A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
G Protein-selective GPCR Conformations Measured Using FRET Sensors in a Live Cell Suspension Fluorometer Assay
In This Article
Summary
Simple methods to detect the selective activation of G proteins by G protein-coupled receptors remain an outstanding challenge in cell signaling. Here, Fӧrster resonance energy transfer (FRET) biosensors have been developed by pairwise tethering a GPCR to G protein peptides to probe conformational changes at controlled concentrations in live cells.
Abstract
Fӧrster resonance energy transfer (FRET)-based studies have become increasingly common in the investigation of GPCR signaling. Our research group developed an intra-molecular FRET sensor to detect the interaction between Gα subunits and GPCRs in live cells following agonist stimulation. Here, we detail the protocol for detecting changes in FRET between the β2-adrenergic receptor and the Gαs C-terminus peptide upon treatment with 100 µM isoproterenol hydrochloride as previously characterized1. Our FRET sensor is a single polypeptide consisting serially of a full-length GPCR, a FRET acceptor fluorophore (mCitrine), an ER/K SPASM (systematic protein affinity strength modulation) linker, a FRET donor fluorophore (mCerulean), and a Gα C-terminal peptide. This protocol will detail cell preparation, transfection conditions, equipment setup, assay execution, and data analysis. This experimental design detects small changes in FRET indicative of protein-protein interactions, and can also be used to compare the strength of interaction across ligands and GPCR-G protein pairings. To enhance the signal-to-noise in our measurements, this protocol requires heightened precision in all steps, and is presented here to enable reproducible execution.
Introduction
G-protein-coupled receptors (GPCRs) are seven-transmembrane receptors. The human genome alone contains approximately 800 genes coding for GPCRs, which are activated by a variety of ligands including light, odorants, hormones, peptides, drugs and other small molecules. Nearly 30% of all pharmaceuticals currently on the market target GPCRs because they play a large role in many disease states2. Despite decades of extensive work done on this receptor family, there remain significant outstanding questions in the field, particularly with regards to the molecular mechanisms that drive GPCR-effector interactions. To date, only one high-resolution crystal structure has been published, providing insight into the interaction between the β2-adrenergic receptor (β2-AR) and the Gs protein3. Together with extensive research in the last three decades, it reiterates one specific structural component that is critical in this interaction: the Gα subunit C-terminus. This structure is important for both G protein activation by the GPCR4 and G protein selection5-6. Hence, the Gα C-terminus provides a crucial link between ligand stimulation of the GPCR and selective G protein activation.
Research over the last decade suggests that GPCRs populate a broad conformational landscape, with ligand-binding stabilizing subsets of GPCR conformations. While several techniques, including crystallography, NMR and fluorescence spectroscopy, and mass spectrometry are available to examine the GPCR conformational landscape, there is a paucity of approaches to elucidate their functional significance in effector selection7. Here, we outline a Fӧrster resonance energy transfer (FRET)-based approach to detect G protein-selective GPCR conformations. FRET relies on the proximity and parallel orientation of two fluorophores with overlapping emission (donor) and excitation (acceptor) spectra8. As the donor and acceptor fluorophores come closer together as a result of either conformational change in the protein or a protein-protein interaction, the FRET between them increases, and can be measured using a range of methods8. FRET-based biosensors have been employed extensively in the GPCR field9. They have been used to probe conformation changes in the GPCR by inserting donor and acceptor in the third intracellular loop and GPCR C-terminus; sensors have been designed to probe GPCR and effector interactions by separately labeling the GPCR and effector (G protein subunits/arrestins) with a FRET pair10; some sensors also detect conformational changes in the G protein11. These biosensors have enabled the field to ask a multitude of outstanding questions including conformational changes in the GPCR and effector, GPCR-effector interaction kinetics, and allosteric ligands12. Our group was particularly interested in creating a biosensor that could detect G protein-specific GPCR conformations under agonist-driven conditions. This biosensor relies on a recently developed technology named SPASM (systematic protein affinity strength modulation)13. SPASM involves tethering interacting protein domains using an ER/K linker, which controls their effective concentrations. Flanking the linker with a FRET pair of fluorophores creates a tool which can report the state of the interaction between proteins12. Previously1 the SPASM module was used to tether the Gα C-terminus to a GPCR and monitor their interactions with FRET fluorophores, mCitrine (referred to in this protocol by its commonly known variant, Yellow Fluorescent Protein (YFP), excitation/emission peak at 490/525 nm) and mCerulean (referred to in this protocol by its commonly known variant Cyan Fluorescent Protein (CFP), excitation/emission peak 430/475 nm). From N- to C-terminus, this genetically encoded single polypeptide contains: a full length GPCR, FRET acceptor (mCitrine/YFP), 10 nm ER/K linker, FRET donor (mCerulean/CFP), and the Gα C-terminus peptide. In this study, sensors are abbreviated as GPCR-linker length-Gα peptide. All components are separated by an unstructured (Gly-Ser-Gly)4 linker which enables free rotation of each domain. The detailed characterization of such sensors was previously performed using two prototypical GPCRs: β2-AR and opsin1.
This sensor is transiently transfected into HEK-293T cells and fluorometer-based live cell experiments measure fluorescence spectra of the FRET pair in arbitrary units of counts per second (CPS) in the presence or absence of ligand. These measurements are used to calculate a FRET ratio between the fluorophores (YFPmax/CFPmax). A change in FRET (ΔFRET) is then calculated by subtracting the average FRET ratio of untreated samples from the FRET ratio of ligand treated samples. ΔFRET can be compared across constructs (β2-AR-10 nm-Gαs peptide versus β2-AR-10 nm-no peptide). Here, we detail the protocol to express these sensors in live HEK-293T cells, monitor their expression, and the setup, execution, and analysis of the fluorometer-based live cell FRET measurement for untreated versus drug treated conditions. While this protocol is specific for the β2-AR-10 nm-Gαs peptide sensor treated with 100 µM isoproterenol bitartrate, it can be optimized for different GPCR-Gα pairs and ligands.
Access restricted. Please log in or start a trial to view this content.
Protocol
1. DNA Preparation
- Design sensor constructs using a modular cloning scheme. Please reference the β2-AR sensor design detailed previously1.
- Prepare DNA according to commercial miniprep kit protocol and elute in 2 mM Tris-HCl solution, pH 8, at concentration ≥ 750 ng/µl, A260/A280 of 1.7 - 1.9, A260/A230 of 2.0 - 2.29.
2. Cell Culture Preparation
- Culture HEK-293T-Flp-n cells in DMEM containing 4.5 g/L D-glucose, supplemented with 10% FBS (heat inactivated) (v/v), 1% L-glutamine supplement, 20 mM HEPES, pH 7.5 at 37 °C in humidified atmosphere at 5% CO2. Handle cells in biological safety hood for subsequent steps.
- Allow cells to grow to a confluent monolayer before passaging into six-well dishes. Time to achieve confluency depends on initial plating density. Use plates that come to confluency within 1 - 2 days of plating for six-well plating. A confluent 10 cm tissue culture-treated dish has cell density of approximately 4 x 106 cells/ml. See Figure 1 for image of cell culture growth.
- Wash cells with 10 ml PBS, and trypsinize with 0.25% trypsin (see Discussion, paragraph 2). Plate 8 x 105 cells/well in 2 ml of media in tissue culture-treated six well dishes and allow to adhere for 16 - 20 hr.
3. Transfection Conditions
- Stagger transfections for constructs that may require different amounts of time to achieve optimal expression (between 20 - 36 hr). Synchronize conditions for a unified experiment time. Also have an untransfected control well at equivalent cell density to be used for background noise and scattering subtraction during analysis.
- Bring transfection reagents to room temperature: reduced serum media, DNA, transfection reagent.
- In a biological safety hood combine reagents in a sterile microcentrifuge tube in the following order: mix 2 µg DNA with 100 µl reduced serum media. Spike 6 µl of transfection reagent into media/DNA mix without touching surface of mixture or the side of the tube. Set up one transfection reaction per well. Transfection conditions can be optimized (1 - 4 µg of DNA, 3 - 6 µl of transfection reagent) to achieve consistent expression levels. See Table 1 for more optimized ratios.
- Incubate mixture at RT in biological safety hood for 15 - 30 min. Do not use reaction if left to incubate for more than 30 min.
- Add reaction to cells in a drop-wise manner across well and gently shake six-well to ensure thorough mixing. Add one reaction per well.
- After 20 hr of expression, monitor fluorescence using tissue culture fluorescence microscope. Assess population expression with 10X objective and protein localization in a cell at 40X. Observe for protein expression at plasma membrane (PM). If substantial internalization is noted, monitor transfection until significant expression is detected at PM.
4. Reagent and Equipment Preparation
- Prepare 100 mM drug stocks and store at -80 °C: isoproterenol bitartrate (100 mM in dH2O containing 300 mM ascorbic acid). Make on ice/in cold room, and flash-freeze immediately. Aliquots can be made and used up to one year.
- Prepare Cell Buffer (~ 2 ml/condition) and store in a 37 °C water bath. Make fresh each day. Reference Table 2 for Cell Buffer constituents.
- Prepare Drug Buffer (10 ml) and store at room temperature. Reference Table 2 for Drug Buffer constituents.
- Acid wash cuvettes using concentrated HCl. Neutralize with a weak base (1 M KOH), and thoroughly wash cuvettes with dH2O.
- Prepare work station around fluorometer with several boxes of 10, 200, and 1,000 µl pipette tips, a timer set with a 10 min countdown, an accessible vacuum line with tips for cuvette cleaning, delicate task wipes, and squirt bottle with ultrapure H2O.
- Heat external water bath for fluorometer and heat block to 37 °C.
- Turn on fluorometer; set fluorescence collection program for CFP collection to excitation 430 nm, bandpass 8 nm; emission range 450 nm - 600 nm, bandpass 4 nm. For YFP collection only as sensor control (see Discussion) set excitation to 490 nm, bandpass 8 nm, emission range 500 - 600 nm, bandpass 4 nm. CFP collection settings will be used to acquire a FRET spectrum in this experiment.
- Place twelve 1.5 ml microcentrifuge tubes in heat block as shown in Figure 2 below. These tubes are holders for cell aliquot tubes (500 µl microcentrifuge tubes.) Place a small piece of tissue in holders 1 and 7 to cushion the cuvettes placed here.
Note: Use separate cuvettes for untreated condition and drug condition to prevent cross-contamination.

Figure 2. Microcentrifuge Tube Set Up and Position Reference in Heat Block. Cuvette for untreated samples is in position 1; cell aliquot tubes are in positions 2 - 6. Cuvette for drug treated samples is in position 7; cell aliquot tubes are in positions 8 - 12. Please click here to view a larger version of this figure.
- Fill holders 2 - 6 and 8 - 12 with ~ 750 µl of water to create a mini 37 °C water bath.
- Place ten 500 µl microcentrifuge tubes for cell aliquots into mini-water baths (holders 2 - 6, 8 - 12). Each tube will be an individual repeat of the condition (5 untreated, 5 drug treated).
- Monitor cells for expression (see step 3.6).
5. Experiment & Data Collection
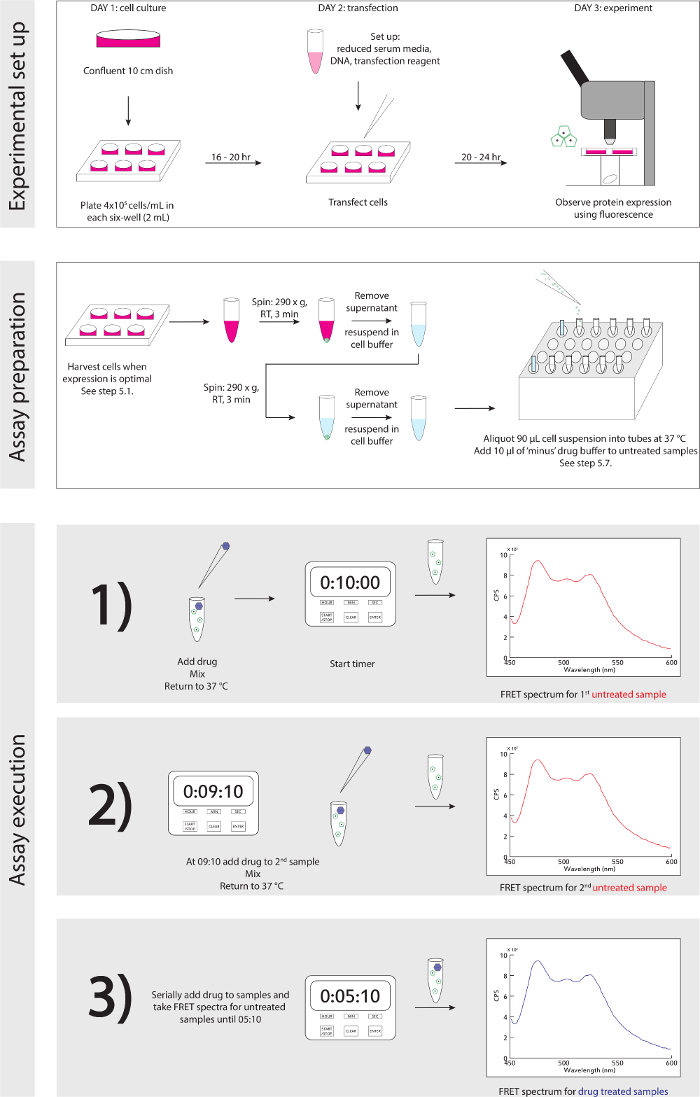
Figure 3. Experimental Schematic. A detailed step-wise guide for experimental set up and execution. Please click here to view a larger version of this figure.
- Reference Figure 3 for experimental schematic. When cells are ready to be harvested, based on protein expression detected with fluorescence microscope (see Discussion, paragraph 4): in biological safety hood, gently remove ~1 ml of media, resuspend cells in their culture with a P1,000 and transfer resuspension into a 1.5 ml microcentrifuge tube.
Note: Avoid using trypsin as it may digest the N-terminus and/or binding pocket of the GPCR sensor. - Count cells to ensure proper cell density in resuspension. Optimize resuspension volume for 4 x 106 cells/ml.
- Spin cells in swinging bucket centrifuge at room temperature, 290 x g for 3 min. Remove supernatant after the centrifugation.
- GENTLY resuspend cells in 1 ml Cell Buffer (stored at 37 °C) and repeat step 5.3. During second spin, gather 100 mM drug stock aliquot from -80 °C. Make 1:100 dilution in Drug Buffer for 1 mM working stock and keep at RT.
- After the second centrifugation, remove supernatant and gently resuspend cells in 1 ml of Cell Buffer (4 x 106 cells/ml). Measure OD600 of the sample in the spectrophotometer using 1 ml of cells and 1 ml of Cell Buffer as a blank. Dispense cells in a disposable plastic cuvette and transfer back to a microcentrifuge tube immediately following spectrophotometry.
- For a control untransfected cell condition spectrum, gently resuspend untransfected cells in 1 ml Cell Buffer with P1,000 pipet, add 90 µl of cells to cuvette and acquire FRET spectrum at excitation 430 nm, bandpass 8 nm, emission 450 - 600 nm, bandpass 4 nm. Collect 3 - 5 repeat spectra with fresh 90 µl of cells. Keep stock of cells at 37 °C between specs, resuspend gently with P1,000 between each sample aliquot, and rinse cuvette with ultrapure H2O between samples.
- For experimental conditions, aliquot 90 µl of transfected cells to each of the 500 µl tubes in holders 2 - 6, 8 - 12 in the heat block. Gently resuspend stock of cells with P1,000 pipet between each aliquot.
- After cells are aliquoted, add 10 µl of Drug Buffer to tubes 2 - 6 for untreated condition samples.
- Begin experiment by adding 10 µl of 1 mM drug solution into tube 8, start the timer to countdown from 10 min, and gently mix tube with P200 pipet. Close tube and return to 37 °C heat block.
- Immediately pick up tube 2, mix gently with P200 (use a new tip), add 90 µl of cell suspension to untreated condition cuvette, and place in fluorometer.
- Acquire FRET spectrum at excitation 430 nm, bandpass 8 nm, emission 450 - 600 nm, bandpass 4 nm.
- At 9 min - 10 sec, spike tube 9 with 10 µl of 1 mM drug solution, gently mix with a P200 (use new tip), and return tube to heat block.
- Repeat steps 5.10 - 5.11 with tube 3 and 5.12 with tube 10.
- Repeat steps 5.10 - 5.13 at 1 min intervals (08:10, 07:10, etc.) until spectra are collected for all untreated condition samples, and drug has been added to all drug condition samples. Use a fresh tip for each pipet step to prevent cross-contamination.
- At 5 min - 10 sec, begin mixing tube 8 (drug condition) gently with P200 pipet, add 90 µl of cell suspension to separate cuvette for drug treated sample and place in fluorometer.
- Acquire FRET spectrum (see step 5.11 for settings).
- Repeat steps 5.15 - 5.16 at 1 min intervals (04:10, 03:10, etc.) for remaining drug condition samples (tubes 9 - 12).
- After experiment ends, save project files, thoroughly wash cuvettes with ultrapure H2O, and re-stock tubes for next condition. Take care to prevent cross-contamination in the wash step. Change the tip on the H2O bottle as well as on the vacuum line in between washes.
6. Data Analysis
- Save and export data files in SPC format to be used for analysis. Analysis programs are available for download from the Sivaramakrishnan Lab publication website.
- Create path files for analysis software which include the analysis programs (v9, v15), untransfected samples files (see step 5.6 for untransfected cell spectrum collection), OUTPUT data file, and comma separated values (CSV) files for data entry.
- Enter following information into CSV file (see sample in Table 3) and designate respective conditions for each sample, including:
File name - individual SPC graph files
Receptor - designate which GPCR construct was tested (e.g., Β2)
Binder - designate which peptide variant of the construct was tested (e.g., S)
Agonist - designate untreated (N) or drug treated (D) conditions
Directory - the path folder in which SPC files are saved, usually organized by date
OD - recorded optical density of sample from spectrophotometer - Enter file names for untransfected samples (step 5.6) to subtract buffer and scattering noise from samples.
- Enter conditions into analysis program.
- Run programs to analyze samples within individual conditions (v9) and across conditions (v15).
- Exclude sample files which are apparent outliers in the data set, or adjust for subtraction by increasing or decreasing OD value of individual files.
- Export data to OUTPUT file for access to calculated FRET ratios (525 nm/475 nm).
- Calculate ΔFRET by subtracting the average FRET ratio for untreated condition from the individual FRET ratios for treated (drug) conditions.
Access restricted. Please log in or start a trial to view this content.
Results
A generalized schematic of the experiment set up and execution is detailed in Figure 3.
In order to detect a FRET change in the narrow dynamic range of the sensor, it is critical to adhere to the nuances of the system be adhered to. Cell quality is imperative to protein expression as well as consistency in sampling. Figure 1 features images of cultured cells growing in a consistent monolayer (10X) that is optimal for six-well plating and transfection F...
Access restricted. Please log in or start a trial to view this content.
Discussion
The tight dynamic range of FRET measurements in this system reinforces the necessity of sensitive quality control in every step of this protocol. The most important steps to ensure a successful FRET experiment are 1) cell culturing, 2) transfection 3) protein expression and 4) timely, precise coordination during the assay execution.
Cell health and maintenance/plating quality can have a significant impact on the signal-to-noise of the experimental system and poor cell health can make it imposs...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors declare that they have no competing interests.
Acknowledgements
R.U.M was funded by the American Heart Association Pre-doctoral Fellowship (14PRE18560010). Research was funded by the American Heart Association Scientist Development Grant (13SDG14270009) & the NIH (1DP2 CA186752-01 & 1-R01-GM-105646-01-A1) to S.S.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| B2-AR-10 nm-Gas peptide sensor | Addgene | 47438 | https://www.addgene.org/Sivaraj_Sivaramakrishnan/ |
| GeneJET Plasmid Miniprep Kit | Fermentas/Fisher Sci | FERK0503 | Elute in 2 mM Tris elution buffer |
| HEK-293T-Flp-n cells | Life Technologies | R78007 | |
| Trypsin (0.25%) | Life Technologies | 25200056 | |
| DMEM- high glucose | Life Technologies | 11960-044 | Warm in 37 °C water bath before use |
| FBS, certified, Heat inactivated, US origin | Life Technologies | 10082147 | |
| Glutamax I 100x | Life Technologies | 35050061 | |
| HEPES | Corning | MT25060CL | |
| Opti-MEM | Life Technologies | 31985-070 | Reduced serum media; Bring to RT before use |
| XtremeGene HP transfection reagenet | Roche | 6366236001 | Highly recommended for its consistency. Bring to RT before use |
| FluoroMax 4 | Horiba | Use with FluorEssence V3.8 software | |
| 3-mm path length quartz cuvette | Starna | NC9729944(16.45F-Q-3/z8.5) | May require cuvette holder/adaptor for use in Fluorometer, available from Starna |
| Sc100-S3 Heated Circulating water bath pump | Fisher Scientific | 13-874-826 | Warm to 37 °C before use |
| Thermomixer Heat Block | Eppendorf | 22670000 | Warm to 37 °C before use |
| Ultrapure DNA/RNAse free water | Life Technologies | 10977015 | Use at RT |
| D(+)-glucose, anhydrous | Sigma | G5767 | |
| aprotinin from bovine lung | Sigma | A1153 | |
| leupeptin hemisulfate | EMD | 10-897 | |
| L-ascorbic acid, reagent grade | Sigma | A0278 | |
| (-)-isoproterenol (+)-bitartrate | Sigma | I2760 | Use fresh aliquot each experiment |
References
- Malik, R. U., et al. Detection of G Protein-selective G Protein-coupled Receptor (GPCR) Conformations in Live Cells. J. Biol. Chem. 288, 17167-17178 (2013).
- Oldham, W. M., Hamm, H. E. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Bio. 9, 60-71 (2008).
- Rasmussen, S. G., et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 477, 549-555 (2011).
- Alexander, N. S., et al. Energetic analysis of the rhodopsin-G-protein complex links the α5 helix to GDP release. Nat. Struct. Mol. Biol. 21 (1), 56-63 (2014).
- Conklin, B. R., Farfel, Z., Lustig, K. D., Julius, D., Bourne, H. R. Substitution of three amino acids switches receptor specificity of Gqα to that of Giα. Nature. 363, 274-276 (1993).
- Conklin, B. R., et al. Carboxyl-Terminal Mutations of Gqα and Gsα That Alter the Fidelity of Receptor Activation. Mol. Pharmacol. 50, 855-890 (1996).
- Onaran, H. O., Costa, T. Where have all the active receptor states gone. Nat. Chem. Bio. 8, 674-677 (2012).
- Jares-Erijman, E. A., Jovin, T. M. FRET imaging. Nat Biotechnol. 21 (11), 1387-1395 (2003).
- Lohse, M. J., Nuber, S., Hoffman, C. Fluorescence/bioluminescence resonance energy transfer techniques to study G-protein-coupled receptor activation and signaling. Pharmacol Rev. 64 (2), 299-336 (2012).
- Vilardaga, J. P., Bünemann, M., Krasel, C., Castro, M., Lohse, M. J. Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nat. Biotechnol. 21, 807-812 (2003).
- Bünemann, M., Frank, M., Lohse, M. J. Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc. Natl. Acad. Sci. USA. 100 (26), 16077-16082 (2003).
- Stumpf, A. D., Hoffman, C. Optical probes based on G protein-coupled receptors - added work or added value. Brit. J. Pharmacol. 173, 255-266 (2016).
- Sivaramakrishnan, S., Spudich, J. A. Systemic control of protein interaction using a modular ER/K α-helix linker. Proc. Natl. Acad. Sci. USA. 108, 20467-20472 (2011).
- Shaner, N. C., Steinbach, P. A., Tsien, R. Y. A guide to choosing fluorescent proteins. Nat. Methods. 2 (12), 905-909 (2005).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved