Standardized Methods for Measuring Induction of the Heat Shock Response in Caenorhabditis elegans
In This Article
Summary
Here, standardized protocols are presented to assess induction of the heat shock response (HSR) in Caenorhabditis elegans using RT-qPCR at the molecular level, fluorescent reporters at the cellular level, and thermorecovery at the organismal level.
Abstract
The heat shock response (HSR) is a cellular stress response induced by cytosolic protein misfolding that functions to restore protein folding homeostasis, or proteostasis. Caenorhabditis elegans occupies a unique and powerful niche for HSR research because the HSR can be assessed at the molecular, cellular, and organismal levels. Therefore, changes at the molecular level can be visualized at the cellular level and their impacts on physiology can be quantitated at the organismal level. While assays for measuring the HSR are straightforward, variations in the timing, temperature, and methodology described in the literature make it challenging to compare results across studies. Furthermore, these issues act as a barrier for anyone seeking to incorporate HSR analysis into their research. Here, a series of protocols is presented for measuring induction of the HSR in a robust and reproducible manner with RT-qPCR, fluorescent reporters, and an organismal thermorecovery assay. Additionally, we show that a widely used thermotolerance assay is not dependent on the well-established master regulator of the HSR, HSF-1, and therefore should not be used for HSR research. Finally, variations in these assays found in the literature are discussed and best practices are proposed to help standardize results across the field, ultimately facilitating neurodegenerative disease, aging, and HSR research.
Introduction
The heat shock response (HSR) is a universal cellular stress response induced by cytosolic protein misfolding caused by temperature increases and other proteotoxic stresses. Activation of the HSR in Caenorhabditis elegans leads to transcriptional upregulation of heat shock genes such as hsp-70 and hsp-16.2. Many heat shock proteins (HSPs) function as molecular chaperones that restore protein folding homeostasis, or proteostasis, by directly interacting with misfolded or damaged proteins. The master regulator of the HSR is the transcription factor Heat Shock Factor 1 (HSF-1), whose activation is elegantly controlled via multiple mechanisms1.
The role of HSF-1 is not restricted to stress. HSF-1 is required for normal growth and development, as deletion of hsf-1 leads to larval arrest2. HSF-1 is also important during aging and age-related neurodegenerative diseases characterized by accumulation of protein aggregates and an inability to maintain proteostasis. Knockdown of hsf-1 causes accumulation of protein aggregates and a shortened lifespan, while overexpression of hsf-1 reduces protein aggregation and extends lifespan3,4. Therefore, regulation of HSF-1 at the molecular level has broad implications for organismal physiology and disease.
C. elegans is a powerful model organism for HSR research because the HSR can be measured at the molecular, cellular, and organismal levels4,5,6. Highlighting the power of this model, key advances in delineating the HSR pathway, such as tissue-specific differences in HSR regulation, have been discovered in C. elegans7,8. Furthermore, C. elegans is widely used for aging research and is an emerging system for modeling diseases linked to proteostasis disruption.
Although heat shock experiments with C. elegans can be quick and reproducible, there are several questions to consider before beginning. For example, which temperature should be used for induction of the HSR and how long should the worms be exposed? Is it better to use a dry incubator or a water bath? Which developmental stage should be used? Unfortunately, the methodologies used to investigate the HSR vary widely from laboratory to laboratory, causing confusion when selecting the best methodologies and making it difficult to compare results across the field.
We present robust and standardized protocols for using RT-qPCR, fluorescent reporters, and thermorecovery to measure the HSR. While these three approaches are complementary, they each have unique advantages and disadvantages. For example, RT-qPCR is the most direct and quantitative measurement of the HSR, and this assay can be easily expanded to include many different heat shock-inducible genes. However, RT-qPCR is the most expensive, can be technically difficult, and requires the use of specialized equipment. In contrast, fluorescent reporters have the advantage of measuring tissue-specific differences in HSR induction. However, they are difficult to quantitate accurately, can only measure induction above a certain threshold, and require the use of a fluorescence microscope. Additionally, the reporter strains described here are developmentally delayed compared to the standard N2 strain. Although newer reporter strains containing single-copy transgenes are available, they have not been tested here9. The third assay, thermorecovery, has the advantage of providing a physiologically relevant readout at the organismal level. However, this assay is arguably the least sensitive and most indirect. Finally, we discuss some common variations found in these assays and propose a set of best practices to facilitate research in this field.
Protocol
1. Maintenance and synchronization of C. elegans
- Maintain worms at 20 °C on Nematode Growth Medium (NGM) plates seeded with OP50 Escherichia coli bacteria by transferring several adults to fresh plates approximately 2x per week10. Care should be taken to prevent worms from running out of food, because this can affect their physiology11.
- Preparation of NGM plates.
- Mix 3 g of NaCl, 2.5 g of Bacto-peptone, 20 g of agar, and deionized (DI) H2O up to 1 L in a flask.
- Autoclave the mixture for sterilization.
- Allow mixture to cool to ~50 °C.
- Add 25 mL of 1 M KH2PO4 (pH = 6), 1 mL of 1 M CaCl2, 1 mL of 1 M MgSO4, and 1 mL of cholesterol (5 mg/mL in 100% ethanol).
- Using sterile technique, pour the mixture into 6 cm plates to yield approximately 100 plates. Pouring plates is easier if the mixture is first transferred to a 300 mL sterile beaker.
- Allow 1 day to solidify at room temperature (RT) before seeding with bacteria or storing at 4 °C.
- Seeding of OP50 bacteria onto NGM plates.
- Grow a saturated overnight OP50 bacterial culture in LB at 30 °C or 37 °C.
- Place approximately 300 µL of the culture onto the center of a 6 cm NGM plate.
- Let plates dry at RT for 1-3 days as needed for the bacterial lawn to adhere to the plate. Plates can then be used or stored at 4 °C.
- Preparation of NGM plates.
- Grow the worms synchronously either by isolating freshly laid eggs (described here) or alternatively by collecting eggs after dissolving worms with bleach.
- Transfer approximately 10 gravid adult worms to a fresh plate using a platinum wire pick. Egg-lay synchronization works best if the adults are in the first day of adulthood.
- After approximately 1 h, remove the worms from the plate. This should result in 40-60 eggs per plate, depending on the conditions and the strain.
2. Fluorescent imaging of HSR reporters
- Synchronize the worms (section 1.2) and maintain at 20 °C until the desired developmental stage. For the AM446 (hsp-70p::gfp) and CL2070 (hsp-16.2p::gfp) fluorescent reporter strains, young adult worms that have not yet reached reproductive maturity are generated 64 h after the egg-laying synchronization.
NOTE: The developmental timing varies with each strain and the temperature at which the worms are raised. Both HSR reporter strains exhibit a slight developmental delay relative to N2. Importantly, the magnitude of HSR induction declines approximately 2-4x after the onset of reproductive maturity (see Discussion). - Heat shock the worms by wrapping plates with paraffin film and submerging in a circulating water bath at 33 °C for 1 h. A thin strip of paraffin film should be wrapped 2x around the plate to seal the edges. Do not cover the bottom of the plate or it could interfere with heat transfer. Submerge the plates upside down using a test tube rack and a lead weight. Remember to include a negative control sample (no heat shock) if necessary.
NOTE: If the paraffin film is not secure, then water will enter the plate and the plate should not be used for data collection. - Recover the worms by removing the plates from the water bath and drying with a paper towel. Remove the paraffin film and incubate the worms at 20 °C for 6-24 h. This recovery period allows sufficient time for GFP synthesis and folding before imaging.
- Prepare slides for imaging. Slides should be prepared fresh for each use.
- Make a 3% agarose solution in water and heat using a microwave until the agarose is dissolved.
- Place a microscope slide for imaging between two other microscope slides that have a strip of laboratory tape on them to create a spacer for the agarose pad.
- Using a 1,000 µL pipette, place a drop (~150 µL) of the heated 3% agarose in the center of the microscope slide.
- Immediately cover the microscope slide with a blank microscope slide perpendicular to the first slide so that the top slide rests on the laboratory tape on the adjacent slides. This spreads out the drop of agarose to create a pad of uniform width.
- Carefully remove the top slide.
- Immobilize the worms by using a 200 µL pipette to add a small drop (~5 µL) of 1 mM levamisole in M9 buffer to the center of the agarose pad. Then transfer 10 worms into the drop of levamisole using a platinum wire pick. Cover with a coverslip. Sealing the coverslip is not necessary for an upright microscope. Optionally, the worms can be aligned when they become paralyzed by spreading the levamisole off, to the outside of the agarose pad, and aligning the worms with a platinum wire pick. Alternatively, the levamisole can be soaked up using a laboratory wipe.
NOTE: Image as soon as possible, because prolonged incubation in levamisole could alter fluorescence. - Image the worms using a fluorescence microscope. The details of image capture vary by microscope and software.
NOTE: To directly compare image intensities, use identical microscope settings in one imaging session. Avoid oversaturating the image.
3. Measurement of HSR gene expression using RT-qPCR
- Synchronize worms (section 1.2) and maintain at 20 °C until the desired developmental stage. For N2 worms, young adult worms that have not yet reached reproductive maturity are generated 60 h after the egg-laying synchronization.
NOTE: The developmental timing varies with each strain and the temperature at which the worms are raised. Importantly, the magnitude of HSR induction declines approximately 2-4x after the onset of reproductive maturity (see Discussion). - Heat shock worms as described in step 2.2.
- Take the plates out of the water bath, remove the paraffin film, and immediately collect the worms. The worms can be collected by washing the plates gently with 1 mL of M9, collecting the liquid in a microcentrifuge tube, and then removing the M9 after centrifugation at 400 x g for 1 min.
- Lyse the worms and purify the RNA using organic extraction.
- Add 250 µL of RNA isolation reagent (see Table of Materials).
- Vortex tubes by hand for 30 s.
- Vortex tubes for 20 min at 4 °C using a microcentrifuge tube attachment (see Table of Materials).
- Add 50 µL of chloroform.
- Vortex for 30 s.
- Incubate the samples at RT for 3 min.
- Centrifuge at ≥14,000 x g for 15 min at 4 °C.
- Transfer the aqueous layer (i.e., top layer, ~125 µL) to a new microcentrifuge tube.
NOTE: Avoid the organic layer and the material in the interface. - Add 50 µL of chloroform.
- Vortex for 30 s.
- Incubate the samples at RT for 3 min.
- Centrifuge at ≥14,000 x g for 5 min at 4 °C.
- Transfer the aqueous layer (~100 µL) to a new microcentrifuge tube.
NOTE: Avoid the organic layer and the material in the interface. - Precipitate RNA with an equal volume (i.e., 100 µL) of isopropanol.
- Incubate at -20 °C for at least 30 min, but preferably overnight.
NOTE: The experiment can be paused here and the RNA can be stored at -20 °C. - Pellet the RNA by centrifugation at ≥14,000 x g for ≥30 min at 4 °C.
- Remove as much of the supernatant as possible without disturbing the pellet.
NOTE: The pellet will be small and may not be visible. The pellet may not adhere tightly to the side of the tube, so caution is necessary to avoid dislodging it. - Wash the pellet with 250 µL of 70% ice-cold ethanol made with RNase-free H2O.
- Centrifuge at ≥14,000 x g for ≥5 min at 4 °C.
- Remove as much supernatant as possible without disturbing the pellet.
- Perform a quick spin at RT to remove any remaining 70% ethanol.
- Dry the pellet by leaving the tubes open at RT as long as needed; typically at least 20 min. Tubes can be covered with a lint-free tissue or aluminum foil to prevent contamination.
- Resuspend the pellet in 20 µL of RNase-free H2O.
- Determine the RNA concentration using a small volume spectrophotometer (2 µL).
NOTE: The experiment can be paused here and the RNA can be temporarily stored at or below -20 °C.
- Remove residual DNA by incubating with DNase I. It is recommended to use a commercially available kit (see Table of Materials) and to follow the manufacturer's instructions.
- With this kit, prepare a 20 µL reaction with 500 ng of RNA and 1 µL of DNase I in a 37 °C water bath for 30 min.
- Add 2.5 µL of DNase inactivation reagent (included in the kit) to each sample and incubate at RT for 5 min with occasional flicking/vortexing.
- Spin down at 14,000 x g for 2 min.
- Without disturbing the white pellet, transfer 15 µL of supernatant to a fresh microtube for cDNA synthesis.
- Conduct cDNA synthesis. It is recommended to use a commercially available kit (see Table of Materials) and to follow the manufacturer's instructions.
- With the kit, prepare a 20 µL reaction with 15 µL of DNase I-treated RNA from the previous step and 1 µL of reverse transcriptase.
- Use the following program for cDNA synthesis: 25 °C for 5 min, 46 °C for 20 min, 95 °C for 1 min, 4 °C hold.
- Dilute cDNA by adding 80 µL of RNase-free H2O directly to the sample.
- Briefly vortex, then spin down and store at -20 °C until needed.
- Perform qPCR. It is recommended to use a commercially available kit (see Table of Materials) and to follow the manufacturer's instructions.
- With the kit, prepare a 25 µL reaction containing 2 µL of cDNA and 200 nM (each) of forward and reverse primers in one well of a 96-well plate.
- Primer sequences for measuring the heat shock genes, hsp-70 and hsp-16.2, and 18S rRNA (for a normalization control) are listed in the Table of Materials. Multiple normalization controls can be used as desired.
- Dilute cDNA samples 50x before measurement of 18S to ensure that the assay is in the linear range. Appropriate qPCR conditions vary with the kit and primers used (see Representative Results).
- Use a real-time PCR detection system (see Table of Materials) for qPCR with 40 cycles of 95 °C for 5 s denaturation, 58 °C for 30 s annealing, and 72 °C for 30 s extension.
NOTE: Optimal annealing temperatures can vary by primers and conditions. - Quantify using either the ΔΔCt or standard curve method12.
4. Thermorecovery assay for measuring HSR at the organismal level
- Synchronize the worms (section 1.2) and maintain at 20 °C until the desired developmental stage. For N2 worms, young adult worms that have not yet reached reproductive maturity are generated 60 h after the egg-laying synchronization.
NOTE: The developmental timing varies with each strain and the temperature at which the worms are raised. Importantly, the magnitude of HSR induction declines approximately 2-4x after the onset of reproductive maturity (see Discussion). - Heat shock the worms as described in step 2.2 for 6 h.
- Remove the plates from the water bath, remove the paraffin film, and allow the worms to recover by incubation at 20 °C for 48 h.
- Count the number of worms that can immediately crawl away after mechanical stimulation without jerky movement or paralysis.
NOTE: The 6 h incubation is optimal for examining conditions that reduce thermorecovery, but longer exposure times may be needed to look for conditions that enhance thermorecovery.
Representative Results
Using the protocols described in this manuscript, HSR induction was measured using fluorescent reporters, RT-qPCR, and thermorecovery assays. In each case, the procedure in section 1.2 was used to generate synchronized, young adult worms that had not reached reproductive maturity.
To visualize HSR induction at the cellular level, the AM446 (hsp-70p::gfp) and CL2070 (hsp-16.2p::gfp) fluorescent reporter strains were analyzed following section 2 of the protocol. In the negative control samples without heat shock, the hsp-16.2 reporter only showed normal autofluorescence, but the hsp-70 reporter had constitutive fluorescence in the anal depressor muscle as previously reported4 (Figure 1A). After 1 h of heat shock at 33 °C, robust fluorescence was observed in both reporters; however, the pattern of expression was distinct depending on which reporter was used (Figure 1B). The hsp-70 reporter was brightest in the intestine and spermatheca, whereas the hsp-16.2 reporter was brightest in the pharynx. Additionally, the hsp-16.2 reporter had a high degree of worm-to-worm variability in the amount of induction as previously described, but the hsp-70 reporter did not13.
A commonly used variation of section 2 is to perform the heat shock in a dry incubator instead of a circulating water bath. Therefore, the difference between the two methodologies was also tested. It was found that both protocols resulted in robust induction of the two fluorescent reporters using our conditions, although a circulating water bath is recommended as a best practice (see Discussion) (Figure 1B).
To test the dependence of the reporters on the transcription factor HSF-1, feeding RNAi was used to knockdown hsf-1 before reporter induction was measured. It was found that fluorescence of both strains was severely reduced upon HSF-1 knockdown, indicating that these reporters are HSF-1-dependent as described in the literature4 (Figure 2). However, it was also observed that pharyngeal fluorescence persisted in both reporters upon hsf-1 knockdown, which is consistent with previous reports that the pharyngeal muscle is resistant to RNAi by feeding14.
To quantitate whole worm induction of the HSR at the molecular level, two endogenous HSPs were measured with RT-qPCR using section 3 of the protocol. Samples were measured in triplicate, a standard curve was generated for each of the primers, and a melt curve was analyzed for each sample for quality control. It was found that a 33 °C heat shock for 1 h resulted in more than a 2,000x increase in relative expression for two heat shock genes, hsp-70 and hsp-16.2 (Figure 3). These results show that both endogenous genes are suitable for measuring HSR induction and that a 33 °C heat shock for 1 h is sufficient to generate a substantial response. However, caution should be used in interpreting the absolute degree of heat shock induction, because the mRNA levels in the absence of heat shock are very low.
To analyze a physiological response to heat shock, an organismal thermorecovery assay was tested using section 4 of the protocol. It was found that exposure of worms to a 6 h heat shock at 33 °C led to a 20% decrease in worms with normal movement after a 48 h recovery (Figure 4A). The dependence of this assay on the HSF-1 transcription factor was tested using feeding RNAi to knockdown hsf-1 before exposing worms to the stress. It was found that knockdown of hsf-1 caused a dramatic decrease in normal movement, with >95% of worms showing jerky movement or paralysis after being prodded with a platinum wire pick.
We compared this thermorecovery assay to a widely used alternative organismal assay commonly referred to as thermotolerance. In the thermotolerance assay, worms are exposed to a continuous 35 °C temperature using a dry incubator, and the percentage of worms alive are measured at various timepoints. Using this assay, it was found that control worms continuously exposed to 35 °C died after approximately 8 h of exposure (Figure 4B). However, when the dependence of this assay on HSF-1 was tested using RNAi knockdown, it was found that inhibition of hsf-1 did not cause a decrease in thermotolerance. Similar results have been previously shown using HSF-1 mutations (see Discussion). Therefore, the use of the thermotolerance assay to measure the HSR is not recommended, and thermorecovery is the preferred method for examining the HSR at the organismal level.

Figure 1: HSR induction measured with fluorescent reporters. (A) The basal and (B) heat-inducible expression of hsp-70p::gfp and hsp-16.2p::gfp reporter strains after 1 h of heat shock at 33 °C in a water bath or incubator. Worms were raised on OP50 bacteria for 64 h, heat shocked, and then recovered at 20 °C for 8 h before imaging. For reference, the no heat-shock worms in (A) were renormalized in (B) to match the range and saturation of the heat-shocked worms. Representative images of two experimental replicates are shown. Scale bar = 250 µm. Please click here to view a larger version of this figure.
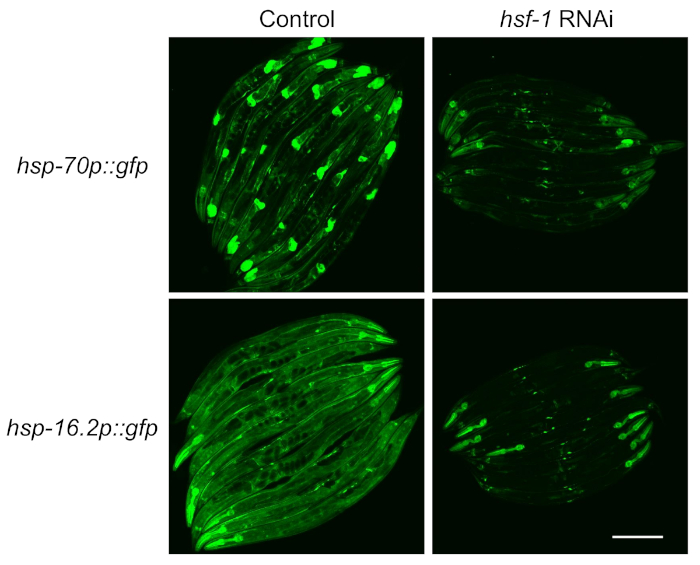
Figure 2: HSR induction measured with fluorescent reporters is dependent on HSF-1. Strains containing the hsp-70p::gfp and hsp-16.2p::gfp reporters were raised on control (L4440 empty vector) or hsf-1 RNAi plates for 64 h, exposed to a 1 h heat shock at 33 °C in a water bath, and then recovered at 20 °C for 8 h before imaging. Representative images of two experimental replicates are shown. Scale bar = 250 µm. Please click here to view a larger version of this figure.
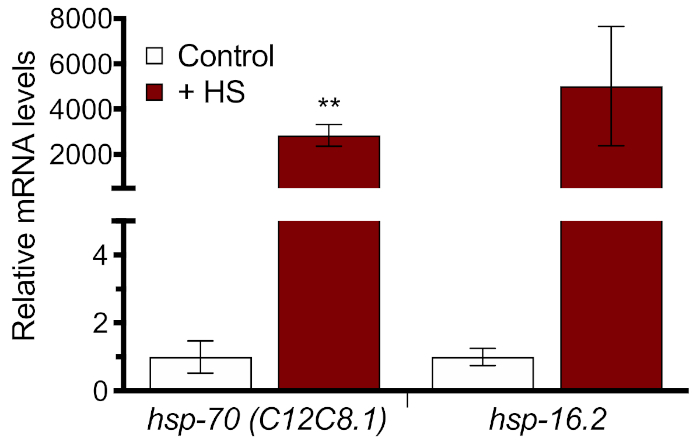
Figure 3: HSR induction measured with RT-qPCR. N2 worms were raised on HT115 bacteria for 60 h and then heat shocked for 1 h in a 33 °C water bath. The relative mRNA levels of hsp-70 (C12C8.1) and hsp-16.2 are shown normalized to the no heat-shock control. Values plotted are the mean of four biological replicates and error bars represent ± SEM. Statistical significance was calculated using an unpaired Student's t-test. **p < 0.01. Please click here to view a larger version of this figure.
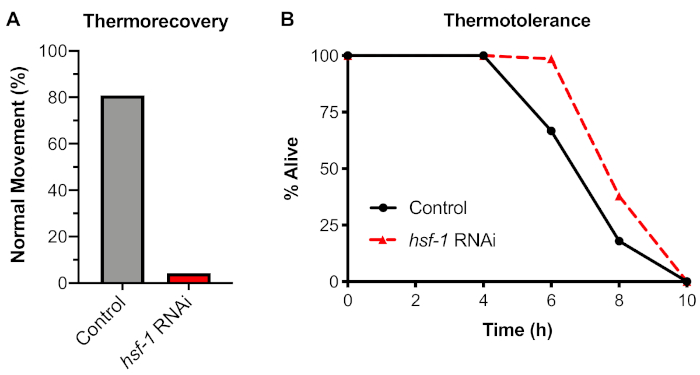
Figure 4: Thermorecovery, but not thermotolerance, is dependent on HSF-1. N2 worms were raised on control (L4440) or hsf-1 RNAi plates for 60 h and then shifted to either: (A) A 33 °C water bath for 6 h and recovered at 20 °C for 48 h before scoring for normal movement (thermorecovery), or (B) A 35 °C dry incubator and removed every 2 h until dead (thermotolerance). Each assay was done with n ≥ 30 individuals on 2 independent days. The average is shown. Please click here to view a larger version of this figure.
Discussion
In the literature a wide variety of temperatures, times, and equipment have been used to assay the HSR, which has introduced unnecessary caveats and led to difficulty in comparing results between laboratories. For example, temperatures ranging anywhere from 32-37 °C and times from 15 min to several hours have been used to induce the HSR15. However, it is reported that lethality occurs as early as 3 h at 37 °C for all stages and 1.5 h for day 1 adults15. Furthermore, we show that exposure of worms to 35 °C causes lethality that is not HSF-1 dependent, making these conditions poorly suited for analysis of the HSR. In contrast, a heat shock of 33 °C for 1 h is robust enough to elicit strong induction of heat shock genes, yet mild enough to not affect worm viability. Indeed, exposure to 33 °C for as long as 6 h only causes 20% of worms to display abnormal movement. Therefore, we propose using a temperature of 33 °C and a time of 1 h as a standardized condition for RT-qPCR and fluorescent reporter assays.
Recent experiments have revealed that developmental staging of worms for HSR experiments is particularly important. It was recently shown that in C. elegans the inducibility of the HSR declines (i.e., collapses) by >50% when hermaphrodites begin egg laying5. Staging the worms correctly is critical because there are often differences in developmental timing in strains carrying mutations. If temperature-sensitive mutants are used, this will also impact results if they are not synchronized by their reproductive age. Therefore, it is recommended to carefully measure the onset of egg laying for every strain to determine when the collapse occurs. The window of time after the L4 molt and before the initiation of reproductive maturity is narrow; therefore, care must be taken so that the HSR collapse does not inadvertently cause variability in results.
In addition to developmental timing, surprisingly small changes in temperature, as little as 1 °C, can have substantial effects on the HSR. For example, thermosensory neurons in C. elegans are sensitive to temperature changes as small as ±0.05 °C16. Thus, it is imperative to use a thermometer that can accurately measure the temperature. Therefore, we propose as best practice the use of a calibrated device for temperature measurement that is precise enough to measure temperatures within ±0.1 °C. Furthermore, a thermometer with a data-logging functionality should be used to measure temperature variations across time. Many incubators are specified to have thermal variations of more than 1 °C in different parts of the incubator and across time, which can have significant effects on HSR experiments. As a best practice, we suggest using incubators that have sufficient insulation and circulation to minimize temperature fluctuations. For conducting heat shock experiments, we propose a best practice of a circulating water bath. The time it takes for an agar plate to reach a desired temperature is approximately 6-7 min in a water bath but much longer in a dry incubator15,17. However, if a circulating water bath is not available, we have shown that robust HSR induction also occurs in a dry incubator using our conditions. If a dry incubator is used, opening of the incubator for the duration of the stress should be minimized.
It is well-established that induction of heat shock genes is dependent on the master regulator of the HSR, HSF-1. Here, we present evidence that the two more indirect assays, fluorescent reporters and thermorecovery, are also dependent on HSF-1. Significantly, we found that a commonly used alternative organismal assay, thermotolerance, is not HSF-1 dependent using hsf-1 RNAi (Figure 4). Similar results have been previously reported using an hsf-1 mutant or a ttx-3 mutant, which blocks the HSR18,19,20. Together, these results indicate that the thermotolerance assay should not be used for HSR research. Furthermore, this suggests that a best practice is to test the HSF-1 dependence for any assay used to measure the HSR.
Taken together, we present a series of standardized protocols and best practices for robust and reproducible measurement of HSR induction in C. elegans. We hope that these methodologies will decrease variability in HSR experiments and increase reproducibility. Facilitating direct comparisons of HSR research between laboratories will serve to accelerate research in the HSR field. Furthermore, standardization will benefit research into aging and neurodegenerative diseases with which the HSR is intimately associated.
Acknowledgements
This work was supported by a donation from Frank Leslie. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Materials
| Name | Company | Catalog Number | Comments |
| 18S-forward primer | TTGCGTCAACTGTGGTCGTG | ||
| 18S-reverse primer | CCAACAAAAAGAACCGAAGT CCTG | ||
| AM446 rmIs223[phsp70::gfp; pRF4(rol-6(su1006))] | Morimoto lab | http://groups.molbiosci.northwestern.edu/morimoto/ | |
| C12C8.1-forward primer | GTACTACGTACTCATGTGTCG GTATTT | ||
| C12C8.1-reverse primer | ACGGGCTTTCCTTGTTTTCC | ||
| CFX Connect Real-Time PCR Detection System | Bio Rad | 1855200 | |
| CL2070 dvIs70 [hsp-16.2p::GFP + rol-6(su1006)] | Caenorhabditis Genetics Center (CGC) | https://cgc.umn.edu/ | |
| EasyLog Thermistor Probe Data Logger with LCD | Lascar | EL-USB-TP-LCD | |
| Greenough Stereo Microscope S9i Series | Leica | ||
| Hard Shell 96 Well PCR Plates | Bio Rad | HSS9601 | |
| hsp-16.2-forward primer | ACTTTACCACTATTTCCGTCC AGC | ||
| hsp-16.2-reverse primer | CCTTGAACCGCTTCTTTCTTTG | ||
| iScript cDNA Synthesis Kit | Bio Rad | 1708891 | |
| iTaq Universal Sybr Green Super Mix | Bio Rad | 1725121 | |
| Laser Scanning Confocal Microscope | Nikon | Eclipse 90i | |
| MultiGene OptiMax Thermo Cycler | Labnet | TC9610 | |
| N2 (WT) | Caenorhabditis Genetics Center (CGC) | https://cgc.umn.edu/ | |
| Nanodrop Lite Spectrophotometer | Thermo Scientific | ND-LITE | |
| Parafilm M Roll | Bemis | 5259-04LC | |
| RapidOut DNA Removal Kit | Thermo Scientific | K2981 | |
| Recirculating Heated Water Bath | Lauda Brinkmann | RE-206 | |
| Traceable Platinum Ultra-Accurate Digital Thermometer | Fisher Scientific | 15-081-103 | |
| TRIzol Reagent | Invitrogen | 15596026 | RNA isolation reagent |
| TurboMix Attachment | Scientific Industries | SI-0564 | |
| Vortex-Genie 2 | Scientific Industries | SI-0236 |
References
- Guisbert, E., Morimoto, R. I. The regulation and function of the heat shock response. Protein Quality Control in Neurodegenerative Diseases. , 1-18 (2013).
- Li, J., Chauve, L., Phelps, G., Brielmann, R. M., Morimoto, R. I. E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response. Genes and Development. 30 (18), 2062-2075 (2016).
- Hsu, A. L., Murphy, C. T., Kenyon, C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 300 (5622), 1142-1145 (2003).
- Morley, J. F., Morimoto, R. I. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Molecular Biology of the Cell. 15 (2), 657-664 (2004).
- Labbadia, J., Morimoto, R. I. Repression of the Heat Shock Response Is a Programmed Event at the Onset of Reproduction. Molecular Cell. 59 (4), 639-650 (2015).
- Link, C. D., Cypser, J. R., Johnson, C. J., Johnson, T. E. Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress & Chaperones. 4 (4), 235 (1999).
- Guisbert, E., Czyz, D. M., Richter, K., McMullen, P. D., Morimoto, R. I. Identification of a Tissue-Selective Heat Shock Response Regulatory Network. PLoS Genetics. 9 (4), 1-12 (2013).
- Ma, J., et al. Cellular Proteomes Drive Tissue-Specific Regulation of the Heat Shock Response. G3: Genes, Genomes, Genetics. 7 (3), 1011-1018 (2017).
- Mendenhall, A. R., et al. Expression of a single-copy hsp-16.2 reporter predicts life span. Journals of Gerontology - Series A Biological Sciences and Medical Sciences. 67 (7), 726-733 (2012).
- Stiernagle, T. Maintenance of C. elegans. WormBook. (1999), 1-11 (2006).
- Avery, L., You, Y. J. C. elegans feeding. WormBook: The Online Review of C. elegans Biology. , 1-23 (2012).
- Livak, K. J., Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 25 (4), 402-408 (2001).
- Rea, S. L., Wu, D., Cypser, J. R., Vaupel, J. W., Johnson, T. E. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nature Genetics. 37 (8), 894-898 (2005).
- Shiu, P. K., Hunter, C. P. Early Developmental Exposure to dsRNA Is Critical for Initiating Efficient Nuclear RNAi in C. elegans. Cell Reports. 18 (12), 2969-2978 (2017).
- Zevian, S. C., Yanowitz, J. L. Methodological considerations for heat shock of the nematode Caenorhabditis elegans. Methods. 68 (3), 450-457 (2014).
- Clark, D. A., Biron, D., Sengupta, P., Samuel, A. D. T. The AFD sensory neurons encode multiple functions underlying thermotactic behavior in Caenorhabditis elegans. Journal of Neuroscience. 26 (28), 7444-7451 (2006).
- Prahlad, V., Cornelius, T., Morimoto, R. I. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 320 (5877), 811-814 (2008).
- McColl, G., et al. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metabolism. 12 (3), 260-272 (2010).
- Douglas, P. M., et al. Heterotypic Signals from Neural HSF-1 Separate Thermotolerance from Longevity. Cell Reports. 12 (7), 1196-1204 (2015).
- Kourtis, N., Nikoletopoulou, V., Tavernarakis, N. Small heat-shock proteins protect from heat-stroke-associated neurodegeneration. Nature. 490 (7419), 213-218 (2012).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved