A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
A Fluorescence-based Assay of Phospholipid Scramblase Activity
In This Article
Summary
We describe a fluorescence-based assay to measure phospholipid scrambling in large unilamellar liposomes reconstituted with opsin.
Abstract
Scramblases translocate phospholipids across the membrane bilayer bidirectionally in an ATP-independent manner. The first scramblase to be identified and biochemically verified was opsin, the apoprotein of the photoreceptor rhodopsin. Rhodopsin is a G protein-coupled receptor localized in rod photoreceptor disc membranes of the retina where it is responsible for the perception of light. Rhodopsin's scramblase activity does not depend on its ligand 11-cis-retinal, i.e., the apoprotein opsin is also active as a scramblase. Although constitutive and regulated phospholipid scrambling play an important role in cell physiology, only a few phospholipid scramblases have been identified so far besides opsin. Here we describe a fluorescence-based assay of opsin's scramblase activity. Opsin is reconstituted into large unilamellar liposomes composed of phosphatidylcholine, phosphatidylglycerol and a trace quantity of fluorescent NBD-labeled PC (1-palmitoyl-2-{6-[7-nitro-2-1,3-benzoxadiazole-4-yl)amino]hexanoyl}-sn-glycero-3-phosphocholine). Scramblase activity is determined by measuring the extent to which NBD-PC molecules located in the inner leaflet of the vesicle are able to access the outer leaflet where their fluorescence is chemically eliminated by a reducing agent that cannot cross the membrane. The methods we describe have general applicability and can be used to identify and characterize scramblase activities of other membrane proteins.
Introduction
The photoreceptor rhodopsin, a prototypical G protein-coupled receptor (reviewed for example in reference 1), is the first phospholipid scramblase to be identified and biochemically verified 2,3. Scramblases are phospholipid transporters that increase the intrinsically slow rate of transbilayer phospholipid movement to physiologically appropriate levels in a bidirectional, ATP-independent manner 4-6. Examples of their actions can be found in the endoplasmic reticulum and bacterial cytoplasmic membrane where constitutive scrambling is needed for membrane homeostasis and growth, as well as for a variety of glycosylation pathways 5. Regulated phospholipid scrambling is needed to expose phosphatidylserine (PS) on the surface of apoptotic cells where it acts as an "eat-me"-signal for macrophages 7 and provides a procoagulant surface on activated blood platelets to catalyze the production of protein factors needed for blood clotting. In photoreceptor disc membranes, rhodopsin's scrambling activity has been suggested to counteract the phospholipid imbalance between the two membrane leaflets of the bilayer that is generated by the ATP-dependent, unidirectional lipid flippase ABCA4 4,8,910-12.
Despite the physiological importance of scramblases, their identity remained elusive until rhodopsin was reported as a scramblase in photoreceptor discs 2, members of the TMEM16 protein family were identified as Ca2+-dependent scramblases needed for PS exposure at the plasma membrane (reviewed in reference 13), and the bacterial protein FtsW was proposed as a Lipid II scramblase required for peptidoglycan synthesis 14. These discoveries were based on the reconstitution of purified proteins in liposomes and demonstration of scramblase activity in the resulting proteoliposomes using the methodology described here. Other potential scramblases 15-21 — the MurJ and AmJ proteins implicated in peptidoglycan biosynthesis, WzxE and related proteins implicated in scrambling O-antigen precursors, MprF protein needed to translocate aminoacylated phosphatidylglycerol across the bacterial cytoplasmic membrane, and Xkr8 family members that have been proposed to expose PS on the surface of apoptotic cells — remain to be tested biochemically. This highlights the importance of a robust assay to identify and characterize scramblase activity.
Here, we describe the reconstitution of purified opsin, the apoprotein of the photoreceptor rhodopsin, into large unilamellar vesicles (LUVs), and subsequent analysis of scramblase activity in the resulting proteoliposomes using a fluorescence-based assay. There are several well-described protocols available in the literature for the heterologous expression and purification of opsin, therefore we will not describe it in this protocol; we use the protocols described in Goren et al. 3 which yields FLAG-tagged, thermostable opsin at about 100 ng/µl in 0.1% (w/v) dodecylmaltoside (DDM).
Reconstitution is achieved by treating LUVs with sufficient detergent so that they swell but do not dissolve. Under these conditions, a membrane protein — supplied in the form of protein-detergent micelles — will integrate into the liposomes and become reconstituted into the liposome membrane upon detergent removal, resulting in proteoliposomes. To reconstitute opsin (obtained as a purified protein in 0.1% (w/v) DDM), LUVs are prepared from a mixture of POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) and POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)]) and saturated with DDM before adding opsin and NBD-PC. The detergent is then removed by treating the sample with polystyrene beads.
The principle underlying the fluorescence-based assay is shown in Figure 1B. LUVs are symmetrically reconstituted with a trace amount of NBD-PC or other NBD-labeled fluorescent phospholipid reporter (Figure 1A). On adding dithionite, a membrane-impermeant dianion, NBD-PC molecules in the outer leaflet of the LUVs are rendered non-fluorescent as the nitro-group of NBD is reduced to a non-fluorescent amino-group. As neither NBD-PC molecules nor dithionite are able to traverse the membrane on the time-scale of the experiment (<10 min), this results in 50% reduction of the fluorescent signal. However, if the liposomes are reconstituted with a scramblase, NBD-PC molecules in the inner leaflet can scramble rapidly to the outside where they are reduced. This results in the total loss of fluorescence in the ideal case (Figure 1C).
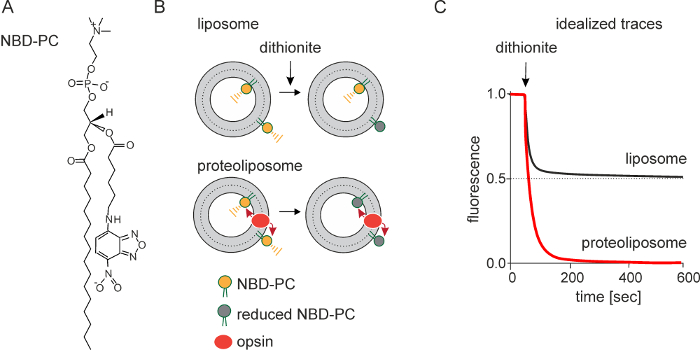
Figure 1: Schematic representation of the scramblase activity assay. The assay uses a fluorescent NBD-labeled reporter lipid; NBD-PC is shown (A). Large unilamellar vesicles are reconstituted with a trace amount of NBD-PC. Reconstitution produces symmetric vesicles, with NBD-PC distributed equally in the outer and inner leaflets. Dithionite (S2O42-) chemically reduces the nitro-group of NBD to a non-fluorescent amino-group. Treatment of protein-free liposomes with dithionite (B, top) causes a 50% reduction of fluorescence since only the NBD-PC molecules in the outer leaflet are reduced: dithionite is negatively charged and cannot cross the membrane to react with NBD-PC molecules in the inner leaflet. Dithionite treatment of opsin-containing proteoliposomes (B, bottom), i.e., scramblase-active proteoliposomes, results in 100% loss of fluorescence as opsin facilitates movement of NBD-PC between the inner and the outer leaflet. (C) shows idealized fluorescence traces obtained on treating protein-free liposomes and opsin-containing proteoliposomes with dithionite. The rate of fluorescence loss is the same in both cases indicating that the chemical reduction of NBD by dithionite is rate-limiting, and that scrambling occurs at a rate equal to or greater than the rate of the chemical reaction. Traces obtained from an actual experiment are shown in Figure 3. Please click here to view a larger version of this figure.
The methods we describe can be used to reconstitute and assay other purified proteins, as well as mixtures of membrane proteins obtained, for example, by extracting microsomes with detergent 22.
Protocol
1. Preparation of Liposomes and Proteoliposomes
- Liposome Formation
- Using a glass syringe, add 1,435 µl POPC (25 mg/ml, in chloroform) and 160 µl POPG (25 mg/ml, in chloroform) to a round bottom flask to obtain 52.5 µmol lipids in a molar ratio of POPC:POPG = 9:1.
- Dry the lipids for 30 min using a rotary evaporator at a rotation speed of 145 rpm (no water bath is needed for this volume of solvent), then transfer the flask to a vacuum desiccator for at least 3 hr, or overnight, at room temperature (RT).
- Hydrate the dried lipid film with 10 ml of 50 mM HEPES pH 7.4, 100 mM NaCl (henceforth referred to as buffer A) by gently swirling the flask until a homogenous, turbid suspension results;
NOTE: The lipid concentration at this stage is expected to be 5.25 mM as no losses should have occurred so far. - Sonicate the suspension in a water bath for 10 min at room temperature with a frequency of 40 kHz. The solution will look somewhat clearer.
- Using an extruder, pass the suspension 10 times through a membrane with a 400 nm pore size, followed by a second cycle of extrusion with 4 passes through a membrane with a 200 nm pore size.
NOTE: The mean diameter of the resulting LUVs is ~175 nm. If necessary, the size and homogeneity of the LUVs can be checked by Dynamic Light Scattering according to the manufacturer's instructions. - Quantify the phospholipid concentration of the LUV suspension as described in section 1.2).
NOTE: Because of losses during extrusion, the concentration is usually around 3.6 mM. - Store the LUVs at 4 °C for about 2 weeks if not used immediately.
- Phospholipid Quantification
NOTE: To determine the phospholipid concentration of the LUV suspension used for reconstitution, as well as that of the proteoliposomes that are eventually generated, an aliquot of the sample is subjected to oxidation by perchloric acid. This procedure breaks down phospholipids to release inorganic phosphate that is then quantified by a colorimetric assay in comparison with standards 23.- Prepare a 40 mM stock solution of sodium phosphate (Na2HPO4) in deionized distilled water.
- Dilute the stock solution with deionized distilled water to obtain 4 mM and 0.4 mM working solutions that will serve as calibration standards.
- Using the working solutions, prepare standards in 13 x 100 mm2 glass tubes ranging from 0 to 80 nmol sodium phosphate in a final volume of 50 µl.
- Take 10 µl each of the LUV and proteoliposome samples to be quantified and dilute with 40 µl of ddH2O in 13 x 100 mm2 glass tubes.
NOTE: As the lipid concentration of the LUVs and proteoliposomes is in the range 2.5-5 µM, the 10 µl of sample should contain 25-50 nmol lipid phosphorus. - Add 300 µl perchloric acid to each of the standards and samples and heat for 1 hr at 145 °C in a heating block. Put marbles on the tubes to prevent evaporation.
- Let the tubes cool to room temperature and add 1 ml of ddH2O.
- Add 400 µl each of freshly prepared 12 g/L ammonium molybdate and 50 g/L sodium ascorbate and vortex to mix.
- Heat for 10 min at 100 °C with marbles on top of the tubes. Remove the tubes from the heating block and let them cool to room temperature.
- Measure the absorbance of the samples against the blank (standard sample containing 0 nmol sodium phosphate) with a spectrometer at a wavelength of 797 nm.
- Determine the phosphate content of samples against the calibration standard curve.
- Reconstitution of Opsin
NOTE: It is necessary to determine optimal swelling conditions for reconstitution as these depend on the nature of the detergent, as well as the lipid composition and concentration of the LUVs. As LUVs change their light scattering properties on swelling the process can be monitored by measuring absorbance (Figure 2) as reviewed by Rigaud and Levy 24 and Geertsma et al. 25.- Pipette 800 µl of LUVs (from section 1.1; as lipid recovery after extrusion is 70%, the expected concentration of LUVs is 3.6 mM phospholipid) into a 2 ml microfuge tube.
- Add 5.3 µl of buffer A and 34.7 µl of 10% (w/v) DDM dissolved in buffer A.
- Incubate for 3 hr at room temperature with end-over-end mixing.
- Meanwhile prepare the polystyrene beads:
- Use 400 mg of beads per sample and weigh them out in a glass beaker.
- Wash them twice with methanol, three times with water and once with buffer A. For each washing step use 5 ml of liquid and stir slowly for 10 min.
NOTE: It is recommended to prepare the polystyrene beads for several samples at once, e.g., weigh in 6 g of beads and wash with 75 ml of liquid. Excess beads can be stored in a refrigerator for several days.
- During the last 30 min of vesicle destabilization dry the NBD-labeled phospholipid in a screw-cap glass tube ('reconstitution glass tube'): Per sample, 9.5 µl of NBD-PC (1 mg/ml in chloroform, yielding 0.4 mol% of total phospholipids) are dried under a stream of nitrogen in a glass tube and subsequently dissolved in 45 µl of 0.1% (w/v) DDM in buffer A.
- After 3 hr of vesicle destabilization add the dissolved-NBD-labeled phospholipid, the DDM-solubilized protein and buffer A such that the final volume of 1 ml contains 0.36% (w/v), i.e., 7 mM, DDM.
- Thus, to generate protein-free liposomes (used as a control sample) add 45 µl of NBD-PC (dissolved in 0.1% DDM), 60 µl of 0.1% DDM and 55 µl of buffer A; for proteoliposomes add, for example, 40 µl of protein (from a typical stock solution of ~110 ng/µl) in 0.1% DDM, 45 µl of NBD-PC (dissolved in 0.1% DDM), 20 µl of 0.1% DDM and 55 µl of buffer A.
NOTE: The order of addition should be as listed.
- Thus, to generate protein-free liposomes (used as a control sample) add 45 µl of NBD-PC (dissolved in 0.1% DDM), 60 µl of 0.1% DDM and 55 µl of buffer A; for proteoliposomes add, for example, 40 µl of protein (from a typical stock solution of ~110 ng/µl) in 0.1% DDM, 45 µl of NBD-PC (dissolved in 0.1% DDM), 20 µl of 0.1% DDM and 55 µl of buffer A.
- Mix the sample for an additional hr end over end at room temperature.
- Add 80 mg of the prepared polystyrene beads and incubate the sample with end-over-end mixing for 1 hr at RT.
- Next add an additional 160 mg of polystyrene beads and incubate with end-over-end mixing for a further 2 hr at RT.
- Transfer the sample (leaving the spent polystyrene beads behind) to a glass screw-cap tube containing 160 mg of fresh polystyrene beads and mix overnight at 4 °C.
NOTE: The easiest way to transfer the sample and avoid sucking up beads is to use a Pasteur pipette that is pushed to the bottom of the glass tube with a little positive pressure. Once the tip of the pipette is firmly at the bottom of the tube, then the sample can be withdrawn easily without interference from the beads. - The next morning, transfer the sample to a microfuge tube without carrying over beads and place on ice in preparation for the scramblase activity assay.
2. Scramblase Activity Assay
NOTE: The fluorescence intensity of liposomes or proteoliposomes diluted with buffer A is monitored over time upon the addition of dithionite in a fluorescence spectrometer. To obtain a stable starting intensity, the fluorescence is recorded for at least 50 sec (or until a stable signal is achieved) before adding dithionite to a constantly stirred sample and is then followed for at least 500 sec after adding dithionite.
- Add 1,950 µl of buffer A to a plastic cuvette containing a mini stir-bar.
- Add 50 µl of the prepared proteo(liposomes) and let the sample equilibrate in the fluorescence spectrometer with constant stirring for several sec.
- Meanwhile prepare a solution of 1 M dithionite in 0.5 M unbuffered Tris (e.g., for two samples weigh out 20 mg of dithionite in a microfuge tube and dissolve in 114 µl ice-cold 0.5 M Tris directly before use and keep on ice for the next sample).
NOTE: The dithionite solution must be prepared freshly and should not be used more than 20 min after preparation; if many measurements are to be made, aliquots of dithionite can be weighed out in advance and dissolved right before use. - Start the fluorescence monitoring (excitation 470 nm, emission 530 nm, slit width 0.5 nm).
- Add 40 µl of the 1 M dithionite solution to the cuvette 50 sec after starting the fluorescence recording (use the septum in the lid of the cuvette chamber if possible) and continue to record the fluorescence for a further 400-600 sec.
- Analyze the data as described in section 3.
3. Data Analysis
- Kinetics of Scrambling
- Characterize the fluorescence trace of each sample obtained by the scramblase activity assay by defining the initial fluorescence, Fi, prior to adding dithionite, and the end-point fluorescence, F, reached after >400 sec. Fi is determined for each sample as the mean value of fluorescence for the 30 sec period prior to addition of dithionite.
- Determine the end-point data corresponding to the extent of fluorescence reduction, R = 100•F/Fi. We use the terms RL for protein-free liposomes and RP for opsin-containing proteoliposomes.
- Determining the Molecular Weight of the Functionally Reconstituted Scramblase
- Convert the fluorescence reduction data according to the following equation:
p(≥1 scramblase) = (RP − RL)/(Rmax − RL) (Equation 1)
NOTE: Where Rmax is the maximum reduction that is obtained when sufficient protein is reconstituted such that all vesicles in the sample possess at least one functional scramblase, and p is the probability that a particular vesicle in a reconstituted sample is 'scramblase-active', i.e., it possesses at least one functional scramblase. The value for RL is typically 45% 3 whereas Rmax is typically 82.5% 3 , instead of the expected 100% (Rmax can be experimentally determined for opsin proteoliposomes with a PPR of >1 mg/mmol). As Rmax <100% it is assumed that a sub-population of vesicles is refractory to reconstitution. For Rmax = 82.5%, the fraction of vesicles that is unable to accept protein is 0.35. - Describe the relationship between p(≥1 scramblase) and PPR (mg protein/mmol phospholipid) by Poisson statistics as follows:
p(≥1 scramblase) = 1 - e-m = 1 - exp(-PPR/α) (Equation 2)
NOTE: Where m = number of scramblases per vesicle and α = mono-exponential fit constant in units of mg protein/mmol phospholipid. - As a fraction of the vesicles does not contribute to scrambling even at high PPR (see discussion), modify the equation to:
p(≥1 scramblase) = 1 - exp(-PPR*/α) (Equation 3)
NOTE: Where PPR* = PPR/(1-f), where f is the refractory population of vesicles or, in this case, PPR* = PPR/0.65 (Equation 4).
NOTE: The fit constant α is determined by fitting a graph of p(≥1 scramblase) versus PPR* with a mono-exponential function. If opsin (molecular weight 41.7 kDa) functionally reconstitutes into 175-nm-diameter vesicles (each vesicle has 280,000 phospholipids 26) as a monomer, α = 0.187 mg mmol−1. If opsin dimerizes prior to reconstitution 3 to yield scramblase-active vesicles, then α = 0.37 mg mmol−1. If PPR rather than PPR* were to be used for the analysis, then the corresponding α values would be 0.122 and 0.244 mg mmol−1. These predicted values for α assume that all opsin molecules are functionally competent. If only a fraction of the molecules is competent to scramble lipids, then the corresponding values of α will be larger.
- Convert the fluorescence reduction data according to the following equation:
Results
We describe the reconstitution of opsin into LUVs to characterize its scramblase activity using a fluorescence-based assay. We analyze the results to place a lower limit on the rate of opsin-mediated phospholipid scrambling and to determine the oligomeric state in which opsin functionally reconstitutes into the vesicles.
To identify optimal reconstitution conditions, it is necessary to determine empirically the amount of deterge...
Discussion
The scramblase activity assay enabled us originally to determine that opsin has phospholipid scramblase activity 2. The assay also allowed us to characterize opsin's scramblase activity by testing specificity (we used a variety of NBD-labeled reporter lipids such as NBD-phosphatidylethanolamine, labeled with NBD on an acyl chain as shown for NBD-PC in Figure 1A, or on the headgroup, NBD-sphingomyelin or NBD- phosphatidylserine 2), the effect of vesicle lipid composition (e.g...
Disclosures
The authors have nothing to disclose.
Acknowledgements
This study was supported by the Velux Stiftung (A.K.M.), NIH grant EY024207 (A.K.M.) and the Austrian Science Fund (FWF) project J3686 (B.P.).
Materials
| Name | Company | Catalog Number | Comments |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine | Avanti Polar Lipids | 850457C | POPC |
| 1-Palmitoyl-2-oleoyl-sn-glycero-3-phospho-rac-(1-glycerol) (sodium salt) | Avanti Polar Lipids | 840457C | POPG |
| 1-palmitoyl-2-{6-[7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphocholine | Avanti Polar Lipids | 810130C | NBD-PC |
| 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid | VWR Scientific | EM-5330 | HEPES |
| NaCl | Sigma | S7653-1KG | NaCl |
| Dodecyl-β-D-maltoside | Anatrace | D310 5 GM | DDM |
| Fluorimeter cuvettes | sigma | C0918-100EA | cuvettes |
| Spectrofluorometer | Photon Technology International, Inc. | fluorimeter | |
| Sodium hydrosulfite technical grade, 85% | Sigma | 157953-5G | dithionite |
| GraphPad Prism 5 software | Prism | ||
| Tris Base | VWR | JTX171-3 | Tris |
| LIPEX 10 ml extruder | Northern Lipids, Inc. | Extruder | |
| Whatman, Drain disc, PE, 25 mm | Sigma | 28156-243 | Disc support |
| Whatman Nuclepore Track-Etched Membranes, 0.4 µm, 25 mm diameter | Sigma | WHA110607 | 400 nm membrane |
| Whatman Nucleopore Track-Etched Membranes, 0.2 µm, 25 mm diameter | Sigma | WHA110606 | 200 nm membrane |
| sodium phosphate | Sigma | S3264-500G | |
| VWR Culture Tubes, Disposable, Borosilicate Glass, 13 x 100 mm | VWR Scientific | 47729-572 | glass tubes |
| Perchloric acid | Sigma | 30755-500ML | |
| Ammonium Molybdate Tetrahydrate | Sigma | A-7302 | ammonium molybdate |
| (+)-Sodium L-ascorbate | Sigma | A7631-25G | sodium ascorbate |
| Bio-Beads SM2 adsorbents | Bio Rad | 1523920 | polystyrene beads |
| 2.0 ml Microtubes clear | VWR Scientific | 10011-742 | Reconstitution tubes |
| Reconstitution glass tube | VWR Scientific | 53283-800 | Reconstitution glass tubes |
| Zetasizer | Malvern | DLS |
References
- Ernst, O. P., et al. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 114, 126-163 (2014).
- Menon, I., et al. Opsin is a phospholipid flippase. Curr. Biol. CB. 21, 149-153 (2011).
- Goren, M. A., et al. Constitutive phospholipid scramblase activity of a G protein-coupled receptor. Nat. Commun. 5, 5115 (2014).
- Ernst, O. P., Menon, A. K. Phospholipid scrambling by rhodopsin. Photochem. Photobiol. Sci. Off. J. Eur. Photochem. Assoc. Eur. Soc. Photobiol. , (2015).
- Sanyal, S., Menon, A. K. Flipping lipids: why an' what's the reason for?. ACS Chem. Biol. 4, 895-909 (2009).
- Bevers, E. M., Williamson, P. L. Phospholipid scramblase: an update. FEBS Lett. 584, 2724-2730 (2010).
- Leventis, P. A., Grinstein, S. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39, 407-427 (2010).
- Quazi, F., Lenevich, S., Molday, R. S. ABCA4 is an N-retinylidene-phosphatidylethanolamine and phosphatidylethanolamine importer. Nat. Commun. 3, 925 (2012).
- Quazi, F., Molday, R. S. ATP-binding cassette transporter ABCA4 and chemical isomerization protect photoreceptor cells from the toxic accumulation of excess 11-cis-retinal. Proc. Natl. Acad. Sci. U. S. A. 111, 5024-5029 (2014).
- Ben-Shabat, S., et al. Formation of a nonaoxirane from A2E, a lipofuscin fluorophore related to macular degeneration, and evidence of singlet oxygen involvement. Angew. Chem. Int. Ed Engl. 41, 814-817 (2002).
- Sparrow, J. R., Wu, Y., Kim, C. Y., Zhou, J. Phospholipid meets all-trans-retinal: the making of RPE bisretinoids. J. Lipid Res. 51 (2010), 247-261 (2010).
- Schütt, F., Davies, S., Kopitz, J., Holz, F. G., Boulton, M. E. Photodamage to human RPE cells by A2-E, a retinoid component of lipofuscin. Invest. Ophthalmol. Vis. Sci. 41, 2303-2308 (2000).
- Picollo, A., Malvezzi, M., Accardi, A. TMEM16 proteins: unknown structure and confusing functions. J. Mol. Biol. 427, 94-105 (2015).
- Mohammadi, T., et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 30, 1425-1432 (2011).
- Meeske, A. J., et al. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc. Natl. Acad. Sci. 112, 6437-6442 (2015).
- Ruiz, N. Lipid Flippases for Bacterial Peptidoglycan Biosynthesis. Lipid Insights. 8, 21-31 (2015).
- Rick, P. D., et al. Evidence that the wzxE gene of Escherichia coli K-12 encodes a protein involved in the transbilayer movement of a trisaccharide-lipid intermediate in the assembly of enterobacterial common antigen. J. Biol. Chem. 278, 16534-16542 (2003).
- Suzuki, J., Imanishi, E., Nagata, S. Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J. Biol. Chem. 289, 30257-30267 (2014).
- Suzuki, J., Nagata, S. Phospholipid scrambling on the plasma membrane. Methods Enzymol. 544, 381-393 (2014).
- Alaimo, C., et al. Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 25, 967-976 (2006).
- Ernst, C. M., Peschel, A. Broad-spectrum antimicrobial peptide restistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol. Microbial. 80 (2), 290-299 (2011).
- Vehring, S., et al. Flip-flop of fluorescently labeled phospholipids in proteoliposomes reconstituted with Saccharomyces cerevisiae microsomal proteins. Eukaryot. Cell. 6, 1625-1634 (2007).
- Rouser, G., et al. Two dimensional then [sic] layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids. 5, 494-496 (1970).
- Rigaud, J. -. L., Lévy, D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 372, 65-86 (2003).
- Geertsma, E. R., Nik Mahmood, N. A. B., Schuurman-Wolters, G. K., Poolman, B. Membrane reconstitution of ABC transporters and assays of translocator function. Nat. Protoc. 3, 256-266 (2008).
- Mimms, L. T., Zampighi, G., Nozaki, Y., Tanford, C., Reynolds, J. A. Phospholipid vesicle formation and transmembrane protein incorporation using octyl glucoside. Biochemistry. 20, 833-840 (1981).
- Malvezzi, M., et al. Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat. Commun. 4, 2367 (2013).
- Eckford, P. D. W., Sharom, F. J. The reconstituted P-glycoprotein multidrug transporter is a flippase for glucosylceramide and other simple glycosphingolipids. Biochem. J. 389, 517-526 (2005).
- Marek, M., Günther-Pomorski, T. Assay of Flippase Activity in Proteoliposomes Using Fluorescent Lipid Derivatives. Methods Mol. Biol. 1377, 181-191 (2016).
- Coleman, J. A., Kwok, M. C. M., Molday, R. S. Localization purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J. Biol. Chem. 284, 32670-32679 (2009).
- Coleman, J. A., Quazi, F., Molday, R. S. Mammalian P4-ATPases and ABC transporters and their role in phospholipid transport. Biochim. Biophys. Acta. 1831, 555-574 (2013).
- Romsicki, Y., Sharom, F. J. Phospholipid flippase activity of the reconstituted P-glycoprotein multidrug transporter. Biochemistry. 40, 6937-6947 (2001).
- Zhou, X., Graham, T. R. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc. Natl. Acad. Sci. U. S. A. 106, 16586-16591 (2009).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved