A subscription to JoVE is required to view this content. Sign in or start your free trial.
Lucifer Yellow - A Robust Paracellular Permeability Marker in a Cell Model of the Human Blood-brain Barrier
In This Article
Summary
We present a fluorescence assay to demonstrate that Lucifer Yellow (LY) is a robust marker to determine the apparent paracellular permeability of hCMEC/D3 cell monolayers, an in vitro model of the human blood-brain barrier. We used this assay to determine the kinetics of a confluent monolayer formation in cultured hCMEC/D3 cells.
Abstract
The blood-brain barrier BBB consists of endothelial cells that form a barrier between the systemic circulation and the brain to prevent the exchange of non-essential ions and toxic substances. Tight junctions (TJ) effectively seal the paracellular space in the monolayers resulting in an intact barrier. This study describes a LY-based fluorescence assay that can be used to determine its apparent permeability coefficient (Papp) and in turn can be used to determine the kinetics of the formation of confluent monolayers and the resulting tight junction barrier integrity in hCMEC/D3 monolayers. We further demonstrate an additional utility of this assay to determine TJ functional integrity in transfected cells. Our data from the LY Papp assay shows that the hCMEC/D3 cells seeded in a transwell setup effectively limit LY paracellular transport 7 days-post culture. As an additional utility of the presented assay, we also demonstrate that the DNA nanoparticle transfection does not alter LY paracellular transport in hCMEC/D3 monolayers.
Introduction
Blood-brain barrier (BBB) is the protective barrier limiting the influx of plasma components into the brain tissue and consists of brain endothelial cells along with supporting cells such as pericytes. The major role of BBB is to serve as a barrier that seals the space between peripheral blood and central nervous system (CNS) to maintain hemostasis of the neural microenvironment1,2. The brain capillary endothelial cells effectively seal the paracellular pathway via formation of intercellular tight junctions (TJs)1. This protective barrier allows glucose and selected nutrients to enter the brain while it prevents the majority of ions, toxic substances and drugs from passing through this tight barrier. Apart from its protective role, the natural barrier function of the BBB poses a severe challenge in the development of drug delivery systems targeting the CNS.
In vitro cell culture models of the BBB are helpful tools to study its biology and to understand the effects of drug treatment on TJ barrier integrity. We used the human cerebral microvascular endothelial cell line (hCMEC/D3) as an in vitro model since it is an accepted model of human brain endothelium3 and recapitulates many functions of the human BBB. hCMEC/D3is one of the most commonly used cell lines for modeling the BBB in vitro4,5,6,7,8,9. Despite its comparatively low values of transendothelial electrical resistance (TEER), a measure of barrier tightness, this cell line retains most of the morphological and functional properties of brain endothelial cells, even as a monoculture in the absence of cocultured glial cells6,7. The hCMEC/D3 cell line expresses multiple BBB markers including active transporters and receptors until approximately passage 35 without undergoing dedifferentiation to unstable phenotypes6,7,9,10,11. The most striking characteristic of hCMEC/D3 cell line as an in vitro BBB model is its ability to form TJs5,9,11,12. It should be noted that although stem cell-derived BBB models showed higher permeability in many studies compared with hCMEC/D3 cell line and they do express some BBB markers, they are yet to evolve as the most common BBB cell model13. Importantly, stem-cell derived BBB models remain to be characterized with respect to maximum passage numbers that allow the cells to maintain stable BBB phenotypes14.
Three primary methods are commonly used to determine the TJ barrier integrity, including the measurement of TEER, measurement of apparent permeability coefficient (Papp) of small hydrophilic tracer molecules such as sucrose, inulin, Lucifer Yellow, etc. and immunostaining of known molecular markers of TJs such as claudin-5, ZO-1, occludin, etc.5. TEER is a relatively simple and quantitative method that measures the electrical resistance across the cell monolayers cultured on a porous membrane substrate5. However, TEER values can be influenced by experimental variables such as composition of the culture medium and the type of measurement instrument. A likely combination of these factors leads to a broad distribution of TEER values ranging from 2 to 1150 Ω cm2 in the hCMEC/D3 cell line cultured for 2-21 days13. Immunostaining is a visual method to determine the presence of TJ proteins by labelling the targeted protein using antibodies. However, immunostaining involves a series of experimental steps, including the need to fix/permeabilize cells that may result in experimental artifacts and the fluorescent signals may fade over time. The above factors may lead to subjective errors affecting data quality.
The primary focus of this work is to present a LY-based apparent permeability assay determine the kinetics of a confluent monolayer formation in cultured hCMEC/D3 cells. Although other advanced in vitro BBB systems, such as co-culture systems, microfluidic systems, are physiologically more relevant mimics with significantly improved barrier function15,16,17, the hCMEC/D3 transwell setup is a simple and reliable model to estimate the kinetics of TJ formation and rapidly screen the effect of different drug formulations on barrier function. In general, Papp values are consistent for various hydrophilic solutes in hCMEC/D3 monolayers. For example, the reported Papp values for various low molecular mass solutes (such as sucrose, mannitol, LY, etc.) in different in vitro BBB models are in the order of 10-4 cm/min5,18,19,20. In our experimental setup, the brain endothelial cells are seeded on a collagen-coated microporous membrane for cell attachment and monolayer formation to mimic the in vivo barrier. The LY added in the apical side is expected to traverse the intercellular tight junctions and accumulate in the basolateral side. Greater concentrations of LY in the basolateral side indicate an immature, not-fully functional barrier while lower concentrations reflect restricted transport due to the presence of functional TJs resulting in a mature barrier.
LY is a hydrophilic dye with distinct excitation/emission peaks and avoids the need to radiolabel tracer molecules such as sucrose, mannitol or inulin. Thus, the fluorescence values of LY can be used to directly calculate its paracellular permeability across the BBB monolayers. Also, compared to many commercially available dyes used in biomedical fields that suffer from small Stokes shifts such as fluorescein21, the Stokes shift of LY is about 108 nm with sufficient spectral separation, thus allowing LY fluorescence data as a robust readout to determine paracellular permeability. We used Western blotting as an orthogonal technique to demonstrate changes in expression of the tight junction marker protein, ZO-1, over culture time. ZO-1 expression detected via Western blotting is used to supplement the LY Papp data and in combination, these data suggest that the observed changes in LY Papp values is reflective of the formation of a monolayer with gradual increase in expression of the tight junction marker, ZO-1.
As pointed out earlier, the central focus of this work is to demonstrate a LY assay as a simple technique to monitor the formation of a confluent monolayer with functional tight junctions. However, to demonstrate an additional utility of the developed assay, we measured the LY Papp in DNA nanoparticle-transfected hCMEC/D3 monolayers. Nucleic acids can be condensed into polyelectrolyte nanoparticles with a diameter of 100-200 nm via electrostatic interaction between the positively charged groups of polymers and the negatively charged phosphate groups of nucleic acids22,23. We refer to these complexes as DNA nanoparticles (DNA NPs) in our work. While our intention is to transfect cells and express the desired protein, we must ensure that the barrier properties of the hCMEC/D3 monolayers are not compromised. Our data suggests that a standard 4 h luciferase gene transfection regime does not measurably change the LY permeability demonstrating the utility of the LY Papp assay to determine changes in TJ barrier integrity.
Protocol
1.General hCMEC/D3 cell culture
- Resuscitation of frozen cells
NOTE: All cell culture maintenance and experiments were performed inside a sterile biosafety hood. Culture media, supplements and reagents were either purchased as sterile products or sterilized via filtration using a 0.22 µm membrane filter to prevent microbial contamination.- Add 8.5 mL of collagen solution (0.15mg/mL) in a tissue culture flask (75 cm2 growth area; referred henceforth as T75) and place it in an incubator (37 °C, 5% CO2) for 1 h.
- Remove the collagen solution and gently wash the flask with sterilized phosphate-buffered saline (PBS). Add 15 mL of complete growth medium to the flask and leave in the CO2 incubator for 15 min.
NOTE: The complete medium (final concentration) contained Endothelial Cell Growth Basal Medium-2 (500 mL) supplemented with fetal bovine serum (5%), penicillin-streptomycin (1%), hydrocortisone (1.4 µM), acid ascorbic (5 µg/mL), chemically defined lipid concentrate (1/100), HEPES (10 mM) and basic fibroblast growth factor (1 ng/mL). - Move a cryovial of frozen hCMEC/D3 cells from the liquid nitrogen tank and rapidly thaw the vial in a 37 °C water bath (< 1 min).
- Once only a tiny flake of ice is visible, quickly aspirate and transfer the cells to the flask containing pre-warmed medium. Gently shake the flask to allow mixing of the cells with the growth medium.
- Place the flask in the incubator (37 °C, 5% CO2) and observe the cells under a light microscope after 2 h to make sure that the cells are attached.
- Once the cells attach to the bottom of flask, remove the old growth medium and add 10 mL of fresh pre-warmed growth medium to replace dimethylsulfoxide in the old growth medium24.
- After 24 h, check under a light microscope to observe spindle-shaped cells and replace the old growth medium with pre-warmed fresh growth medium.
- Cell culture maintenance
- Replenish the growth medium every other day until 100% confluence. Check the cells under the microscope before removing the old growth medium and also after adding fresh growth medium. Take out the flask from the incubator and examine hCMEC/D3 cells under phase contrast microscope to ensure they appear healthy.
NOTE: The majority of cells should be attached to the bottom of the flask, have a spindle-shaped morphology and often times, light refracting around their membranes is also seen. Growth medium should be transparent (non-cloudy) and pinkish-orange in color. - Remove old growth medium from the flask and transfer 10 mL of pre-warmed fresh medium into the flask.
NOTE: The medium should be added to the top side of the flask and not directly on the surface of the cells to avoid affecting cell attachment. - Turn the flask back into horizontal position and gently rock it several times and check hCMEC/D3 cells under the microscope before returning the flask to the incubator (37 °C, 5% CO2).
- Observe the cells under an inverted light microscope each time before and after handling the cells, both during regular culture work and during experiments. Record any noticeable changes in cell number or morphology in the laboratory notebook.
- Replenish the growth medium every other day until 100% confluence. Check the cells under the microscope before removing the old growth medium and also after adding fresh growth medium. Take out the flask from the incubator and examine hCMEC/D3 cells under phase contrast microscope to ensure they appear healthy.
- Cell passaging
- Incubate a new T75 flask with 8.5 mL of collagen solution for 1 h in the incubator (37 °C, 5% CO2).
- Remove the collagen solution and gently wash the flask with sterilized PBS. Add 10 mL of pre-warmed hCMEC/D3 medium to the new flask and place the flask in the incubator (37 °C, 5% CO2).
- Take out the flask from the incubator and examine hCMEC/D3 cells under a phase contrast microscope to check if the cells are 100% confluent.
- Remove hCMEC/D3 cell medium from the flask containing cells and wash hCMEC/D3 cells with 10 mL of PBS.
NOTE: FBS added to growth medium contains protease inhibitors such as α1-antitrypsin and α2-macroglobulin. These inhibit the trypsinization process. Thus, it is essential to wash the cells with PBS to remove traces of FBS to prevent the inhibition of the trypsinization process. - Add 1 mL of 0.25% trypsin solution containing 0.02% EDTA and trypsinize for 2-5 min in the incubator (37 °C, 5% CO2) (tap the flask gently on the sides to help detachment).
NOTE: Never leave the cells on trypsin/EDTA for more than 6 min. - Add 10 mL of pre-warmed hCMEC/D3 medium to stop the trypsinization process and resuspend hCMEC/D3 cell by pipetting up and down several times. Then, remove the entire cell suspension from the flask into a 15 mL tube.
- Transfer 1 mL of the cell suspension from the 15 mL tube to the new flask with pre-warmed fresh medium (splitting cells 1:10) and return the new flask back to the incubator.
NOTE: Before transferring to the new flask, pipette the cell suspension up and down several times to minimize cell concentration gradients.
2. Cell plating
- Place tissue culture inserts with microporous membranes (pore size: 0.4 µm, material: polyethylene terephthalate (PET)) into a 24-well culture plate.
- Add 400 µL of collagen type I (0.15 mg/mL) in each tissue culture insert and incubate for 1 h in the CO2 incubator (37 °C, 5% CO2). Rock the 24-well plate gently to allow even spreading of the collagen solution over the microporous membrane in the tissue culture inserts.
- Remove the collagen solution and gently wash the microporous membrane with 0.4 mL of 1x PBS buffer.
- Plate hCMEC/D3 cells with the density of 50,000 cells/cm2 in the cell inserts (15,000 cells in 500 µL of medium).
NOTE: In order to minimize the differences in cell number in each tissue culture insert, the cell suspension was resuspended with a 10 mL pipette before adding cells to the inserts. - Place the 24-well plate with tissue culture setup in an incubator (37 °C, 5% CO2) to allow cell attachment and proliferation.
- Incubate the plate for 7 days to allow the cells to reach 100% confluency. Remove the growth medium every other day and transfer 0.5 mL of pre-warmed fresh media into tissue culture inserts.
- Repeat the plating procedure (steps 2.2-2.6) on a 12-well plate, 48-well plate and 96-well plate. Use the 12-well plate for Western blotting to determine changes in ZO-1 expression. Use the 48-well plate for DNA NP transfection. Use the 96-well plate for the ATP assay to determine cell viability in transfected cells.
3. Kinetics of cell growth.
- Seed the cells at a density of 50,000 cell/cm2 in a collagen-coated 24-well tissue culture plate.
- On each day of the experiment, remove the growth medium and gently wash the cells twice with 500 µL of 1x PBS. Then, add 30 µL of 0.25% trypsin solution containing 0.02% EDTA and leave the plate for about 2-5 min in an incubator (37 °C, 5% CO2).
NOTE: Gradual formation of confluent monolayer may affect the extent of cell detachment and it is necessary to increase the volumes of trypsin/EDTA as indicated here: 30 µL for 1-5 days post-seeding, 60 µL for 6-7 days post-seeding and 100 µL for 8-10 days post-seeding. - Add either 470 µL, 440 µL or 400 µL of growth medium based on the volume of trypsin/EDTA solution added in the step 3.2 to prepare 500 µL of cell suspension in each well.
- Suspend the cells by pipetting up and down in each well several times and observe the cells under a microscope to make sure all cells are suspended in the growth medium. If some cells are still attached to the plate bottom after pipetting several times, gently scrape the cells using a plastic cell scraper to facilitate cell detachment.
- Remove 0.1 mL of cell suspension from 500 µL cell suspension in step 3.3 and add to a 1.5 mL tube. Then, add 0.1 mL of 0.4% Trypan blue solution to the cell suspension and mix well.
- Clean a hemacytometer with 70% isopropyl alcohol. Add 20 µL of mixture from step 3.5 on each side in the V-groove and locate the 16 squares under the microscope. The 16 squares are considered as one grid. Locate two random grids on each side of the hemacytometer and count all the living, non-blue cells.
NOTE: Cells that appeared blue in color from excluded from counting, <1% of the cells stained blue at all time points. - Calculate the cell density (cells/cm2) based on following formulas.
Average # of cells/grid =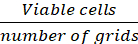 (Eq.1)
(Eq.1)
Dilution Factor=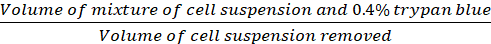 (Eq.2)
(Eq.2)
Cell density (viable cells/cm2) =
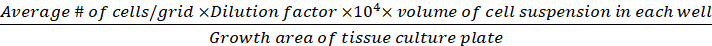
(Eq.3)
Equations 1-3. Viable cells are the number of cells counted in each grid, number of grids correspond to the number of grids located under the microscope, Volume of mixture of cell suspension and 0.4% Trypan blue is the volume prepared in step 3.5, volume of cell suspension removed is the volume removed from 500 µL cell suspension in step 3.5, the volume of cell suspension in each well is the 500 µL cell suspension from step 3.3, growth area of tissue culture plate is the growth area of single well in 24-well plate.
4. Lucifer yellow apparent permeability (LY P app ) assay
- For determining LY Papp on each day post-seeding, follow steps starting 4.3. For determining the Papp in transfected hCMEC/D3 cells, add 8.3 µL of the transfection formulation (Figure 1) mixed with 50 µL of complete growth medium and incubate for 4 h. Transfection formulations are described in section 5.
- After the 4 h transfection, gently wash the apical side twice using sterile 1x PBS buffer to remove any residual transfection mixture. Varying volumes of PBS buffer left behind after removal can affect the concentration of LY in the apical side. Take care to ensure that residual PBS buffer in tissue culture inserts is completely removed. Carefully aspirate residual reagents and medium to minimize cell detachment.
NOTE: This step is skipped when measuring every-day apparent permeability (Papp) of hCMEC/D3 cells. - Remove the growth medium and add 1.5 mL of pre-warmed (37 °C) transport buffer (25 mM HEPES, 145 mM NaCl, 3 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, 1 mM NaH2PO4, 5 mM glucose, pH 7.4) to the basolateral side.
NOTE: The volume of transport buffer in all basolateral compartments should be equal to ensure accuracy of permeability coefficient calculation. - Add 58.3 µL of 20 µM LY solution to the apical side of each transwell insert. Save 50 µL of the 20 µM LY solution for fluorescence measurements. After removing residual PBS buffer from apical side completely, add LY solution as quick as possible to avoid drying hCMEC/D3 cells. Ensure accurate volumes of LY solution in the apical side.
NOTE: To minimize the decay of LY fluorescence intensity, light exposure should be limited. Once the LY powder is reconstituted, the solution should be stored at 4 °C, protected from light. - Incubate in a rotary plate shaker (37 °C, 100 rpm) for 60 min. Then, remove 30 µL of the LY sample from each apical compartment. Then transfer the 20 µM LY solution and the apical side samples to pre-labeled tubes and dilute the sample 10-fold using transport buffer.
NOTE: It is required to dilute the 20 µM LY stock solution and the apical side samples because the high fluorescence intensity of these samples may potentially overload and damage the fluorescence detector of the fluorescence microplate reader. - Remove 500 µL from each basolateral compartment and transfer the sample to pre-labeled tubes.
NOTE: Samples are removed from separate transwell at indicated time points. The time points are each day post-seeding starting on day 1 until day 10. - Prepare a series of LY standards for the standard curve (39.00 nM, 78.13 nM, 156.25 nM, 312 nM, 625 nM, 1250 nM, 2500 nM).
- Add 100 µL of each standard (in duplicate), apical and basolateral sample to each well in a black 96-well plate (Figure 1).
NOTE: Black plates absorb light and reduce background and fluorescence crossover among wells. - Use a fluorescence microplate reader (set points: excitation 428 nm, emission 536 nm) to measure the LY fluorescence intensity to calculate the Papp. A fluorescence plate reader is used for this study.
- Calculate the Papp and %LY recovery values as described in the manuscript text.
Equation 4. Formulae for calculating Papp values.
Calculate the apparent permeability (Papp) coefficient and the %LY recovery using following equations. It should be noted that the Papp values can be calculated either based on mass25 or concentration of LY.
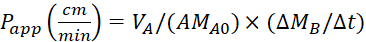 or
or 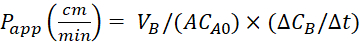
VA - volume in the apical compartment
VB - volume in the basolateral compartment
A - the surface area of the transwell insert membrane (0.3 cm2)
MA0 - the initial mass in the apical compartment
ΔMB/Δt - the change of mass over time in the basolateral compartment
CA0 - the initial concentration in the apical compartment
ΔCB/Δt - the change in concentration over time in the basolateral compartment.
Equation 5. Formulae for calculating % LY recovery.
Recovery (%) =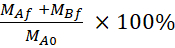
MAf is the mass in the apical compartment at the end time point, MBf is the mass in the basolateral compartment at the end time point, MA0 is the initial mass in the apical compartment.26 Note: The initial mass is calculated based on the volume of the 20 µM LY solution in the step 4.5. This experiment was always done using cells under passage number 35 and was conducted four independent times.
5. Calcium depletion
- Remove the growth medium from the transwell inserts and 12-well plate and gently wash the apical side and 12-well plate using pre-warmed calcium-free medium (Minimum Essential Medium Eagle Spinner (S-MEM) medium) to remove calcium ions from the transwell inserts and 12-well plate.
- Add 500 µL or 2 mL of pre-warmed S-MEM 1xmedium to the inserts or 12-well plate and incubate for 24 h in the incubator (37 °C, 5% CO2). After incubation, remove the S-MEM 1x medium and wash the cells once using pre-warmed 1x PBS buffer.
- Follow steps 4.4-4.10 described in section 4 for LY treatment and subsequent steps. Follow steps 8.1.2-8.2.4 described in section 8 for Western blotting and subsequent steps.
6. Transfection
- Preparation of DNA nanoparticles (DNA NPs).
- Dilute the stock solution of gWIZ-Luc plasmid in 10 mM sodium acetate (NaAc) buffer (pH 5.0) and allow the DNA solution to stand for 10 min at room temperature (RT).
NOTE: The DNA NP containing predominantly single DNA molecules can be prepared at DNA concentrations 20-40 µg/mL23. Thus, gWIZ-Luc plasmid stock solution needs to be diluted. The frozen gWIZ-Luc plasmid stock needs to be thawed completely on ice to minimize temperature stress. Gently vortex the diluted DNA stock solution for 30 s on a standard benchtop vortexer set at knob position 3-5. - Calculate the desired N/P ratios, used here as a numerical parameter to reflect NP composition.

Equation 6. N/P ratio calculation: the ratio of moles of the amine groups of cationic polymers to those of the phosphate groups of DNA. Mass of cationic polymers means total weighed amount of cationic polymer; (Mass/Charge)cationic polymers refers to the molecular weight of cationic polymer (poly (ethyleneglycol)5k-block-polyaspartamide with 48 diethylenetriamine side chains (PEG-DET)) normalized to the number of charged primary amines (48) per cationic polymer (mol/mol), this value for our polymer is 306 Da; Mass of DNA means total amount of DNA used in the formulation obtained by multiplying the volume and concentration in mg/mL; (Mass/Charge)DNA refers to the molecular weight of DNA normalized to number of phosphate group per double-stranded DNA (325 Da per nucleobase).
NOTE: The DNA NP preparation table (Table 1) contains the formulation recipe for the different samples tested in our experiments. DNA NP were prepared using a rapid titration technique. The PEG-DET polymer solution was added along the walls of the tube while holding the tube in a horizontal position. Then the tube was switched to a vertical position, followed by quickly vortexing at maximum speed for 10s. The DNA NP were allowed to stand for 30 min at RT prior to use. The rule-of-thumb for DNA NP dosing is 0.5 µg DNA for ca. 1 cm2 growth area. So, for each transwell insert/each well in a 48-well plate/each well in a 96-well plate, prepare DNA NP at N/P 10 containing 0.157/0.5/0.195 µg/well of gWIZ-Luc DNA. - For samples containing indicated concentration of Poloxamer P84 (P84), add P84 to DNA NP and vortex for 5 s. The final concentration of P84 in each sample is either 0.01% or 0.03% wt.
- Dilute the stock solution of gWIZ-Luc plasmid in 10 mM sodium acetate (NaAc) buffer (pH 5.0) and allow the DNA solution to stand for 10 min at room temperature (RT).
- DNA NP transfection in a 48-well plate setup
- Seed the cells with the density of 50,000 cell/cm2 in a 48 well plate and grow until confluence in the incubator (37 °C, 5% CO2).
- For each treatment group, mix 25 µL of the indicated sample (transfection formulation) and 150 µL of complete growth medium and 175 µL of this mixture to each well.
- Observe the hCMEC/D3 cells under a microscope to ensure the cells appear healthy and are 100% confluent at the time of the experiment.
- Remove the growth medium from wells and 175 µL transfection mixture to each well. Then, incubate the plate for 4 h in the incubator (37 °C, 5% CO2).
- After 4 h, remove the transfection mixture and wash the hCMEC/D3 cells with pre-warmed sterile 1x PBS buffer.
NOTE: To minimize the accidental detachment of hCMEC/D3 cells from the plate/insert surface during the washing steps, carefully pipette enough sterile PBS along the walls of the wells and remove any nanoparticles in the residual culture media. - Gently rock the plate a few times and carefully aspirate and discard the PBS wash and add 500 µL of hCMEC/D3 pre-warmed culture medium.
- Microscopically examine the cells and record observations on cell morphology and any possible effects of transfection.
- Incubate for 24 h in the 37 °C cell culture incubator to allow luciferase production. After 24 h, remove growth medium completely and wash the cells once with pre-warmed 1x PBS.
- Lyse the transfected cells by adding 100 µL of ice-cold luciferase cell culture lysis 1x reagent per well.
- For measurement of the luciferase protein content, add 20 µL of cell lysate and 100 µL of luciferase assay buffer (20 mM glycylglycine (pH 8), 1 mM MgCl2, 0.1 mM EDTA, 3.5 mM DTT, 0.5 mM ATP, 0.27 mM coenzyme A) into a 1.5 mL tube.
- Read the luminescence of the sample described in the step 6.2.10 on a Luminometer with a Single Auto-injector.
NOTE: The luminescence should be integrated over 10 s before reading. - Measure the total amount of cellular protein in the lysate using a bicinchoninic acid assay (BCA assay) kit by following the manufacturer's protocol.
- Calculate and express luciferase gene expression as Relative Light Units (RLU) per total cellular protein.
7. Luminescent ATP assay
- Seed the cells with the density of 50,000 cell/cm2 in a 96 well plate and grow until confluence in the incubator (37 °C, 5% CO2).
- Transfect the cells with 9.7 µL of the transfection formulation (preparation details are in section 6) and 58.4 µL of complete growth medium for 4 h.
- Remove the transfection mixture and gently wash the cells with pre-warmed PBS 1x buffer twice to remove the treatment reagents completely.
NOTE: Different volumes of residual buffer could dilute the ATP assay reagents to different extent and can potentially affect the data. - Mix 75 µL of fresh pre-warmed medium and ATP assay reagent in a 1:1 dilution using a multichannel pipette. Make sure that the liquid level in all the multichannel pipette tips is the same.
- Place the plate on a nutating shaker for 15 min at room temperature. After 15 min of adding the reagents, transfer 60 µL of each sample into a white 96-well plate.
NOTE: White plates are better to reflect output light than clear or black plates. - Pop any air bubbles using a needle prior to reading the plate. Read the plate on a luminometer with a 1 s integration time. Read the plate within 20 min after adding of ATP assay reagents. Timing is critical for comparison across different plates, because the luminescence signal is transient with a fast decay rate.
- Calculate the percent (%) cell viability using this formula: (luminescence of transfected cells/luminescence of control, untreated cells) x 100.
8. Western blotting for measurement of tight junction protein ZO-1
- Cell lysis and protein extraction
NOTE: All the steps for protein extraction from cells must be carried out at 2-8 °C.- Seed the cells at a density of 50,000 cell/cm2 in a collagen-coated 12-well tissue culture plate.
- On day 3, day 5, day 7, day 10 post-seeding and on day 7 (cells pre-incubated with calcium free medium), remove the growth medium and gently wash the cells twice with 2 mL of ice-cold 1x PBS. Then, add 300 µL mixture of ice-cold 1x RIPA lysis buffer containing 3 µg/mL aprotinin in each well.
NOTE: The mixture should be freshly made and kept on ice. Aprotinin is used to inhibit proteases present in lysates from degrading the protein of interest. - After two freeze-thaw cycles (-80 °C), scrape the cells using a cold plastic cell scraper. Collect the cell lysates in microfuge tubes. Then, centrifuge the tubes at 200 x g for 30 min at 4 °C.
- Collect the supernatant into clean tubes and place them on ice. Measure the total amount of cellular protein in the lysates using the BCA assay kit by following the manufacturer's protocol.
- Western blotting for detecting of tight junction protein ZO-1
- Denature aliquots of total homogenates containing 40 µg of total protein with 1x Laemmli buffer at 95 °C, 5 min and subject to electrophoresis in a reducing 6-7.5% sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) (90 V, 10 min through stacking gel, 120 V through resolving gels).
NOTE: When loading the samples or standards, remember to load slowly and carefully into each lane, being careful not to break the well in the process. - Transfer the separated proteins onto a nitrocellulose membrane with pH 8.5 transfer buffer which contains 192 mM Glycine, 25 mM Tris Base, 10% methanol and 0.1% SDS (75 V, 110 min at room temperature).
NOTE: Do not touch the membrane. Use 70% isopropanol-washed plastic forceps to handle the membrane. In order to successfully transfer the ZO-1 protein (MW 200 kDa) to the membrane, pH should be around 8.3-8.5. If the transfer buffer is more acidic than that, transfer would not happen. If the ladder bands are visible still on the gel, it would be helpful to increase transfer time and SDS concentration. - After washing the membrane by using Tris-buffered saline containing 0.1%Tween 20 (T-TBS), use blocking solution (1:1 LiCOR-Odyssey Block: 1x Tris buffered saline) to block the membranes for 60 min.
- Carefully cut the membrane into two strips. Incubate with two primary antibodies (ZO-1 monoclonal antibody, dilution, 1: 900, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody, dilution, 1: 10,000) overnight at 4 °C. Then, incubate the ZO-1 and GAPDH membranes with donkey anti-mouse IgG (dilution, 1:50,000). After washing the membranes using T-TBS, image the membranes in the 700 channel on a 16-bit imager.
- Denature aliquots of total homogenates containing 40 µg of total protein with 1x Laemmli buffer at 95 °C, 5 min and subject to electrophoresis in a reducing 6-7.5% sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) (90 V, 10 min through stacking gel, 120 V through resolving gels).
Results
First, we determined the effect of culturing time on LY permeability to determine the apparent kinetics of TJ formation. The mean LY Papp values from day 1 to 10-post seeding are shown in Figure 2a. On day 1, the mean Papp was 4.25 x 10-4 cm/min and slightly dropped to 3.32 x 10-4 cm/min on day 2. The mean Papp value slightly increased to 3.93 x 10-4 cm/min on day 3 and fluctuate...
Discussion
A key role of the BBB is to prevent the exchange of non-essential ions and toxic substances between the systemic circulation and the brain to maintain hemostasis of neural microenvironment. One of the characteristic features of the BBB is the ability of the capillary endothelial cells to form tight junctions (TJs) that effectively seal the paracellular route of transport. We demonstrated a LY Papp assay as a quantitative method to determine the apparent kinetics of TJ barrier formation in cultured hCMEC/D3 mon...
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors are thankful for the financial support from the 2017 New Investigator Award from the American Association of Pharmacy, a Hunkele Dreaded Disease award from Duquesne University and the School of Pharmacy start-up funds for the Manickam laboratory. We would like to thank the Leak laboratory (Duquesne University) for western blotting assistance and allowing use of their Odyssey 16-bit imager. We would also like to include a special note of appreciation for Kandarp Dave (Manickam laboratory) for help with western blotting.
Materials
| Name | Company | Catalog Number | Comments |
| hCMEC/D3 cell line | Cedarlane Laboratories | 102114.3C-P25 | human cerebral microvascular endothelial cell line |
| gWizLuc | Aldevron | 5000-5001 | Plasmid DNA encoding luciferase gene |
| lucifer yellow CH dilithium salt | Invitrogen | 155267 | |
| Transwell inserts with polyethylene terephthalate (PET) track-etched membranes | Falcon | 353095 | |
| Tissue culture flask | Olympus Plastics | 25-207 | |
| 24-well Flat Bottom | Olympus Plastics | 25-107 | |
| Black 96-Well Immuno Plates | Thermo Scientific | 437111 | |
| S-MEM 1X | Gibco | 1951695 | Spinner-minimum essential medium (S-MEM) |
| EBM-2 | Clonetics | CC-3156 | Endothelial cell basal medium-2(EBM-2) |
| phosphate-buffered saline 1X | HyClone | SH3025601 | |
| Collagen Type I | Discovery Labware, Inc. | 354236 | |
| Pierce BCA Protein Assay Kit | Thermo Scientific | 23227 | |
| Cell Culture Lysis 5X Reagent | Promega | E1531 | |
| Beetle Luciferin, Potassium Salt | Promega | E1601 | |
| SpectraMax i3 | Molecular Devices | Fluorescence Plate Reader | |
| Trypan Blue Solution, 0.4% | Gibco | 15250061 | |
| ZO-1 Polyclonal Antibody | ThermoFisher | 61-7300 | |
| anti-GAPDH antibody | abcam | ab8245 | |
| Alexa Fluor680-conjugated AffiniPure Donkey Anti-Mouse LgG(H+L) | Jackson ImmunoResearch Inc | 128817 | |
| 12-well, Flat Bottom | Olympus Plastics | 25-106 | |
| RIPA buffer (5X) | Alfa Aesar | J62524 | |
| Aprotinin | Fisher BioReagents | BP2503-10 | |
| Odyssey CLx imager | LI-COR Biosciences | for scanning western blot membranes |
References
- Abbott, N. J., Patabendige, A. A., Dolman, D. E., Yusof, S. R., Begley, D. J. Structure and function of the blood-brain barrier. Neurobiology Of Disease. 37 (1), 13-25 (2010).
- Griep, L. M., et al. BBB on chip: microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomedical Microdevices. 15 (1), 145-150 (2013).
- Camos, S., Mallolas, J. Experimental models for assaying microvascular endothelial cell pathophysiology in stroke. Molecules. 15 (12), 9104-9134 (2010).
- Cucullo, L., et al. Immortalized human brain endothelial cells and flow-based vascular modeling: a marriage of convenience for rational neurovascular studies. Journal of Cerebral Blood Flow & Metabolism. 28 (2), 312-328 (2008).
- Wolff, A., Antfolk, M., Brodin, B., Tenje, M. In vitro Blood-Brain Barrier Models-An Overview of Established Models and New Microfluidic Approaches. Journal of Pharmaceutical Sciences. 104 (9), 2727-2746 (2015).
- Weksler, B., Romero, I. A., Couraud, P. O. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids and Barriers of the CNS. 10 (1), 16 (2013).
- Ohtsuki, S., et al. Quantitative targeted absolute proteomic analysis of transporters, receptors and junction proteins for validation of human cerebral microvascular endothelial cell line hCMEC/D3 as a human blood-brain barrier model. Molecular Pharmaceutics. 10 (1), 289-296 (2013).
- Tornabene, E., Brodin, B. Stroke and Drug Delivery--In vitro Models of the Ischemic Blood-Brain Barrier. Journal of Pharmaceutical Sciences. 105 (2), 398-405 (2016).
- Weksler, B., Romero, I. A., Couraud, P. O. The hCMEC/D3 cell line as a model of the human blood brain barrier. Fluids and Barriers of the CNS. 10 (1), 16 (2013).
- Llombart, V., et al. Characterization of secretomes from a human blood brain barrier endothelial cells in-vitro model after ischemia by stable isotope labeling with aminoacids in cell culture (SILAC). Journal of Proteomics. 133, 100-112 (2016).
- Weksler, B. B., et al. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. The FASEB Journal. 19 (13), 1872-1874 (2005).
- Avdeef, A. How well can in vitro brain microcapillary endothelial cell models predict rodent in vivo blood-brain barrier permeability?. European Journal of Pharmaceutical Sciences. 43 (3), 109-124 (2011).
- Rahman, N. A., et al. Immortalized endothelial cell lines for in vitro blood-brain barrier models: A systematic review. Brain Research. 1642, 532-545 (2016).
- Cecchelli, R., et al. A stable and reproducible human blood-brain barrier model derived from hematopoietic stem cells. PLoS One. 9 (6), e99733 (2014).
- Shao, X., et al. Development of a blood-brain barrier model in a membrane-based microchip for characterization of drug permeability and cytotoxicity for drug screening. Analytica Chimica Acta. 934, 186-193 (2016).
- Walter, F. R., et al. A versatile lab-on-a-chip tool for modeling biological barriers. Sensors and Actuators B: Chemical. 222, 1209-1219 (2016).
- Cecchelli, R., et al. In vitro model for evaluating drug transport across the blood–brain barrier. Advanced Drug Delivery Reviews. 36, (1999).
- Cecchelli, R., et al. Modelling of the blood–brain barrier in drug discovery and development. Nature reviews Drug discovery. 6 (8), 650 (2007).
- Reichel, A., Begley, D. J., Abbott, N. J. . The Blood-Brain Barrier. , 307-324 (2003).
- Deli, M. A., Ábrahám, C. S., Kataoka, Y., Niwa, M. Permeability Studies on In vitro Blood–Brain Barrier Models: Physiology, Pathology, and Pharmacology. Cellular and Molecular Neurobiology. 25 (1), 59-127 (2005).
- Ren, T. B., et al. A General Method To Increase Stokes Shift by Introducing Alternating Vibronic Structures. Journal of the American Chemical Society. 140 (24), 7716-7722 (2018).
- Pack, D. W., Hoffman, A. S., Pun, S., Stayton, P. S. Design and development of polymers for gene delivery. Nature Reviews Drug Discovery. 4 (7), 581-593 (2005).
- Oupický, D., Konák, C., Ulbrich, K., Wolfert, M. A., Seymour, L. W. DNA delivery systems based on complexes of DNA with synthetic polycations and their copolymers. Journal of Controlled Release. 65, (2000).
- Couraud, P. O. . The hCMEC/D3 CELL LINE: IMMORTALIZED HUMAN CEREBRAL MICROVASCULAR ENDOTHELIAL CELLS As a model of human Blood-Brain Barrier. , (2012).
- Youdim, K. u. r. e. s. h. A., A, A. A. a. N. J. In vitro trans-monolayer permeability calculations: often forgotten assumptions. research focus reviews. 8, (2003).
- Eigenmann, D. E., Xue, G., Kim, K. S., Moses, A. V., Hamburger, M., Oufir, M. Comparative study of four immortalized human brain capillary endothelial cell lines, hCMEC/D3, hBMEC, TY10, and BB19, and optimization of culture conditions, for an in vitro blood-brain barrier model for drug permeability studies. Fluid and Barriers of the CNS. 10 (33), (2013).
- Hubatsch, I., Ragnarsson, E. G. E., Artursson, P. Determination of drug permeability and prediction of drug absorption in Caco-2 monolayers. Nature Protocols. 2 (9), 2111-2119 (2007).
- Balda, M. S., Anderson, J. M. Two classes of tight junctions are revealed by ZO-1 isoforms. The American Physiological Society. , (1992).
- Brown, R. C., Davis, T. P. Calcium Modulation of Adherens and Tight Junction Function: A Potential Mechanism for Blood-Brain Barrier Disruption After Stroke. Stroke. 33 (6), 1706-1711 (2002).
- Gorodeski, G., Jin, W., Hopfer, U. Extracellular Ca2+ directly regulates tight junctional permeability in the human cervical cell line CaSki. American Journal of Physiology-Cell Physiology. 272 (2), C511-C524 (1997).
- Stuart, R. O., Sun, A., Panichas, M., Hebert, S. C., Brenner, B. M., Nigam, S. K. Critical Role for lntracellular Calcium in Tight Junction Biogenesis. Journal of Cellular Physiology. 159, (1994).
- Tobey, N. A. Calcium-switch technique and junctional permeability in native rabbit esophageal epithelium. American Journal of Physiology-Gastrointestinal and Liver Physiology. , (2004).
- Tobey, N. A., Argote, C. M., Hosseini, S. S., Orlando, R. C. Calcium-switch technique and junctional permeability in native rabbit esophageal epithelium. American Journal of Physiology-Gastrointestinal and Liver Physiology. 286, (2004).
- Posimo, J. M., et al. Viability assays for cells in culture. Journal of visualized experiments : JoVE. (83), e50645 (2014).
- Cipolla, M. J., Crete, R., Vitullo, L., Rix, R. D. Transcellular transport as a mechanism of blood-brain barrier disruption during stroke. Frontiers in Bioscience. 9 (3), 777-785 (2004).
- Kreuter, J. Influence of the surface properties on nanoparticle-mediated transport of drugs to the brain. Journal of nanoscience and nanotechnology. 4 (5), 484-488 (2004).
- Markoutsa, E., et al. Uptake and permeability studies of BBB-targeting immunoliposomes using the hCMEC/D3 cell line. European Journal of Pharmaceutics and Biopharmaceutics. 77 (2), 265-274 (2011).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved