Characterization of Immune Cells in Human Adipose Tissue by Using Flow Cytometry
* These authors contributed equally
In This Article
Summary
This article describes a method to analyze immune cell content of adipose tissue by isolation of immune cells from adipose tissue and subsequent analysis using flow cytometry.
Abstract
Infiltration of immune cells in the subcutaneous and visceral adipose tissue (AT) deposits leads to a low-grade inflammation contributing to the development of obesity-associated complications such as type 2 diabetes. To quantitatively and qualitatively investigate the immune cell subsets in human AT deposits, we have developed a flow cytometry approach. The stromal vascular fraction (SVF), containing the immune cells, is isolated from subcutaneous and visceral AT biopsies by collagenase digestion. Adipocytes are removed after centrifugation. The SVF cells are stained for multiple membrane-bound markers selected to differentiate between immune cell subsets and analyzed using flow cytometry. As a result of this approach, pro- and anti-inflammatory macrophage subsets, dendritic cells (DCs), B-cells, CD4+ and CD8+ T-cells, and NK cells can be detected and quantified. This method gives detailed information about immune cells in AT and the amount of each specific subset. Since there are numerous fluorescent antibodies available, our flow cytometry approach can be adjusted to measure various other cellular and intracellular markers of interest.
Introduction
Obesity is characterized with low-grade AT inflammation1 and infiltration of pro-inflammatory immune cells in both visceral and subcutaneous AT (vAT, sAT). Accumulation of pro-inflammatory immune cells in the vAT leads to insulin resistance which is a primary risk factor for developing type 2 diabetes2. Immune cells of both the innate and adaptive immune system are found in the obese AT, such as macrophages, mast cells, neutrophils, CD4+ and CD8+ T-cells, and B-cells3,4,5,6,7. These immune cells, together with endothelial cells, stromal cells, adipocyte progenitors, fibroblasts, and pericytes, constitute the SVF8 and are the main source of pro-inflammatory substances in the AT9.
The inflammatory status of AT is commonly investigated by techniques including Western blot10, qPCR11, and immunohistochemistry11. However, when using these techniques, the entire AT, adipocytes, and SVF, is used. This makes it difficult to determine the amount and subsets of immune cells present in the AT. Immune cells have various cell markers to define and categorize them, such as macrophages. Macrophages show significant heterogeneity in both function and cell surface marker expression12. Therefore, they are often categorized into two macrophage populations: M1 and M2. M2 macrophages are usually called alternatively activated macrophages12,13 and reside in the AT of lean, metabolically normal humans14. However, during obesity, a phenotypic switch occurs from M2 macrophages to M1 macrophages. These classically activated M1 macrophages express CD11C12 and accumulate around dead adipocytes to form crown-like structures13. It has been shown that CD11C+ macrophages in the AT impair insulin action and are associated with insulin resistance in obese humans15. To identify M1 and M2 macrophages in the AT, immunohistochemistry is an option. This technique gives information about the location of the macrophages in the tissue. However, it will limit the number of markers that can be used in one staining. Moreover, it is also difficult to quantify. Therefore, to investigate the different immune cell subsets in the vAT and sAT deposits, we have developed a flow cytometry approach. This approach gives us the opportunity to use multiple markers per cell with one flow cytometry analysis to define cell subsets and count the numbers of each subset present in the AT deposits.
Protocol
Visceral and subcutaneous AT samples were taken from subjects enrolled in the study approved by the Medical Ethical committee Jessa Hospital, Hasselt, and Hasselt University, Belgium, in accordance with the Declaration of Helsinki.
1. Preparation of Reagents
- Collagenase solution
- Dissolve 1 g of Collagenase I in 10 mL of phosphate buffered saline (PBS, without calcium and magnesium) to make a 100 mg/mL stock solution. Prepare 200 µL aliquots and store at -20 °C.
- Dissolve 1 g of Collagenase XI in 10 mL of PBS to make a 100 mg/mL stock solution. Prepare 200 µL aliquots and store at -20 °C.
- Dissolve 10 mg of DNase I in 10 mL of PBS to make a 10 mg/mL stock solution. Prepare 180 µL aliquots and store at -20 °C.
- Add 100 µL Collagenase I (100 mg/mL), 100 µL Collagenase XI (100 mg/mL), and 90 µL DNase I (10 mg/mL) to 10 mL of DMEM Ham's F12. Make collagenase solution fresh for each isolation.
- Erythrocyte lysis buffer
- Dissolve 0.84 g NH4Cl in 100 mL of ultrapure water.
- Set the pH at 7.4 before use. Store in a glass flask at 4 °C.
- Place the erythrocyte lysis buffer on ice before use.
- FACS buffer
- Dissolve 0.5 g bovine serum albumin (BSA) in 100 mL of PBS to obtain 0.5% BSA PBS.
- Dissolve 65 mg of NaN3 in 100 mL 0.5% BSA PBS to obtain 10 mM NaN3 0.5% BSA PBS. Store solution in a glass flask at 4 °C.
- Place FACS buffer on ice before use.
CAUTION: NaN3 is highly toxic. Work in a fume hood and wear safety glasses and gloves for protection while handling NaN3.
- Human IgG block
- Dissolve 10 mg of human IgG in 10 mL PBS to obtain 1 mg/mL. Prepare 100 µL aliquots and store at -20 °C.
- Place the human IgG block on ice before use.
2. Isolation of SVF from AT
- Cut 1 g of AT biopsy into small pieces (± 2 mm2) with a scalpel and transfer to a 50-mL centrifuge tube (e.g., Falcon tube). Add 10 mL of collagenase solution to each AT sample.
NOTE: Close the lid of the tube completely and turn the lid ¼ turn back. - Incubate for 60 min at 37 °C in a water-bath under gentle shaking (60 cycles/min).
- Filter the resulting suspension with a 200 µM filter and collect the sample in a new 50-mL centrifuge tube. Add 7 mL PBS on top of the filter to rinse the filter and obtain all the cells.
- Centrifuge the sample at 280 x g for 5 min at 4 °C.
- Remove the floating adipocyte fraction by pipetting. The cell pellet is the SVF.
NOTE: Remove the adipocyte fraction to obtain SVF. Avoid submerging the entire tip in the sample because this will only remove the PBS and not the floating adipocytes. - Resuspend the SVF in 5 mL of PBS to remove collagenase, filter the suspension with a 70 µM filter, rinse the filter with 5 mL PBS, and centrifuge the sample at 280 x g for 5 min at 4 °C.
- Remove the supernatant and resuspend the pellet in 3 mL of erythrocyte lysis buffer.
- Incubate for 5 min on ice. Add 7 mL of PBS after incubation.
- Centrifuge the sample at 280 x g for 5 min at 4 °C.
3. Staining of SVF for Flow Cytometry Analysis
- Dissolve the cell pellet in 90 µL 4 °C FACS buffer and add 10 µL of 1 mg/mL human IgG block. Divide the cell suspension in 2 wells of a 96 v-shape well plate. Place the plate on ice and let the human IgG block incubate for 15 min.
- Add 100 µL FACS buffer to each sample to wash and centrifuge the plate for 5 min with 280 x g at 4 °C. Remove the supernatant by tipping the plate upside down in one smooth movement without tapping the plate.
NOTE: Make sure to remove any remaining liquid from the top of the plate with a tissue while keeping the plate upside down. - Prepare antibody cocktails for macrophage and DC subsets (FACS panel 1) and for T- and B-cell subsets (FACS panel 2) as described in Table 1 and Table 2. The volumes described in Table 1 and Table 2 are selected after optimizing the antibody concentrations and are sufficient for one vAT or sAT sample.
NOTE: In FACS panel 1, use the markers CD303 and CD141 to confirm that CD11C+ CD11Blow cells are DCs. However, it is recommended to exclude these markers from the panel to include a live/dead staining. Both FACS panel 1 and 2 can be combined with the LIVE/DEAD Fixable Red Dead Cell Stain Kit viability staining when excluding CD303 in panel 1 as the PE channel will be unused. Perform viability staining according to the manufacturer's instructions. - Resuspend the pellet in 29.5 µL antibody cocktail for FACS panel 1, and 23 µL antibody cocktail for FACS panel 2. Incubate for 30 min in the dark on ice.
- Add 150 µL FACS buffer to each well and resuspend the cell pellet to perform a second wash step. Centrifuge the plate for 5 min at 280 x g and 4 °C. Remove the supernatant by tipping the plate upside down.
- Add 150 µL 1% formaldehyde solution to each well to fix the cells. Transfer the cell suspension from each well to the corresponding FACS tube by pipetting with a P200 pipette. Store FACS tubes at 4 °C in the dark up to 7 days.
NOTE: Direct measurement is also possible. Add 150 µL FACS buffer to each well instead of 1% formaldehyde, transfer the cells by pipetting with a P200 pipette to the corresponding FACS tubes, and analyze the cells.
CAUTION: Formaldehyde is very toxic. Prepare formaldehyde solutions while working in a fume hood to avoid inhalation and wear gloves and safety glasses for protection.
4. Flow Cytometry Analysis
- Before the first measurement, use an unstained negative control to set the forward scatter (FSC) and side scatter (SSC). Adjust the voltages of the flow cytometer according to the manufacturer's instructions so that all populations of interest are visible in the FSC and SSC graph and a distinction between debris and live cells can be made.
- Perform multi-color compensation analysis with antibody capture beads following the manufacturer's protocol.
- Prepare fluorescence minus one (FMO) controls by making the antibody mix but exclude one antibody from the mix. Do this for every antibody, creating 8 antibody mixes for FACS panel 1, and 6 antibody mixes for FACS panel 2. These FMO antibody mixes are used to stain SVF as described previously in this protocol.
- Measure all FMO controls and set the gating strategy based on FMO controls. Use the FMO controls to detect possible auto-fluorescence of the cells.
NOTE: By removing one antibody from the mix, any fluorescence level detected in this channel is a background/autofluorescent signal. Hence, by comparing the different FMO control FACS results, gates can be drawn on specific populations ensuring that the gatings are based on positive cells and not based on auto-fluorescence. - Vortex the FACS tubes at 800 rpm before placing them in the flow cytometer and starting the measurement.
NOTE: A minimum of 50,000 events in the live gate is recommended to ensure enough cells are measured from each subpopulation.
Representative Results
The SVF isolated from vAT and sAT was measured using flow cytometry. Flow cytometry measurements generate plots showing different cell populations based on cellular markers (Figure 1A and 1B). First, by plotting the forward scatter width (FSC-W) and forward scatter area (FSC-A), cell aggregates can be eliminated from further analysis by gating the single cells as low FSC-W. Next, live cells are selected, and cellular debris is excluded by gating the cells of the correct size and complexity using FSC-A and the side scatter area (SSC-A), respectively. Dead cells are small and therefore visible as a distinct population with a small FSC-A. Next, immune cells were selected by use of the pan-leukocyte marker CD45 (Panel 1 and 2, Figure 1A). To analyze macrophages, other immune cells such as T-cells (CD3), B-cells (CD19), neutrophils (CD66b+ CD11b+), and NK-cells (CD56) were excluded from further analysis by using distinct antibodies targeting these cells, but with the same fluorochrome. Further subdivision of the remaining cells was based on CD11b and CD11c expression. This resulted in the following populations: CD11b+ CD11c+ macrophages, CD11b+ CD11c- macrophages, and CD11blow/- CD11c+ DCs (FACS panel 1, Figure 1B). Measurement of mean fluorescence intensity (MFI) allowed the quantification of the expression of CD303 (plasmacytoid DC marker) and CD141 (DC marker), on CD11b+ CD11c+ macrophages and CD11blow/- CD11c+ DCs. Expression of both these markers were higher in CD11blow/- CD11c+ cells confirming that CD11blow/- CD11c+ cells were DCs (Figure 1C).
The CD45+ cells (Figure 1A) were divided into T-cells and B-cells using CD3 and CD19, respectively. T-cells were subdivided into T-helper cells (CD4+) and cytotoxic T-cells (CD8+). Lastly, CD3-CD19-cells were plotted to quantify NK-cells using the marker CD56 (FACS panel 2, Figure 1D). The number of cells in each gate is quantified and can be used to calculate the percentage of this cell type of all living cells (Table 3).
The percentage of living cells can be calculated for each subject allowing the calculation of an average of all subjects in a group of for example lean or obese men displaying the abundance of a specific immune cell, i.e., the pro-inflammatory CD11b+ CD11c+ macrophage in visceral AT (Figure 2).
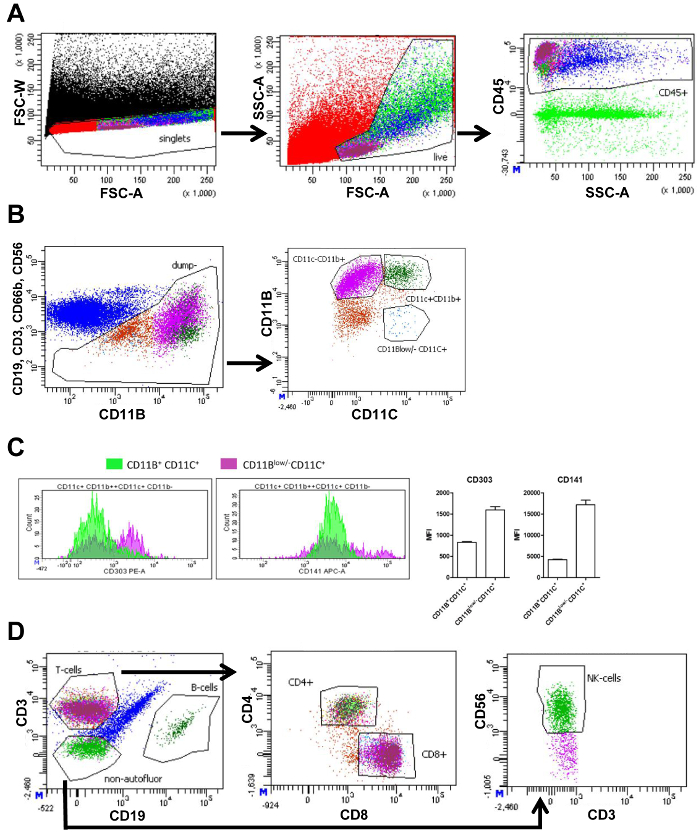
Figure 1. FACS gating strategy of visceral adipose tissue. (A) FACS plot of all events (black) with forward scatter width intensity (FSC-W) and forward scatter area intensity (FSC-A) containing a gate to select only single cells (red) followed by a FACS plot based on FSC-A and side scatter area intensity (SSC-A) containing a gate selecting live cells (light green). Next plot with SSC-A and CD45 fluorescence intensity contains a gate selecting all CD45+ (immune) cells (blue). (B) FACS plot of CD19, CD3, CD66b, and CD56 fluorescence intensity versus CD11b fluorescence intensity, and a gate selecting all cells that are CD19, CD3, CD66b, and CD56 negative (brown) for further division into populations. Further subdivision in the next plot based on CD11b and CD11c fluorescence intensity. Gates are displayed containing CD11b+ CD11c+ macrophages (dark green), CD11b+ CD11c- macrophages (purple), and CD11blow/- CD11c+ dendritic cells (blue). (C) Amount of CD11b+, CD11c+, or CD11blow/- CD11c+ cells (Y-axis) displaying their levels of fluorescence intensity (X-axis) for CD303 and CD141, and the corresponding quantification of the mean fluorescence intensity (MFI). (D) FACS plot displaying the previous CD45+ population (blue) based on CD3 and CD19 fluorescence intensity containing gates selecting T-cells (magenta), B-cells (dark green), and non-autofluorescent cells (green) negative for both CD3 and CD19. The following plot is based on CD4 and CD8 fluorescence with gates selecting CD4+ (light green) and CD8+ (magenta) T-cells. An identical gating strategy is used for subcutaneous adipose tissue. An identical gating strategy is used for subcutaneous adipose tissue. This figure has been modified from Wouters et al.16 Please click here to view a larger version of this figure.
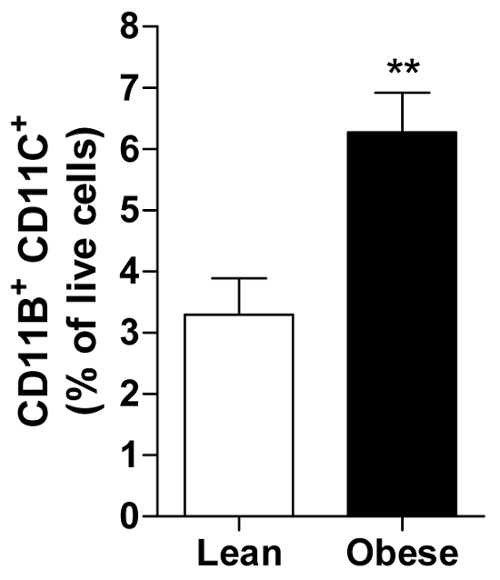
Figure 2. Obese vAT contains more pro-inflammatory macrophages. The amount of CD11b+ CD11c+ macrophages presented as percentage of all living cells in vAT of lean and obese men. All data are means ± SEM; n = 20 for lean and n = 31 for obese. **p ≤ 0.01 vs. lean. Please click here to view a larger version of this figure.
| Target | Definition target | Present on | Fluorochrome | Quantity | Clone |
| CD11B | myeloid integrin marker | granulocytes, monocytes/macrophages, dendritic cellslow and natural killer cells | BV421 | 2.5 µL | ICRF44 |
| CD19 | common B cell antigen | B cell development from pro-B cell to blastoid B cell and plasma B cell | Fitc | 3 µL | HIB19 |
| CD3 | common T cell antigen | T lymphocytes, natural killer T cells and thymocytes | Fitc | 3 µL | UCHT1 |
| CD66B | member of the carcinoembryonic antigen (CEA)-like glycoprotein family | granulocytes | Fitc | 5 µL | G10F5 |
| CD56 | heavily glycosylated adhesion protein | natural killer cells and natural killer T cells | Fitc | 5 µL | B159 |
| CD303 | type II transmembrane glycoprotein | plasmacytoid dendritic cells | PE | 1 µL | 201A |
| CD141 | thrombomodulin | monocytes/macrophageslow, subpopulation of dendritic cells | APC | 1 µL | M80 |
| CD11C | type I transmembrane glycoprotein; integrin αx | monocytes/macrophages, dendritic cells, granulocytes, natural killer cells, subset of B and T cells | APC-Cy7 | 0.5 µL | Bu15 |
| CD45 | common leukocyte antigen | all human leukocytes including lymphocytes, monocytes, granulocytes, eosinophils, and thymocytes | PE-Cy7 | 1 µL | HI30 |
| FACS buffer | - | - | - | 7.5 µl | - |
Table 1. Antibody cocktail for FACS panel 1 to identify macrophage subsets and dendritic cells populations. Amount of antibody described is for the analysis of one sample.
| Target | Definition target | Present on | Fluorochrome | Quantity | Clone |
| CD19 | common B cell antigen | B cell development from pro-B cell to blastoid B cell and plasma B cell | BV421 | 1 µL | HIB19 |
| CD3 | common T cell antigen | T lymphocytes, natural killer T cells and thymocytes | V500 | 3 µL | UCHT1 |
| CD56 | heavily glycosylated adhesion protein | natural killer cells and natural killer T cells | APC | 5 µL | HCD56 |
| CD4 | Ig superfamily, type I transmembrane glycoprotein | T helper cell, thymocytes, monocytes/macrophages, type II natural killer T cells | PerCP-Cy5.5 | 1 µL | RPA-T4 |
| CD8 | α-subunit of a disulfide-linked bimolecular complex | cytotoxic T cells, thymocytes, subset of natural killer cells | APC-H7 | 2 µL | SK1 |
| CD45 | common leukocyte antigen | all human leukocytes including lymphocytes, monocytes, granulocytes, eosinophils, and thymocytes | PE-Cy7 | 1 µL | HI30 |
| FACS buffer | - | - | - | 10 µl | - |
Table 2. Antibody cocktail for FACS panel 2to identify T- and B-cell populations. Amount of antibody described is for the analysis of one sample.
| Macrophage panel | ||
| Gate name | # Cells | % of live |
| Single cells | 183054 | |
| live | 10477 | 100 |
| CD45 | 4100 | 39.13 |
| dump- | 771 | 7.36 |
| CD11C- CD11B+ | 430 | 4.1 |
| CD11C+ CD11B+ | 104 | 0.99 |
| CD11C+ CD11Blow/- | 15 | 0.14 |
| T-cell, B-cell, NK-cell panel | ||
| Gate name | #Cells | % of live |
| Single cells | 34616 | |
| Live | 3728 | 100 |
| CD45+ | 1589 | 42.62 |
| NK-cells | 29 | 0.78 |
| CD3+ | 953 | 25.56 |
| CD4+ | 601 | 16.12 |
| CD8+ | 328 | 8.8 |
| CD19+ | 20 | 0.54 |
Table 3. Immune cell abundance of different cell types in vAT. Number of cells in each gate and the percentage of the different cell types based on the total amount of living cells.
Discussion
These methods describe how to isolate the SVF from vAT and sAT and quantify the relative amounts of immune cells within these tissues. Furthermore, the methods state how to determine the expression of markers on specific cell types.
Flow cytometry of tissue immune cells is a powerful technique to phenotype the immunological state of tissues. The quantification of tissue immune cells can have many applications. As described in the results, it is possible to compare the presence of specific immune cells between groups of patients (e.g., lean vs. obese). In addition, by also performing flow cytometry on blood of the same patients, associations between circulating cells and tissue cells can be investigated. With this application we were able to determine that a specific subset of circulating monocytes is associated with pro-inflammatory CD11C+ adipose tissue macrophages16.
Adjustments to the described protocol will expand its applications as numerous available fluorescent antibodies make flow cytometry very versatile. With different antibodies nearly all cell types can be distinguished and the expression of many markers can be detected. Furthermore, it is possible to stain markers intracellularly by permeabilizing the cell membrane to allow intracellular binding of the fluorescent antibodies. These characteristics allow distinction of the very diverse macrophage populations beyond the overly simplified M1 and M2 macrophage subtypes. Besides measurement of surface marker expression, proteins (i.e., cytokines) can be stained intracellularly providing information on macrophage functionality. In addition, proliferation markers such as Ki67 are used to quantify proliferation rates. As described, distinction between macrophages and DCs was based on MFI levels of DC markers. A general macrophage marker, such as CD68 can be incorporated into the macrophage panel (FACS panel 1). However, CD68 needs to be stained intracellularly requiring permeabilization of the cell membrane which is not preferable and would extend the protocol. Other macrophage markers are subset markers such as CD163 and CD206 or CD11c, the latter being integrated in the macrophage panel presented here.
In our FACS panels, a marker to distinguish live and dead cells was not included, which would be preferable because it allows a more accurate exclusion of dead cells than the use of FSC and SSC. Frequently used are the DNA staining viability dyes propidium iodide (PI) or 4',6-diamidino-2-phenylindole (DAPI) as well as free amine reacting dyes such as the LIVE/DEAD Fixable Dead Cell Stain Kit, which is available in different dye colors. However, PI and DAPI cannot be used when fixing the cells. As described in the protocol, the LIVE/DEAD Fixable Red Dead Cell viability staining can be integrated into both panels without affecting the overall FACS gating strategy.
In addition, the data are expressed as a percentage of live cells meaning all data are relative. Only by entering an exact, known number of cells into the flow cytometer, would it be possible to determine the exact numbers of each cell type. An approximate number of cells could be calculated after counting the cells in the SVF fraction by using a counting chamber. However, this number would have to be adjusted for the amount of biopsy tissue used to isolate the SVF, but this has limitations when comparing lean to obese AT. A similar mass of obese AT consists of less adipocytes as they are filled with lipids and have expanded greatly. This could lead to an underestimation of immune cell number if presented as number of immune cells per gram of AT or per adipocyte.
In human studies, inclusion of patients is usually done over a longer period of time making standardization of experimental procedures of great importance. For comparison of flow cytometry data between patients, there are several options. As described in this protocol, cells can be fixed before measurement allowing analysis of several samples on the same day. This can also be achieved by freezing the SVF before staining them, which allows the staining procedure to be equal between all samples, but the viability of cells might be affected. Lastly, also employed in this study, are fluorescent beads to install compensation levels, and cytometer tracking beads were used bi-weekly to standardize daily measurements of the cytometer. This last option is the most efficient when measuring samples from a study spanning a long period of time.
A limiting factor for flow cytometry in general is the use of fluorescence. The number of fluorescent labels that can be detected simultaneously is limited due to overlap in emission spectra. However, with smart FACS panel development and the use of several antibody cocktails per vAT or sAT sample, this issue can be overcome as described in this protocol. An important aspect of FACS panel development is FMO controls. By using all antibodies of the panel except for one, potential autofluorescence levels can be appreciated when comparing the FMO with the full panel. This allows accurate gating of populations and these procedures should be performed when setting up a new FACS panel. In addition, new generations of FACS devices can detect up to 50 parameters allowing simultaneous detection of many characteristics per cell. Another issue related to the fluorescence aspect is the autofluorescence of cells, particularly macrophages. After excitation of the cells with the FACS laser (mainly with 488 nm wavelength excitation), these cells emit a fluorescent signal (mainly <640 nm) that can overlap with the emission spectra of the antibody labels17,18. To account for this, unstained cells should be measured to determine the autofluorescence in each channel. With this knowledge, fluorochromes should be selected that display a signal strength that exceeds the autofluorescent signal. This autofluorescent background signal should be taken into account when determining the gating strategy of the populations. Therefore, by application of this protocol and intelligent FACS panel design it is possible to in depth phenotype macrophage subtypes. New distinct AT macrophages and their function can be characterized.
Disclosures
The authors declare no conflicts of interest.
Acknowledgements
We would like to thank J. van de Gaar and M. Vroomen (Maastricht University, The Netherlands) for their technical support. In addition, we would like to thank K. Verboven, D. Hansen, J. Jocken, and E. Blaak for providing the blood and tissue biopsies used for setting up this protocol and the subsequent experiments.
Materials
| Name | Company | Catalog Number | Comments |
| Centrifuge | Hettich Rotanta 460R | 5660 | |
| Dulbecco’s Modified Eagle’s Medium (DMEM)-Ham’s F12 | Gibco ThermoFisher | 31330-095 | |
| Collagenase XI from Clostridium histolyticum | Sigma Aldrich | C7657-1g | |
| Collagenase I from Clostridium histolyticum | Sigma Aldrich | C0130-1g | |
| DNAse I from bovine pancreas | Sigma Aldrich | DN25 | |
| NH4Cl | Merck | 1.01145.0500 | |
| Bovine Serum Albumine | Sigma Aldrich | A4503-100 | |
| NaN3 | Merck | 1.06688.0100 | |
| 200 µM syringe Filcons | BD Biosciences | 340613 | |
| 70 µM filter | Greiner Bio-one | 542070 | |
| Fc block / human IgG | Sigma Aldrich | 14506 | |
| Formaldehyde | Merck | 1.04003.2500 | |
| CD11b-BV421 | Biolegend | 301324 | |
| CD19-Fitc | BD Biosciences | 555412 | |
| CD3-Fitc | BD Biosciences | 561807 | |
| CD66b-Fitc | BD Biosciences | 555724 | |
| CD56-Fitc | BD Biosciences | 562794 | |
| CD11c-APC-Cy7 | Biolegend | 337218 | |
| CD45-PE-Cy7 | BD Biosciences | 557748 | |
| CD19-BV421 | Biolegend | 302234 | |
| CD3-V500 | BD Biosciences | 561416 | |
| CD56-APC | Biolegend | 318310 | |
| CD4-PerCP-Cy5.5 | Biolegend | 300528 | |
| CD8-APC-H7 | BD Biosciences | 641400 | |
| CD303-PE | Biolegend | 354204 | |
| CD141-APC | Biolegend | 344106 | |
| FACS-Canto II | BD Biosciences | ||
| 96 v-shape well plate | Greiner Bio-one | 651101 | |
| PBS PH 7.4 | Gibco ThermoFisher | 10010023 | |
| FACS tube 5 mL polystyrene round-bottom tube | Corning | 352052 | |
| IgG from human serum | Sigma Aldrich | I4506 | |
| Anti-Rat Ig, κ/Negative Control Compensation Particles Set | BD Biosciences | 552844 | |
| Anti-Mouse Ig, κ/Negative Control Compensation Particles Set | BD Biosciences | 552843 | |
| LIVE/DEAD Fixable Red Dead Cell Stain Kit | ThermoFisher | L23102 | |
| IKA MS 3 basic shaker | Sigma Aldrich | Z645028-1EA |
References
- McNelis, J. C., Olefsky, J. M. Macrophages, immunity, and metabolic disease. Immunity. 41 (1), 36-48 (2014).
- Makki, K., Froguel, P., Wolowczuk, I. Adipose tissue in obesity-related inflammation and insulin resistance: cells, cytokines, and chemokines. ISRN Inflamm. 2013, 139239 (2013).
- DeFuria, J., et al. B cells promote inflammation in obesity and type 2 diabetes through regulation of T-cell function and an inflammatory cytokine profile. Proc Natl Acad Sci U S A. 110 (13), 5133-5138 (2013).
- Elgazar-Carmon, V., Rudich, A., Hadad, N., Levy, R. Neutrophils transiently infiltrate intra-abdominal fat early in the course of high-fat feeding. J Lipid Res. 49 (9), 1894-1903 (2008).
- Han, J. M., Levings, M. K. Immune regulation in obesity-associated adipose inflammation. J Immunol. 191 (2), 527-532 (2013).
- Liu, J., et al. Genetic deficiency and pharmacological stabilization of mast cells reduce diet-induced obesity and diabetes in mice. Nat Med. 15 (8), 940-945 (2009).
- Nishimura, S., et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 15 (8), 914-920 (2009).
- Koh, Y. J., et al. Stromal vascular fraction from adipose tissue forms profound vascular network through the dynamic reassembly of blood endothelial cells. Arterioscler Thromb Vasc Biol. 31 (5), 1141-1150 (2011).
- Peinado, J. R., et al. The stromal-vascular fraction of adipose tissue contributes to major differences between subcutaneous and visceral fat depots. Proteomics. 10 (18), 3356-3366 (2010).
- Leggate, M., et al. Determination of inflammatory and prominent proteomic changes in plasma and adipose tissue after high-intensity intermittent training in overweight and obese males. Journal of Applied Physiology. 112 (8), 1353-1360 (2012).
- Divoux, A., et al. Mast cells in human adipose tissue: link with morbid obesity, inflammatory status, and diabetes. J Clin Endocrinol Metab. 97 (9), E1677-E1685 (2012).
- Harford, K. A., Reynolds, C. M., McGillicuddy, F. C., Roche, H. M. Fats, inflammation and insulin resistance: insights to the role of macrophage and T-cell accumulation in adipose tissue. Proc Nutr Soc. 70 (4), 408-417 (2011).
- Lumeng, C. N., Bodzin, J. L., Saltiel, A. R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 117 (1), 175-184 (2007).
- Morris, D. L., Singer, K., Lumeng, C. N. Adipose tissue macrophages: phenotypic plasticity and diversity in lean and obese states. Curr Opin Clin Nutr Metab Care. 14 (4), 341-346 (2011).
- Wentworth, J. M., et al. Pro-inflammatory CD11c+CD206+ adipose tissue macrophages are associated with insulin resistance in human obesity. Diabetes. 59 (7), 1648-1656 (2010).
- Wouters, K., et al. Circulating classical monocytes are associated with CD11c+ macrophages in human visceral adipose tissue. Sci Rep. 7, 42665 (2017).
- Duan, M., et al. Distinct macrophage subpopulations characterize acute infection and chronic inflammatory lung disease. J Immunol. 189 (2), 946-955 (2012).
- Li, F., et al. Autofluorescence contributes to false-positive intracellular Foxp3 staining in macrophages: a lesson learned from flow cytometry. J Immunol Methods. 386 (1-2), 101-107 (2012).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2025 MyJoVE Corporation. All rights reserved