, 4371-4375 (2014).
- Jespersen, C., et al. Chromatin structure determines accessibility of a hairpin polyamide–chlorambucil conjugate at histone H4 genes in pancreatic cancer cells. Bioorg. Med. Chem. Lett. 22, 4068-4071 (2012).
- Chou, C. J., et al. Small molecules targeting histone H4 as potential therapeutics for chronic myelogenous leukemia. Molecular Cancer Therapeutics. 7, 769-778 (2008).
- Nickols, N. G., Dervan, P. B. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc. Natl. Acad. Sci. U.S.A. 104, 10418-10423 (2007).
- Minoshima, M., Bando, T., Sasaki, S., Fujimoto, J., Sugiyama, H. Pyrrole-imidazole hairpin polyamides with high affinity at 5CGCG3 DNA sequence; influence of cytosine methylation on binding. Nucleic Acids Res. 36, 2889-2894 (2008).
- Warren, C. L., et al. Fabrication of duplex DNA microarrays incorporating methyl-5-cytosine. Lab on a Chip. 12, 376-380 (2012).
- Dudouet, B., et al. Accessibility of nuclear chromatin by DNA binding polyamides. Chem. Biol. 10, 859-867 (2003).
- Carlson, C. D., et al. Specificity landscapes of DNA binding molecules elucidate biological function. Proc. Natl. Acad. Sci. U.S.A. 107, 4544-4549 (2010).
- Warren, C. L., et al. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl. Acad. Sci. U.S.A. 103, 867-872 (2006).
- Tietjen, J. R., Donato, L. J., Bhimisaria, D., Ansari, A. Z., Voigt, C. Chapter One - Sequence-Specificity and Energy Landscapes of DNA-Binding Molecules. Methods Enzymol. 497, 3-30 (2011).
- Puckett, J. W., et al. Quantitative microarray profiling of DNA-binding molecules. J. Am. Chem. Soc. 129, 12310-12319 (2007).
- Keles, S., Warren, C. L., Carlson, C. D., Ansari, A. Z. CSI-Tree: a regression tree approach for modeling binding properties of DNA-binding molecules based on cognate site identification (CSI) data. Nucleic Acids Res. 36, 3171-3184 (2008).
- Hauschild, K. E., Stover, J. S., Boger, D. L., Ansari, A. Z. CSI-FID: High throughput label-free detection of DNA binding molecules. Bioorg. Med. Chem. Lett. 19, 3779-3782 (2009).
- Lee, M., Roldan, M. C., Haskell, M. K., McAdam, S. R., Hartley, J. A. . In vitro Photoinduced Cytotoxicity and DNA Binding Properties of Psoralen and Coumarin Conjugates of Netropsin Analogs: DNA Sequence-Directed Alkylation and Cross-Link. 37, 1208-1213 (1994).
- Wurtz, N. R., Dervan, P. B. Sequence specific alkylation of DNA by hairpin pyrrole–imidazole polyamide conjugates. Chem. Biol. 7, 153-161 (2000).
- Tung, S. -. Y., Hong, J. -. Y., Walz, T., Moazed, D., Liou, G. -. G. Chromatin affinity-precipitation using a small metabolic molecule: its application to analysis of O-acetyl-ADP-ribose. Cell. Mol. Life Sci. 69, 641-650 (2012).
- Rodriguez, R., Miller, K. M. Unravelling the genomic targets of small molecules using high-throughput sequencing. Nat Rev Genet. 15, 783-796 (2014).
- Guan, L., Disney, M. D. Covalent Small-Molecule–RNA Complex Formation Enables Cellular Profiling of Small-Molecule–RNA Interactions. Angew. Chem. Int. Ed. 52, 10010-10013 (2013).
- White, J. D., et al. Picazoplatin, an Azide-Containing Platinum(II) Derivative for Target Analysis by Click Chemistry. J. Am. Chem. Soc. 135, 11680-11683 (2013).
- Rodriguez, R., et al. Small-molecule–induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 8, 301-310 (2012).
- Bando, T., Sugiyama, H. Synthesis and Biological Properties of Sequence-Specific DNA-Alkylating Pyrrole−Imidazole Polyamides. Acc. Chem. Res. 39, 935-944 (2006).
- Anders, L., et al. Genome-wide localization of small molecules. Nat. Biotechnol. 32, 92-96 (2014).
- Jin, C., et al. Chem-seq permits identification of genomic targets of drugs against androgen receptor regulation selected by functional phenotypic screens. Proc. Natl. Acad. Sci. U.S.A. 111, 9235-9240 (2014).
- Landt, S. G., et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Research. 22, 1813-1831 (2012).
- Erwin, G. S., Bhimsaria, D., Eguchi, A., Ansari, A. Z. Mapping Polyamide–DNA Interactions in Human Cells Reveals a New Design Strategy for Effective Targeting of Genomic Sites. Angew. Chem. Int. Ed. 53, 10124-10128 (2014).
- Hyde, J. E., Hearst, J. E. Binding of psoralen derivatives to DNA and chromatin: influence of the ionic environment on dark binding and photoreactivity. Biochemistry. 17, 1251-1257 (1978).
- Erwin, G. S., Bhimsaria, D., Rodríguez-Martínez, J. A., Grieshop, M. P., Ansari, A. Z. Genome-wide localization of polyamide-based genome readers reveals sequence-based binding to repressive heterochromatin. In preparation. , (2015).
- Chen, G., et al. Chemically defined conditions for human iPSC derivation and culture. Nat Meth. 8, 424-429 (2011).
- Deliard, S., Zhao, J., Xia, Q., Grant, S. F. A. Generation of High Quality Chromatin Immunoprecipitation DNA Template for High-throughput Sequencing (ChIP-seq). J Vis Exp. (74), e50286 (2013).
- Shi, Y. B., Spielmann, H. P., Hearst, J. E. Base-catalyzed reversal of a psoralen-DNA cross-link. Biochemistry. 27, 5174-5178 (1988).
- Kumaresan, K. R., Hang, B., Lambert, M. W. Human Endonucleolytic Incision of DNA 3′ and 5′ to a Site-directed Psoralen Monoadduct and Interstrand. J. Biol. Chem. 270, 30709-30716 (1995).
- Cimino, G. D., Shi, Y. B., Hearst, J. E. Wavelength dependence for the photoreversal of a psoralen-DNA crosslink. Biochemistry. 25, 3013-3020 (1986).
- Heinz, S., et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell. 38, 576-589 (2010).
- Zhang, Y., et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
- Kharchenko, P. V., Tolstorukov, M. Y., Park, P. J. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotech. 26, 1351-1359 (2008).
- Diamandis, E. P., Christopoulos, T. K. The biotin-(strept)avidin system: principles and applications in biotechnology. Clin. Chem. 37, 625-636 (1991).
- Martinson, H. G., True, R. J. On the mechanism of nucleosome unfolding. Biochemistry. 18, 1089-1094 (1979).
- Gloss, L. M., Placek, B. J. The Effect of Salts on the Stability of the H2A−H2B Histone Dimer. Biochemistry. 41, 14951-14959 (2002).
- Jackson, V. Formaldehyde Cross-Linking for Studying Nucleosomal Dynamics. Methods. 17, 125-139 (1999).
- Kasinathan, S., Orsi, G. A., Zentner, G. E., Ahmad, K., Henikoff, S. High-resolution mapping of transcription factor binding sites on native chromatin. Nat Meth. 11, 203-209 (2014).
- Teytelman, L., Thurtle, D. M., Rine, J., van Oudenaarden, A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 18602-18607 (2013).
- . Phantompeakqualtools home page Available from: https://www.encodeproject.org/software/phantompeakqualtools/ (2010)
- Wang, D., Lippard, S. J. Cellular processing of platinum anticancer drugs. Nature Reviews Drug Discovery. 4, 307-320 (2005).
- Hurley, L. H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2, 188-200 (2002).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved