CRISPR/Cas9 Ribonucleoprotein-mediated Precise Gene Editing by Tube Electroporation
In This Article
Summary
Presented here is a protocol for efficient CRISPR/Cas9 ribonucleoprotein-mediated gene editing in mammalian cells using tube electroporation.
Abstract
Gene editing nucleases, represented by CRISPR-associated protein 9 (Cas9), are becoming mainstream tools in biomedical research. Successful delivery of CRISPR/Cas9 elements into the target cells by transfection is a prerequisite for efficient gene editing. This protocol demonstrates that tube electroporation (TE) machine-mediated delivery of CRISPR/Cas9 ribonucleoprotein (RNP), along with single-stranded oligodeoxynucleotide (ssODN) donor templates to different types of mammalian cells, leads to robust precise gene editing events. First, TE was applied to deliver CRISPR/Cas9 RNP and ssODNs to induce disease-causing mutations in the interleukin 2 receptor subunit gamma (IL2RG) gene and sepiapterin reductase (SPR) gene in rabbit fibroblast cells. Precise mutation rates of 3.57%-20% were achieved as determined by bacterial TA cloning sequencing. The same strategy was then used in human iPSCs on several clinically relevant genes including epidermal growth factor receptor (EGFR), myosin binding protein C, cardiac (Mybpc3), and hemoglobin subunit beta (HBB). Consistently, highly precise mutation rates were achieved (11.65%-37.92%) as determined by deep sequencing (DeepSeq). The present work demonstrates that tube electroporation of CRISPR/Cas9 RNP represents an efficient transfection protocol for gene editing in mammalian cells.
Introduction
CRISPR/Cas9 is the most commonly used programmable nuclease for gene editing. It works through single guide RNA (sgRNA)-mediated recognition of both target sequences and an adjacent protospacer adjacent motif (PAM) sequence in the genome. The Cas9 nuclease generates a double-stranded DNA break (DSB) located three nucleotides upstream of the PAM sequence1. The DSBs are repaired either through error-prone non-homologous end joining (NHEJ) or homology-directed repair (HDR) pathways. To achieve precise gene editing through the HDR pathway, donor templates are often provided in the format of plasmid DNA (pDNA) or single-stranded oligodeoxynucleotide (ssODN).
CRISPR/Cas9 and the sgRNA can be delivered to the cells in three formats: the ribonucleoprotein (RNP) complex of Cas9 protein and gRNA2,3; Cas9 mRNA and sgRNA4,5; or plasmid DNA (pDNA) that contains the necessary promoters, driven sgRNA, and Cas9 coding region6,7,8. Many groups have demonstrated that when CRISPR/Cas9 is delivered as RNP, the gene editing efficiency often outperforms those achieved in pDNA or mRNA formats, attributable to the much smaller size of RNP compared to the nucleic acids9. Furthermore, it has been previously shown that a novel tube electroporation (TE) machine is particularly effective in gene editing applications in several cell types9.
Presented in the present work is a step-by-step protocol in utilizing TE for the delivery of CRISPR/Cas9 RNP to mammalian cells of different species at several clinically relevant loci. This novel TE transfection technique and high HDR rate phenomenon may find broad applications in biomedical research.
Protocol
All animal maintenance, care, and use procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Michigan.
1. Preparation of Cells
- Acquire human iPSCs (ACS-1030) from the American Type Culture Collection (ATCC). Culture iPScs on artificial extracellular matrix with feeder-free cell culture medium (see Table of Materials) in a cell culture incubator (5% CO2 at 37 °C) following the supplier's instructions.
- 2 h prior to transfection, treat the iPSCs with 10 µM Rho-associated, coiled-coil containing protein kinase (ROCK) inhibitor Y27632 (use of which reduces apoptosis of dissociated human hiPSCs and increases survival and cloning efficiency of hiPSCs without affecting their pluripotency).
- When transfecting, dissociate iPSCs with cell detachment solution (see Table of Materials) to single cells at 37 °C for 5 min. Count the cell number.
- Establish a rabbit fibroblast cell culture using a primary culture of rabbit ear skin tissue biopsies, as previously described10.
- A 0.5 cm x 0.5 cm ear skin biopsy is obtained from the tip of the rabbit ear. Shave the hair off the ear tissue.
- Rinse 2x with Dulbecco's phosphate-buffered saline (DPBS) with 5% penicillin-streptomycin. Transfer the ear tissue to a new 6 cm tissue culture dish, then cut the tissue into small pieces (~1.0 mm x 1.0 mm). Add a few drops of fetal bovine serum to prevent the tissue from drying out.
- Spread the shredded tissue to a 10 cm tissue culture dish, then add 10 mL of culture medium. Rabbit fibroblast cells are cultured in Dulbecco's Modified Eagle's Medium (DMEM) with 10% fetal bovine serum. Put the tissue culture dish in the cell culture incubator (5% CO2 at 37 °C).
- Three to five days after plating, use trypsin-EDTA to digest cells at 37 °C for 2 min. Count the cell number.
2. Design and Synthesis of gRNAs and Donor Oligos
- For each gene, design guide RNA based on the sequence of the targeted locus using an online tool (for example, <http://crispor.tefor.net/>).
- Paste in the DNA sequence of interest.
- Select a genome and protospacer-adjacent motif (PAM). Possible guide sequences in input DNA sequences will be displayed on the output page. It is recommended to select gRNA with higher predicted efficiency and lower off-target potentials.
- Synthesize DNA by a commercial vendor for transcribing gRNAs. Perform in vitro transcription of gRNA using a gRNA synthesis kit according to the manufacturer's instructions.
- Purify the gRNA using an RNA purification micro column included in the gRNA synthesis kit. Measure the concentration, then store the gRNAs at -80 °C.
- Design an ssODN donor template for each mutation site. The ssODNs can be synthesized by commercial vendors such as IDT. In general, each ssODN is 120-160 nucleotides (nt) in length, consisting of 60-80 nt in the left homology arm and 60-80 nt in the right homology arm. To prevent recutting of the edited DNA, a silent mutation at the PAM should be introduced in the ssODN whenever possible. The CRISPR cut site should be located as close to the intended genomic change as possible.
3. Tube Electroporation of Cas9 RNP and ssODNs
- Prepare the cells as described in section 1.
- Resuspend 2-3 x 105 cells in 20 µL of electroporation buffer. Pipette up and down carefully to produce a single-cell suspension.
- For Cas9 RNP transfection, premix 2 µg of Cas9-NLS protein with 0.67 µg of gRNA at room temperature (RT) for 10-15 min. Next, gently mix the formed RNP complex along with 2 µg of ssODN with cells.
- Transfer the cell mixture to a 20 µL electroporation tube using universal fit pipette tips provided by the tube electroporation kit. To achieve better electroporation, try to avoid the formation of air bubbles during transfer.
- Place the electroporation tube into the slot of the electroporator and press "Go" to finish. Follow manufacturer's suggested parameters for each cell type. For example, for human iPSCs and rabbit fibroblast cells, the voltage set is 420 V and pulse time is 30 ms. A successful electroporation cycle is indicated by the pulse report on the display screen of the electroporator.
- After the electroporation, transfer the human iPS cells to 1 mL of pre-warmed Y-27632-containing culture medium described in cell culture part. For rabbit fibroblast cells, transfer them to DMEM with 10% fetal bovine serum.
- Plate the resuspended cells to one well of a 12 well cell culture plate.
- Change the culture medium every day. Y-27632 is removed from the human iPSC culture medium 24 h post-electroporation.
4. Analysis of Gene Editing Events
- Harvest cells 72 h after electroporation. Digest cells from the culture plate using trypsin-EDTA for rabbit fibroblast cells or cell detachment solution for human iPSCs. After centrifuge, resuspend cells with 350 mL of lysis buffer (1 M Tris HCl, 5 M NaCl, 0.5 M EDTA; pH 8.0, 10% SDS, add 20 µL of 20 mg/mL proteinase K stock per 1 mL of lysis buffer), then incubate at 55 °C overnight.
- Extract the genomic DNA with phenol-chloroform using standard procedures.
- Amplify 100-200 bp DNA fragments containing targeted region using high-fidelity DNA polymerase, then purify the DNA fragments from gels using a gel extraction kit or directly from PCR products using a PCR SV mini kit.
- To determine gene editing efficiency by bacterial colony sequencing, ligase the purified PCR products into a pCR4-TOPO vector using a TOPO TA cloning kit. Randomly pick up bacterial clones, then sequence the inserts using a universal sequencing primer provided by the TOPO TA cloning kit.
- To determine gene editing efficiency by deep sequencing, send the purified PCR products (~100-200 bp) from step 4.3 for CRISPR amplicon sequencing in a DNA sequencing core.
Representative Results
TE of Cas9 RNP and ssODNs to rabbit fibroblast cells
The overall process of TE-mediated delivery of Cas9 RNP to mammalian cells is illustrated in Figure 1. First, C231Y and Q235X mutations were produced in the IL2RG gene, and the R150G mutation was produced in the SPR gene in rabbit fibroblast cells. Loss-of-function mutations in IL2RG and SPR genes are known to cause primary immunodeficiency11 and motor and cognitive deficits12, respectively.
The specific sgRNA designs are illustrated in Figure 2A. The primers used to amplify the targeted regions are listed in Table 3. Sequences of ssODNs are shown in Table 1. The gene editing rates were determined by bacterial TA cloning (Figure 2B). At the IL2RG C231 locus, out of the 28 clones that were sequenced, one (3.57%) carried the precise C231Y mutation, four (14.28%) carried insertion or deletion (indel) mutations, and the remaining 23 (82%) were wild-type. At the IL2RG Q235 locus, out of the 27 clones that were sequenced, two (7.41%) carried the precise Q235X mutation, three carried indel mutations (11.11%) and the remaining were wild-type. At the SPG R150 locus, of the 20 clones sequenced, five (25%) carried the precise R150G mutation, 10 (50%) carried indel mutations, and the remaining were wild-type.
TE of Cas9 RNP and ssODNs to human iPSCs
TE was then used to deliver Cas9 RNP and ssODNs to human iPSCs and target clinically relevant loci in EGFR, Mybpc3, and HBB genes. Point mutations in the EGFR T790 proximal region confer resistance to EGFR tyrosine kinase inhibitors in patients of non-small cell lung cancer (NSCLC) harboring activating mutations of EGFR13. A frameshift mutation in exon 16 in Mybpc3 is implicated in hypertrophic cardiomyopathy14. The E6V point mutation in the HBB gene leads to sickle cell disease15.
The specific sgRNA designs are illustrated in Figure 3A. The primers used to amplify the targeted regions are listed in Table 3. Sequences of ssODNs are shown in Table 1. The gene editing rates were determined by DeepSeq (Figure 3B). At the EGFR locus, 15.68% of alleles carried the precise point mutations (6,315 reads), 22.75% carried indel mutations (9,162 reads), and the remaining 61.57% were wild-type (24,797 reads). At the Mybpc3 locus, 37.92% carried the precise 4-bp TGAA deletion (11,654 reads), 2.24% carried indel mutations (410 reads) and the remaining 59.84% were wild-type (18,692 reads). At the HBB locus, 11.65% carried the precise E6V mutation (6,565 reads), 23.35% carried indel mutations (13,163 reads) and the remaining 65% were wild-type (36,644 reads).

Figure 1: Flow chart of tube electroporation of Cas9 RNP.

Figure 2: Gene editing of rabbit fibroblast cells. (A) Illustration of target sequences. Boxes indicate targeted loci. Underlined letters correspond to gRNA sequences. Red colored letters indicate PAM sequences. (B) TA cloning results of gene editing events. Boxes indicate precisely mutated loci. Indel sequence shown is only representative of one allele type. Other indel sequences are not shown. Please click here to view a larger version of this figure.
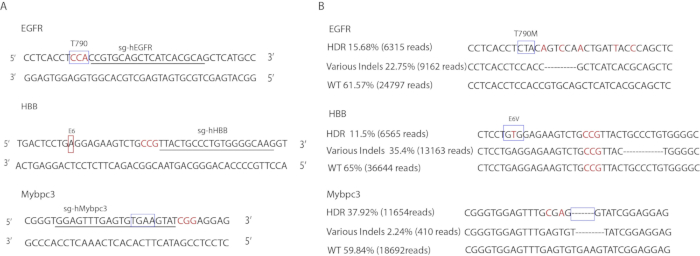
Figure 3: Gene editing of human iPSCs. (A) Illustration of target sequences. Boxes indicate targeted loci. Underlined letters correspond to gRNA sequence. Red colored letters indicate PAM sequences. (B) Deepseq results of gene editing events. Boxes indicate precisely mutated loci. Red colored letters indicate silent mutations that were introduced in the donor templates. Indel sequence shown is only representative of one allele type. Other indel sequences are not shown. Please click here to view a larger version of this figure.
| Locus | Oligo sequence | |||||||
| (targeted mutation) | ||||||||
| Rabbit IL2RG (C231Y) | AGCGTGGATGGGCAGAAACTCTACACGTTCCGAGTCCGGAGCCGTTTTAACCCTTTGTATGGGAGTGCTCAGCATTGGAGT GAATGGAGCCACCCGATCCACTGGGGGAGCAAAACTTCAAAGGGTAAAATGGGCCT | |||||||
| Rabbit IL2RG (Q235X) | AGCGTGGATGGGCAGAAACTCTACACGTTCCGAGTCCGGAGCCGTTTTAACCCTTTGTGTGGGAGTGCTTAGCATTGGAGT GAATGGAGCCACCCGATCCACTGGGGGAGCAAAACTTCAAAGGGTAAAATGGGCCT | |||||||
| Rabbit SPR | gacctccatgctctgcctgacctcctgcatcctgaaggcgtttcctgccagtcctggCctcagcgggactgtggtgaacatctcgtcgctgtgtgccctgcagcccttcaagggctggg cgctgtac | |||||||
| (R150G) | ||||||||
| Human EGFR | ACGTGATGGCCAGCGTGGACAACCCCCACGTGTGCCGCCTGCTGGGCATCTGCCTCACCTCTACAGTCCAACTGATTACCC AGCTCATGCCCTTCGGCTGCCTCCTGGACTATGTCCGGGAACACAAAGACAATATTGGCTCCCAGTAC | |||||||
| (Point mutations proximate to T790) | ||||||||
| Human Mybpc3 | GCCCCCTGTGCTCATCACGCGCCCCTTGGAGGACCAGCTGGTGATGGTGGGGCAGCGGGTGGAGTTTGCGAGGTATCGGA GGAGGGGGCGCAAGTCAAATGGTGAGTTCCAGAAGCACGGGGCATGGGTGTTGGGGGCAT | |||||||
| (4-bp deletion) | ||||||||
| Human HBB | TCTGACACAACTGTGTTCACTAGCAACCTCAAACAGACACCATGGTGCATCTGACTCCTGTGGAGAAGTCTGCAGTTACTGCC CTGTGGGGCAAGGTGAACGTGGATGAAGTTGGTGGTGAGGCCCTGGGCAG | |||||||
| (E6V) | ||||||||
Table 1: Sequences of ssODNs.
| Step | Problem | Possible reasons | Solutions |
| 2.1 | Low indel rate | Poor guide RNA design, Guide RNA stocks >6 months, low guide RNA concentration | Redesign guide RNA, produce/order new guide RNA. |
| 2.3 | Low PGE efficiency | poor donor DNA design, low efficient guide RNA, incorrect amount of donor DNA or poor quality DNA | Increase homology arm length, introduce PAM mutation, introduce silent mutations to the donor DNA, use a more efficient guide RNA, Optimize the ratio of Cas9 protein over guide RNA. |
| 3.4 | Failed transfection | Air bubbles formed during transferring cells-buffer mixture to electroporation tube, incorrect voltage/duration setting | Try to avoid the formation of air bubbles, adjust the voltage/duration setting. |
| 3.6 | low cell viability after electroporation | Low survival of single human ipsc | Add ROCK inhibitor after electroporation, increase number of cells. |
| 4.1 | Failed PCR | High GC contents, or repetitive sequence | Optimize PCR condition, add DMSO to PCR system. |
Table 2: Troubleshooting guides for frequent problems.
| Primer name | sequence | note |
| Rb-IL2RG-F | CATGACAGTGACAGGGTCCC | For amplifying rabbit IL2RG DNA fragment |
| Rb-IL2RG-R | TGCCAGAGACACAAGCGAAC | |
| Rb-SPR-F | GTACTTTGGAGGGACAGAGG | For amplifying rabbit SPR DNA fragment |
| RB-SPR-R | CTCAGCACCCTGACACTGGG | |
| H-EGFR-F | TGATGGCCAGCGTGGACAAC | For amplifying human EGFR DNA fragment |
| H-EGFR-R | ACCAGTTGAGCAGGTACTGGG | |
| H-Mybpc3-F | ATGCCCCGTGCTTCTGGAAC | For amplifying human Mybpc3 DNA fragment |
| H-Mybpc3-R | TCAGGGGAGCCAACCCTCAT | |
| H-HBB-F | TAACCTTGATACCAACCTGC | For amplifying human HBB DNA fragment |
| H-HBB-R | CATTTGCTTCTGACACAACT |
Table 3: Primers used in step 4.3.
Discussion
The tube electroporation method was effective in delivering CRISPR/Cas9 RNP and ssODNs to rabbit and human cells, leading to robust precise gene editing (PGE). The primary difference between TE and other conventional electroporation devices is the use of a tube, in which two electrodes are on the top and bottom of the tube and the sample is loaded in full then sealed upon electroporation (Figure 1). In contrast, in a conventional cuvette, the electrodes are on the sides and the sample is not fully sealed during electroporation. This new design reduces air bubble generation and compresses air bubble size, which consequently improves even distribution of electric voltage, and as a result leads to reduced cell death and high transfection efficiency9. In the present work, high PGE rates (15%-37%) were achieved targeting EGFR, Mybpc3 and HBB genes in human iPSCs. These results are consistent with a prior report in which high PGE rates were achieved in human stem cells9.
Disease-causing mutations were targeted in IL2RG and SPR genes in rabbit cells. Recently, IL2RG-knockout rabbits have been produced as models for human X-linked severe combined immunodeficiency (SCID-X1)16,17. The present work shows that patient IL2RG mutations (e.g., C231Y and Q235X) can be efficiently generated in rabbit cells, demonstrating the feasibility of creating SCID-X1 rabbit models carrying patient mutations. It was also demonstrated that SPR R150G mutations can be efficiently created in rabbit cells. This mutation causes motor and cognitive deficits in children12. These IL2RG and SPR mutation rabbit models, once generated, may serve as valuable preclinical models for translational studies. They may also be used to establish gene editing-based therapeutics for these monogenic diseases.
One concern for CRISPR/Cas9-mediated gene editing applications is the off-target editing events. Indel rates were analyzed at predicted top off-target sites for sgRNAs used in this study (Table S1), using methods previously described9. In total, seven potential top off-target loci were analyzed for sg-rb-IL2RG-01, five for sg-rb-SPR, seven for sg-hEGFR, five for sg-hMybpc3, and seven for sg-hHBB), using the primers listed in Table S2. No off-target indels were revealed by the T7E1 assays (Figure S1), indicating minimal off-target risks for CRISPR/Cas9-mediated gene editing using these sgRNAs. It also indicates that the tube electroporation method itself does not cause or increase off-target edits. Nevertheless, efforts should be dedicated to reduce or eliminate undesirable off-target edits. Whole-genome sequencing may be necessary to exclude such events for cells that are intended to be used in clinical applications.
At the technical level, the following are considered key factors to achieving efficient precise genome editing by CRISPR/Cas9 RNP tube electroporation. First, it is advised to select an efficient sgRNA with predicted low off-target potential. It is important to validate the indel efficiency of the selected sgRNA before using it for PEG applications. It is not rare that a software predicted good sgRNA fails at the validation step.
Second, to achieve high PGE, it is recommended to induce a PAM mutation to the ssODN donor whenever possible. The rationale is that by doing so, CRISPR/Cas9 re-cutting after donor template integration is prevented. In certain cases, the PGE itself introduces PAM mutations. In other cases, it is possible to introduce silent mutations to the PAM sequence. In the event that a PAM mutation is not possible, it is advised to try to include several silent mutations in the donor that corresponds to the sgRNA sequence.
Thirdly, particularly relevant to TE, it is important to avoid the formation of air bubbles when transferring cells and RNP mixture to the electroporation tube. While the design of a TE tube already minimizes air bubble formation, careful handling will further reduce and may even complete avoid air bubble formation. A trouble shooting guide for frequent problems that may be encountered in the application of tube electroporation for CRISPR/Cas9 ribonucleoprotein mediated precise gene editing is provided in Table 2.
In conclusion, it is demonstrated here that tube electroporation is an effective means for the delivery of CRISPR/Cas9 RNP and ssODNs to mammalian cells to achieve high PGE rates. This new TE transfection technique and its robust precise gene editing rate may facilitate the development of gene editing applications.
Disclosures
J. C. works at Celetrix LLC, manufacturer of the tube electroporator. L. M., L. J., J. S., D. Y., J. Z., Y. E. C., and J. X. declare no competing interests.
Acknowledgements
This work was supported by the National Institutes of Health (R21OD023194 to JX). This work utilized Core Services supported by Center for Advanced Models for Translational Sciences and Therapeutics (CAMTraST) at the University of Michigan Medical Center.
Materials
| Name | Company | Catalog Number | Comments |
| Accutase | STEMCELL Technologies | 792 | Cell detachment solution for human iPSCs, first used in Step 1.1.2. |
| Cas9 Nuclease 3NLS | IDT | 1074182 | Cas9 protein, first used in Step 3.3. |
| DMEM | Thermo Fisher | 11965092 | For cell culture, first used in Step 1.2.3. |
| DPBS | Thermo Fisher | 1708075 | For preparing cell culture, first used in Step 1.2.2. |
| EDTA | Lonza | 51201 | For making lysis buffer, first used in Step 4.1. |
| Electroporation buffer | Celetrix | 13–0104 | The electroporation buffer, first used in Step 3.2. |
| Electroporation tubes | Celetrix | 20 μL: 12–0107; 120 μL: 12–0104 | The electroporation tube, first used in Step 3.4. |
| Electroporator | Celetrix | CTX-1500A LE | The tube electroporation machine, first used in Step 3.5 |
| Fetal bovine serum | Sigma Aldrich | 12003C | For cell culture, first used in Step 1.2.2. |
| Forma CO2 Incubators | Thermo Fisher | Model 370 | For cell culture, first used in Step 1.1. |
| Gel Extraction Kit | Qiagen | 28115 | For gel purification, first used in Step 4.3. |
| Human induced pluripotent stem cells | American Type Culture Collection | ACS-1030 | Human iPSCs, first used in Step 1.1. |
| Matrigel | Corning | 354277 | Artificial extracellular matrix; for precoating cell culture plate, first used in Step 1.1. |
| mTeSR 1 medium | STEMCELL Technologies | 85850 | Feeder-free cell culture medium for human iPSCs, first used in Step 1.1. |
| PCR SV mini | GeneAll | 103-102 | For PCR product purification, first used in Step 4.3. |
| Penicillin-Streptomycin | Thermo Fisher | 15140163 | For preparing cell culture, first used in Step 1.2.2. |
| Phenol-chloroform | Thermo Fisher | 15593031 | For DNA extraction, first used in Step 4.2. |
| Precision gRNA Synthesis Kit | Invitrogen | A29377 | For the generation of full length gRNA (guide RNA), first used in Step 2.4. |
| Proteinase K Solution | Thermo Fisher | AM2548 | For DNA extraction, first used in Step 4.1. |
| Q5 high-fidelity DNA polymerase | NEB | M0491 | For PCR amplification, first used in Step 4.3. |
| Sodium dodecyl sulfate | Sigma Aldrich | L3771 | For making lysis buffer, first used in Step 4.1. |
| TA Cloning Kit | Thermo Fisher | K457502 | For TA clone sequencing, first used in Step 4.4. |
| Tissue Culture Dish (10 cm) | FALCON | 353003 | For cell culture, first used in Step 1.2.3. |
| Tissue Culture Dish (12 well) | FALCON | 353043 | For cell culture, first used in Step 3.7. |
| Tissue Culture Dish (6 cm) | FALCON | 353004 | For cell culture, first used in Step 1.2.2. |
| Tris HCl | Thermo Fisher | BP1757-500 | For making lysis buffer, first used in Step 4.1. |
| Trypsin-EDTA | Thermo Fisher | 25200056 | For cell digestion, first used in Step 1.2. 4. |
| Universal Fit Pipette Tips | Celetrix | 14-0101 | For electroporation, first used in Step 3.4. |
| Y27632 | LC Labs | Y-5301 | The apoptosis inhibotor, first used in Step 1.1.1. |
References
- Cong, L., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 339 (6121), 819-823 (2013).
- Mout, R., et al. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 11 (3), 2452-2458 (2017).
- Zuris, J. A., et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nature Biotechnology. 33 (1), 73-80 (2015).
- Miller, J. B., et al. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew Chem Int Ed Engl. 56 (4), 1059-1063 (2017).
- Finn, J. D., et al. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Reports. 22 (9), 2227-2235 (2018).
- Liang, C., et al. Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma. Biomaterials. 147, 68-85 (2017).
- Luo, Y. L., et al. Macrophage-Specific in Vivo Gene Editing Using Cationic Lipid-Assisted Polymeric Nanoparticles. ACS Nano. 12 (2), 994-1005 (2018).
- Wang, H. X., et al. Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proceedings of the National Academy of Sciences of the United States of America. 115 (19), 4903-4908 (2018).
- Xu, X., et al. Efficient homology-directed gene editing by CRISPR/Cas9 in human stem and primary cells using tube electroporation. Scientific Reports. 8 (1), 11649 (2018).
- Du, F., et al. Beneficial effect of young oocytes for rabbit somatic cell nuclear transfer. Cloning Stem Cells. 11 (1), 131-140 (2009).
- Allenspach, E., Rawlings, D. J., Scharenberg, A. M., Adam, M. P., et al. . GeneReviews(R). , (1993).
- Friedman, J., et al., Adam, M. P., et al. . GeneReviews(R). , (1993).
- Hidaka, N., et al. Most T790M mutations are present on the same EGFR allele as activating mutations in patients with non-small cell lung cancer. Lung Cancer. 108, 75-82 (2017).
- Ma, H., et al. Correction of a pathogenic gene mutation in human embryos. Nature. 548 (7668), 413-419 (2017).
- Vakulskas, C. A., et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nature Medicine. 24 (8), 1216-1224 (2018).
- Song, J., et al. Bacterial and Pneumocystis Infections in the Lungs of Gene-Knockout Rabbits with Severe Combined Immunodeficiency. Frontiers in Immunology. 9, 429 (2018).
- Song, J., et al. Production of immunodeficient rabbits by multiplex embryo transfer and multiplex gene targeting. Scientific Reports. 7 (1), 12202 (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved