A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
HPLC-based Assay to Monitor Extracellular Nucleotide/Nucleoside Metabolism in Human Chronic Lymphocytic Leukemia Cells
In This Article
Summary
The protocol described here represents an easy and reproducible method that employs reverse phase high-performance liquid chromatography (RP-HPLC) to measure purine metabolism on chronic lymphocytic leukemia (CLL) cells cultured under different conditions.
Abstract
This method describes a sensitive, specific, reliable and reproducible reverse phase high-performance liquid chromatography (RP-HPLC) assay developed and validated for the quantification of extracellular purine nucleotides and nucleosides produced by purified chronic lymphocytic leukemia (CLL) cells under different culture conditions. The chromatographic separation of adenosine 5'-monophosphate (AMP), adenosine (ADO) and inosine (INO) is performed at RT on a silica-based, reversed-phase column that is used for polar compound retention. The method includes a binary mobile phase, which consists of 7 mM ammonium acetate and acetonitrile with a flow rate of 1.00 ml/min. The eluates are monitored using a Photodiode Array UV detector set at 260 nm. A standard calibration curve is generated to calculate the equation for the analytical quantification of each purine compound. System control, data acquisition and analysis are then performed. Applying this protocol, AMP, INO and ADO elute at 7, 11 and 11.9 min, respectively, and the total run time for each sample is 20 min. This protocol may be applied to different cell types and cell lines (both suspension and adherent), using culture media as matrix. The advantages are easy and fast sample preparation and the requirement of a small amount of supernatant for analysis. Furthermore, the use of a serum-free medium allows skipping the protein precipitation step with acetonitrile that impacts the final concentration of purine compounds. One of the limitations of the method is the requirement of the equilibration column run before each single sample run, making the total run time of the experiment longer and preventing high throughput screening applications.
Introduction
Adenosine (ADO) is a purine nucleoside with an adenine molecule attached to a ribose sugar molecule moiety through a glycosidic bond. When present in the extracellular environment, it protects cells from excessive damage by the action of the immune system. This role has been highlighted using different disease models, such as colitis1, diabetes2, asthma3, sepsis4, and ischemic injury5. One of the main ADO functions is the inhibition of immune responses in the tumor microenvironment, contributing to tumor immune evasion6. For this reason, the mechanisms involved in ADO formation and signaling are of considerable therapeutic interest7.
ADO levels in the tissue microenvironment are relatively low under normal physiologic conditions and certainly below the sensitivity threshold of immune cells. However, during hypoxia, ischemia, inflammation, infection, metabolic stress and tumor transformation they rapidly increase8. The elevated extracellular ADO levels in response to tissue-perturbing signals have a dual function: to report tissue injury in an autocrine and paracrine way and to generate tissue responses that can be generally viewed as cytoprotective.
Extracellular ADO can be formed through a variety of mechanisms, which include release from intracellular compartments mediated by nucleoside transporters9 or accumulation because of impaired degradation operated by adenosine deaminase. The main pathway leading to increased extracellular ADO levels involves the action of a cascade of ectonucleotidases, which are membrane associated ectoenzymes generating ADO by phosphohydrolysis of nucleotides released from dead or dying cells. This pathway proceeds through the sequential action of CD39 (ectonucleoside triphosphate diphosphohydrolase-1) that converts extracellular adenosine 5'-triphosphate (ATP) or adenosine 5'-diphosphate (ADP) to adenosine 5'-monophosphate (AMP) and of CD73 (5'-nucleotidase), which converts AMP to ADO10.
Extracellular ADO elicits its physiological responses by binding to four transmembrane ADO receptors, namely A1, A2A, A2B and A3. Each receptor has different affinities for ADO and specific tissue distribution. All the receptors have seven transmembrane domains and are G-protein coupled to intracellular GTP-binding proteins (G proteins), that can induce (Gs protein) or inhibit (Gi protein) adenylate cyclase activity and, subsequently, the production of intracellular cAMP. Therefore, changes in cytoplasmic cAMP levels impact on intracellular protein kinase activity during physiological responses11. Under physiological conditions extracellular ADO is below 1 µM, which can activate indiscriminately A1, A2A and A3 receptors. However, the activation of A2B subtype requires considerably higher concentrations of the nucleoside, such as those generated under pathophysiological conditions. Alternatively, extracellular ADO can be degraded to inosine (INO) by adenosine deaminase (ADA) and CD26, an ADA complexing protein localizing ADA on the cell surface. Another possibility is that ADO is internalized by the cell through the equilibrative nucleoside transporters (ENT) and phosphorylated to AMP by ADO kinase protein12,13.
The aim of this protocol is to describe an analytical method of reverse phase high-performance liquid chromatography (RP-HPLC) to quantify in a single run the substrate AMP and the products ADO and INO, as generated by human lymphocytes. Our experience was initially obtained using cells from chronic lymphocytic leukemia (CLL) patients, which are characterized by the expansion of a mature population of CD19+/CD5+ B lymphocytes constitutively expressing CD3914,15. We showed approximately 30% of CLL patients express the CD73 ectoenzyme and that this phenotype correlates with a poor prognosis16. This subpopulation of leukemic cells co-expressing CD39 and CD73 can actively produce extracellular ADO from ADP and/or AMP. Preincubation of CD73+ CLL cells with α,β-methylene-ADP (APCP), a known inhibitor of CD73 enzymatic activity, completely blocks extracellular ADO synthesis confirming that CD73 represents the bottle-neck enzyme of that cascade16.
CLL cells also express ADA and the ADA complexing protein CD26, which are responsible for the conversion of ADO into INO. By using specific ADA inhibitors, such as erythro-9-(2-Hydroxy-3-nonyl)I wiadenine (EHNA) hydrochloride and deoxycoformycin (dCF), it is possible to block extracellular ADO degradation into INO. Furthermore, pretreatment with an ADA inhibitor in combination with dipyridamole, that blocks nucleoside transporters, enhances ADO accumulation in cell supernatants.
We have then extended this protocol to cells derived from other lineages, including T lymphocytes and myeloid cells, confirming CD73-dependent ADO production. These findings suggest that this HPLC protocol is highly versatile and that it can be applied to different cell lineages and to different culture conditions (Figure 1).
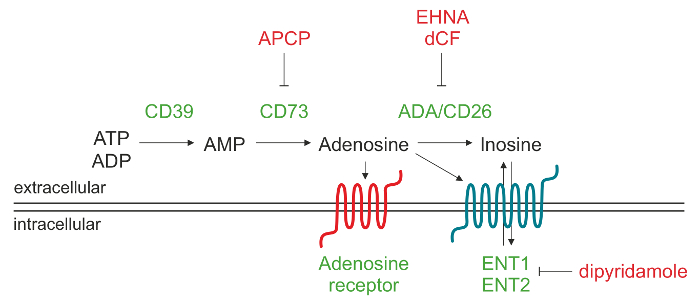
Figure 1. Schematic representation of the enzymatic machinery responsible for extracellular ADO production. Adenosine 5'-triphosphate (ATP) and/or adenosine 5'-diphosphate (ADP) can be degraded by CD39 to adenosine 5'-monophosphate (AMP), which in turn is converted by CD73 into the nucleoside adenosine (ADO). Once ADO is produced in the extracellular space, it may reenter the cell through the nucleoside transporters (ENT), be converted into inosine (INO) or bind to different types of P1 ADO receptors. Please click here to view a larger version of this figure.
Access restricted. Please log in or start a trial to view this content.
Protocol
CLL blood samples are obtained in accordance with Institutional Guidelines and Declaration of Helsinki.
1. Isolation of Leukemic Lymphocytes from Blood Samples of CLL Patients
- Collect blood sample in sodium heparin (green-top) tube17.
- Make 1: 3 dilution of whole blood with RT 1x phosphate buffered saline (PBS).
- Purify peripheral blood mononuclear cells (PBMC) from blood samples by density gradient centrifugation.
- Underlay 5 ml of density centrifugation media (e.g., Ficoll) in a 15 ml centrifuge tube and carefully transfer 10 ml of diluted blood. Immediately centrifuge at 1,500 x g for 20 min at RT.
- Aspirate part of the supernatant with a glass Pasteur pipette connected to a vacuum pump and collect the PBMC ring at the interphase between density centrifugation media and diluted plasma with a 5 ml pipette. Transfer to a 50 ml centrifuge tube and wash with PBS (50 ml final volume). Centrifuge at 500 x g for 5 min at 4 °C.
- Aspirate the supernatant with a glass Pasteur pipette, resuspend the pellet in 50 ml of PBS and count the cells using a hemocytometer.
- Stain PBMCs with anti-CD19 and -CD5 antibodies for flow cytometry following a standard protocol for immunostaining and check for PBMC subpopulation composition18.
Note: CLL lymphocytes are CD19+/CD5+. - Check the % of CD19+/CD5+ cells using a flow cytometer18. Purify B lymphocytes from patients with a ≥95% of CD19+/CD5+ leukemic cells following the instructions detailed in step 2 for the negative isolation of B cells19.
2. Purification of Leukemic B Cells by Negative Isolation
- Resuspend 107 PBMCs/ml in PBS 0.1 % bovine serum albumin (BSA) 2 mM EDTA, pH 7.4 (isolation buffer).
- Add 10 µg of mouse monoclonal anti-CD3, -CD14 and -CD16 primary antibodies for 107 cells and incubate for 30 min at 4 °C.
- Wash the cells with the isolation buffer and centrifuge at 500 x g for 5 min at 4 °C.
- Resuspend the cells in the isolation buffer at 107 PBMCs/ml.
- Before incubating with the cells, resuspend sheep anti-mouse IgG magnetic beads in the vial. Transfer 100 µl of beads per 107 cells to a tube.
- Add 1 mL of isolation buffer and place the tube in a magnet holder for 1 min. Discard the supernatant with a 5 ml pipette.
- Remove the tube from the magnet and resuspend the washed beads in 100 µl of isolation buffer.
- Incubate 107 cells with 100 µl of magnetic beads for 30 min at 4 °C with gentle tilting and rotation.
- Place the tube in the magnet holder for 2 min and transfer the supernatant with the CD19+/CD5+ unbound cells to a fresh tube with a 5 ml pipette.
- At the end of the isolation protocol, stain 100 µl of cells with anti-CD19 and -CD5 antibodies for flow cytometry and incubate 30 min at 4 °C. Wash the cells with 3 ml of PBS 1% BSA, discard the excess of supernatant and check B cell purity again by flow cytometry19.
Note: Perform an additional negative isolation step if B cell purity is lower than 95%.
3. Preparation of Standard and Inhibitors Stock Solutions
- To prepare AMP, weigh out 0.00345 g of AMP and dissolve it in 5 ml of serum-free medium to have a 2 mM standard solution of AMP.
- To prepare ADO, weigh out 0.00265 g of ADO and dissolve it in 5 ml of serum-free medium to have a 2 mM standard solution of ADO.
- To prepare INO, weigh out 0.00268 g of INO and dissolve it in 5 ml of serum-free medium to have a 2 mM standard solution of INO.
- Prepare a 400 µM solution of AMP, ADO and INO by mixing 1 ml of the 2 mM stock solution in 4 ml of serum-free medium (5 ml final volume).
- Make 1:4 dilution of each 400 µM standard solution to obtain a 100 µM concentration by pipetting 500 µl of the 400 µM solution in 1,500 µl of serum-free medium (2 ml final volume).
- Prepare serial dilutions of each compound in serum-free medium to obtain the following concentrations: 100 µM - 50 µM - 25 µM - 10 µM - 5 µM - 2.5 µM - 1 µM - 0.5 µM - 0.25 µM.
Note: For example, pipette 1 ml of the 100 µM solution in 1 ml of serum-free medium (1:2 dilution) to obtain 50 µM concentration and proceed with the remaining dilutions.
- Prepare a 10 mM stock solution of APCP dissolved in PBS; aliquot and stock at - 30 °C.
- Prepare 10 mM stock solutions of EHNA hydrochloride, dCF and dipyridamole dissolved in dimethyl sulfoxide (DMSO); aliquot and stock at -30 °C.
4. Program the HPLC Method
- Prepare a 7 mM ammonium acetate buffer by dissolving 0.770 g of ammonium acetate in 2 L of double deionised water (Buffer A). Adjust to pH 3.0 with hydrochloric acid.
- Prepare a glass bottle containing at least 2 L of ultrapure acetonitrile for HPLC (Buffer B) and connect the guard column and the column to the HPLC.
Note: The columns and buffers are at RT while in use. - Log into the HPLC software and select the "browse project" button. Go to the "file" menu and select "new method" and then "instrument method".
- Program the equilibration column method as per Table 1. Set a flow rate of 1.00 ml/min and program the UV detector to read at 260 nm. Save the instrument method and select again “new method” from the “file” menu and then “method set”. Choose the instrument method previously saved and save the current method set with the same name.
| Time | Flow rate (ml/min) | %A | %B |
| 1.00 | 100 | 0 | |
| 1.24 | 1.00 | 100 | 0 |
| 6.22 | 1.00 | 2 | 98 |
| 18.65 | 1.00 | 2 | 98 |
Table 1: Equilibration column method. Schematic representation of solvent changes for the equilibration of the column. Buffer A: 7 mM ammonium acetate, pH 3.0. Buffer B: acetonitrile.
- Repeat the step 4.3 to program the run sample method indicated in Table 2. Set a flow rate of 1.00 mL/min and program the UV detector to read at 260 nm. Save the instrument method and repeat the same procedure as described in step 4.4 to save the method set..
| Time | Flow rate (ml/min) | %A | %B |
| 1.00 | 0 | 100 | |
| 3.74 | 1.00 | 0 | 100 |
| 13.71 | 1.00 | 15 | 85 |
| 17.00 | 1.00 | 100 | 0 |
| 20.00 | 1.00 | 100 | 0 |
Table 2: Run sample method. Schematic representation of solvent changes for HPLC measurement of purine compounds. Buffer A: 7 mM ammonium acetate, pH 3.0. Buffer B: acetonitrile.
- Select the "run samples" button, choose "new sample set method" from the "file" menu.
- Select the "empty" option and enter the number of the vial (in the autosampler), the sample name, the injection volume (50 µl). Select the method set saved before for the "method set" column and enter the total run time (min).
Note: Enter the equilibration column method (Table 1) in the "method set" column before each sample run and enter the total run time (min).
5. Generation of a Standard Calibration Curve for Each Compound
- Inject 50 µl of blank (specific serum-free medium, which does not contain adenosine deaminase) and of each standard sample following the methods described in Table 1 and Table 2.
- Use the equilibration column method (Table 1) prior to each sample run and set the gradient conditions described in Table 2 to obtain adequate peak separation. Refer to Table 3 for reported retention times of AMP, ADO and INO under these HPLC conditions.
| Retention Time | λmax | |
| AMP | 8.00 min | 260 |
| INO | 11.00 min | 260 |
| ADO | 11.90 min | 260 |
Table 3: Retention times of purine compounds. Typical retention times observed for AMP, ADO and INO. The UV detector is programmed to read at 260 nm.
- Determine the peak area of AMP, ADO and INO at different concentrations choosing the "process" button in the HPLC software and next the "integrate" option. Alternatively select manually the starting and end points of each peak.
- Plot the peak areas against the nominal concentration of each standard to obtain the nine-point calibration curve (100, 50, 25, 10, 5, 2.5, 1, 0.5, 0.25 µM).
- Calculate the equation of a straight line for AMP, ADO and INO: y = mx + b,
where x is the µM concentration and y correspond to the peak area.
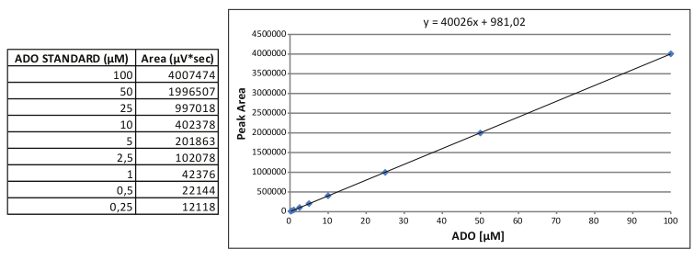
Figure 2. Generation of an internal standard curve. Representative calibration standard curve for ADO and the relative equation obtained. Please click here to view a larger version of this figure.
6. Pretreatment with the Inhibitors and Incubation with the Substrate (AMP)
- To test AMP consumption, ADO and INO generation without inhibition of CD73, ADA and nucleoside transporters, resuspend 2 x 106 CD19+/CD5+ CLL cells (isolated and purified in steps 1 and 2) in 250 µl of serum-free medium and plate cells in a 48 well plate (or in a 1.5 ml microcentrifuge tube).
- To block CD73 enzymatic activity, dilute 1:1,000 the stock solution of APCP (10 mM) in the culture medium to obtain a 10 µM final concentration. Resuspend 2 x 106 CLL cells in 250 µl of culture medium containing APCP and pretreat 60 min at 37 °C in a cell culture incubator.
- To block the conversion of ADO into INO, dilute 1:1,000 the stock solution of EHNA hydrochloride and/or dCF (both 10 mM) in serum-free medium to have a 10 µM final concentration. Resuspend 2 x 106 CLL cells in 250 µl of culture medium containing EHNA and/or dCF. Incubate for 30 min at 37 °C in the cell culture incubator.
- To block the uptake of ADO through the nucleoside transporters, dilute 1:1,000 the stock solution of dipyridamole (10 mM) in serum-free medium to have a 10 µM final concentration. Resuspend 2 x 106 CLL cells in 250 µl of culture medium containing dipyridamole and incubate for 30 min at 37 °C in the cell culture incubator.
- Prepare single solutions of APCP, EHNA, dCF and dipyridamole diluted 1:1,000 in 400 µM AMP.
- Add 250 µl of 400 µM AMP or AMP plus inhibitors to obtain a final concentration of 200 µM AMP. Include a condition in the absence of the substrate to be used as blank sample.
- Incubate from 30 to 60 min at 37 °C.
Note: The optimal concentration of AMP and the incubation time were experimentally determined. - At the end of the incubation time, collect 500 µl of the supernatants in cold microcentrifuge tubes (on ice) and immediately centrifuge at 17,000 x g at 4 °C for 5 min.
- Transfer the supernatants in new 1.5 ml microcentrifuge tubes and immediately store at -80 °C or proceed to sample preparation for the HPLC runs.
7. Samples Preparation for HPLC
- Filter the supernatants in new 1.5 ml microcentrifuge tubes with the 0.2 µm syringe filters. Use 1 ml syringes for filtering.
- If the HPLC system is provided with an autosampler, prepare the glass vials for HPLC and use the 0.1 ml micro-inserts for small volumes of sample.
- Transfer at least 100 µl of sample in the glass vial with a micropipette or a glass Pasteur pipette. Be careful to transfer without bubbles and close the vial with the screw cap.
- Samples are now ready for analysis by RP-HPLC.
8. HPLC Measurements of Purines
- Select the "run samples" button; choose "new sample set method" from the "file" menu.
- Select the "empty" option and indicate: the number of the vial (in the autosampler), the sample name, the injection volume (50 µl).
- Select the equilibration column method and the run sample method described in Table 1 and Table 2, respectively, for all the blank and sample runs that follow.
Note: Set the equilibration column method (Table 1) before each sample run. - Inject 50 µl of blank (serum-free medium) and of each sample.
- Determine the concentration of purines in each sample, quantify the AMP, ADO and INO by obtaining the peak areas at the retention times described in Table 3.
- Select the "process" button and next the "integrate" option. Alternatively choose manually the start and end points of each single peak to obtain the area measurement. This is done by tracing a line between the start and end points of the peak.
- Determine the µM concentration applying the equation obtained for the standard curve: y = mx + b, where x represents the µM concentration and y is the area of the peak measured from the unknown sample.
Note: An example of the ADO calibration standard curve is reported in Figure 2, where: x = (y area - 981.02)/40026
- Considering that 2 x 106 cells were resuspended in 0.500 ml of medium, calculate the µmoles of AMP, ADO and INO consumed and/or produced by 106 CLL cells applying the following proportion:
µmoles : 1,000 ml = x µmoles : 0.250 ml
Note: The µmoles in 1,000 ml (number µmoles in 1 L) of are equivalent to the µM concentration and x are the µmoles of nucleotide or nucleoside produced by 106 cells.- Lastly, convert the µmoles into nmoles consumed and/or produced by 106 CLL cells:
nmoles = µmoles x 1,000
- Lastly, convert the µmoles into nmoles consumed and/or produced by 106 CLL cells:
Access restricted. Please log in or start a trial to view this content.
Results
To evaluate the percentage (%) of leukemic cells in freshly purified PBMCs from a representative CLL patient, cells are marked with anti-CD19 and anti-CD5 antibodies. The left panel of Figure 3 represents a cytofluorimetric dot plot with a selective gate on live cells. Figure 3 shows an example of PBMC from a CLL patient before (middle panel) and after (right panel) B cell purification.
An examp...
Access restricted. Please log in or start a trial to view this content.
Discussion
The protocol described here permits to evaluate the activity of the CD39/CD73 adenosinergic machinery in cell culture media from purified human leukemic cells. Through this HPLC method we can follow and quantitatively measure the enzymatic generation of ADO (CD73-dependent) and its subsequent degradation to INO (CD26/ADA dependent). The use of enzyme inhibitors allows to control the protocol and to have internal controls. The advantages and novelties of this protocol are that i) it may be applied to cells that are growin...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work is supported by Associazione Italiana Ricerca Cancro (IG #12754).
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Human blood | |||
| Milli-Q water | Millipore | double deionised water | |
| Ficoll-Paque Plus | GE-Healthcare | 17-1440-03 | |
| purified anti-CD3, -CD14, -CD16 | made in-house | mouse monoclonal | |
| PE-labeled anti-CD19 | Miltenyi Biotec | 120-014-229 | |
| FITC-labeled anti-CD5 | Miltenyi Biotec | 130-096-574 | |
| Dynabeads sheep anti-mouse IgG | Invitrogen | 11031 | |
| Phosphate-buffered saline (PBS) | Amresco | E404-200TABS | tablets |
| bovine serum albumin (BSA) | ID bio | 1000-70 | standard grade |
| isolation buffer | PBS 0.1% BSA 2 mM EDTA, pH 7.4 | ||
| AIM V serum free medium | GIBCO | 12055-091 | liquid (research grade) |
| adenosine 5’-diphosphate (ADP) | Sigma-Aldrich | A2754 | |
| adenosine 5’-monosphate (AMP) | Sigma-Aldrich | A1752 | |
| adenosine (ADO) | Sigma-Aldrich | A9251 | |
| inosine (INO) | Sigma-Aldrich | I4125 | |
| α,β-methylene-ADP (APCP) | Sigma-Aldrich | M8386 | CD73 inhibitor |
| EHNA hydrochloride | Sigma-Aldrich | E114 | adenosine deaminase inhibitor |
| Deoxycoformycin (dCF) | Tocris | 2033 | adenosine deaminase inhibitor |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D2650 | |
| Dipyridamole | Sigma-Aldrich | D9766 | nucleoside transporter inhibitor |
| acetonitrile (CHROMASOLV Plus) | Sigma-Aldrich | 34998 | HPLC-grade |
| ammonium acetate | Sigma-Aldrich | 9688 | 7 mM, pH 3.0 |
| hydrochloric acid | Sigma-Aldrich | 30721-1L | min. 37% |
| Name | Company | Catalog Number | Comments |
| Equipment | |||
| Bürker cell counter | VWR | 631-0920 | hemocytometer |
| DynaMag-15 Magnet | Invitrogen | 12301D | Dynal magnetic bead separator |
| microcentrifuge safe-lock tubes | Eppendorf | 030-120-0086 | 1.5 ml |
| PET centrifuge tubes | Corning | 430053/430304 | 15 – 50 ml |
| Minisart RC4 syringe filters | Sartorius Stedim Biotech | 17821 | membrane 0.2 µm |
| short thread vials | VWR | 548-0029 | 1.5 ml/glass |
| micro-inserts | VWR | 548-0006 | 0.1 ml/glass |
| screw caps | VWR | 548-0085 | 9 mm/PP blue |
| Atlantis dC18 Column | Waters | 186001344 | 5 µm, 4.6 mm x 150 mm |
| Atlantis dC18 Guard Column | Waters | 186001323 | 5 µm, 4.6 mm x 20 mm |
| Waters Alliance 2965 Separations Module | Waters | HPLC separation module | |
| Waters 2998 Photodiode Array (PDA) Detector | Waters | UV detector | |
| Waters Empower2 software | Waters |
References
- Naganuma, M., Wiznerowicz, E. B., Lappas, C. M., Linden, J., Worthington, M. T., Ernst, P. B. Cutting edge: Critical role for A2A adenosine receptors in the T cell-mediated regulation of colitis. J Immunology. 177 (5), 2765-2769 (2006).
- Nemeth, Z. H., et al. Adenosine receptor activation ameliorates type 1 diabetes. FASEB J. 21 (10), 2379-2388 (2007).
- Fan, M., Jamal Mustafa, S. Role of adenosine in airway inflammation in an allergic mouse model of asthma. Int Immunopharmacol. 6 (1), 36-45 (2006).
- Csoka, B., et al. A2B adenosine receptors protect against sepsis-induced mortality by dampening excessive inflammation. J Immunol. 185 (1), 542-550 (2010).
- Peart, J. N., Headrick, J. P. Adenosinergic cardioprotection: multiple receptors, multiple pathways. Pharmacol Ther. 114 (2), 208-221 (2007).
- Ohta, A., et al. A2A adenosine receptor protects tumors from antitumor T cells. Proc Natl Acad Sci U S A. 103 (35), 13132-13137 (2006).
- Hasko, G., Linden, J., Cronstein, B., Pacher, P. Adenosine receptors: therapeutic aspects for inflammatory and immune diseases. Nat Rev Drug Discov. 7 (9), 759-770 (2008).
- Cronstein, B. N. Adenosine, an endogenous anti-inflammatory agent. J Appl Physiol (1985). 76 (1), 5-13 (1994).
- Molina-Arcas, M., Casado, F. J., Pastor-Anglada, M. Nucleoside transporter proteins. Curr Vasc Pharmacol. 7 (4), 426-434 (2009).
- Deaglio, S., et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 204 (6), 1257-1265 (2007).
- Linden, J. Regulation of leukocyte function by adenosine receptors. Adv Pharmacol. 61, 95-114 (2011).
- Antonioli, L., Blandizzi, C., Pacher, P., Hasko, G. Immunity, inflammation and cancer: a leading role for adenosine. Nat Rev Cancer. 13 (12), 842-857 (2013).
- Antonioli, L., Csoka, B., Fornai, M., et al. Adenosine and inflammation: what's new on the horizon. Drug Discov Today. 19 (8), 1051-1068 (1051).
- Chiorazzi, N., Rai, K. R., Ferrarini, M. Chronic lymphocytic leukemia. N Engl J Med. 352 (8), 804-815 (2005).
- Abousamra, N. K., Salah El-Din, M., Hamza Elzahaf, E., Esmael, M. E. Ectonucleoside triphosphate diphosphohydrolase-1 (E-NTPDase1/CD39) as a new prognostic marker in chronic lymphocytic leukemia. Leuk Lymphoma. 56 (1), 113-119 (2015).
- Serra, S., et al. CD73-generated extracellular adenosine in chronic lymphocytic leukemia creates local conditions counteracting drug-induced cell death. Blood. 118 (23), 6141-6152 (2011).
- Chen, L. S., Keating, M. J., Gandhi, V. Blood collection methods affect cellular protein integrity: implications for clinical trial biomarkers and ZAP-70 inn CLL. Blood. 124 (7), 1192-1195 (2014).
- Kalina, T., et al. EuroFlow standardization of flow cytometer instrument settings and immunophenotyping protocols. Leukemia. 26 (9), 1986-2010 (2012).
- Deaglio, S., et al. CD38 and ZAP-70 are functionally linked and mark CLL cells with high migratory potential. Blood. 110 (12), 4012-4021 (2007).
- Sachsenmeier, K. F., et al. Development of a novel ectonucleotidase assay suitable for high-throughput screening. J Biomol Screen. 17 (7), 993-998 (2012).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved