A subscription to JoVE is required to view this content. Sign in or start your free trial.
Live-cell Imaging of Single-Cell Arrays (LISCA) - a Versatile Technique to Quantify Cellular Kinetics
In This Article
Summary
We present a method for the acquisition of fluorescence reporter time courses from single cells using micropatterned arrays. The protocol describes the preparation of single-cell arrays, the setup and operation of live-cell scanning time-lapse microscopy and an open-source image analysis tool for automated preselection, visual control and tracking of cell-integrated fluorescence time courses per adhesion site.
Abstract
Live-cell Imaging of Single-Cell Arrays (LISCA) is a versatile method to collect time courses of fluorescence signals from individual cells in high throughput. In general, the acquisition of single-cell time courses from cultured cells is hampered by cell motility and diversity of cell shapes. Adhesive micro-arrays standardize single-cell conditions and facilitate image analysis. LISCA combines single-cell microarrays with scanning time-lapse microscopy and automated image processing. Here, we describe the experimental steps of taking single-cell fluorescence time courses in a LISCA format. We transfect cells adherent to a micropatterned array using mRNA encoding for enhanced green fluorescent protein (eGFP) and monitor the eGFP expression kinetics of hundreds of cells in parallel via scanning time-lapse microscopy. The image data stacks are automatically processed by newly developed software that integrates fluorescence intensity over selected cell contours to generate single-cell fluorescence time courses. We demonstrate that eGFP expression time courses after mRNA transfection are well described by a simple kinetic translation model that reveals expression and degradation rates of mRNA. Further applications of LISCA for event time correlations of multiple markers in the context of signaling apoptosis are discussed.
Introduction
In recent years, the importance of single-cell experiments has become apparent. Data from single cells allow the investigation of cell-to-cell variability, the resolution of intracellular parameter correlations and the detection of cellular kinetics that remain hidden in ensemble measurements1,2,3. In order to investigate cellular kinetics of thousands of single cells in parallel, new approaches are needed that enable monitoring the cells under standardized conditions over a time period of several hours up to several days followed by a quantitative data analysis
Protocol
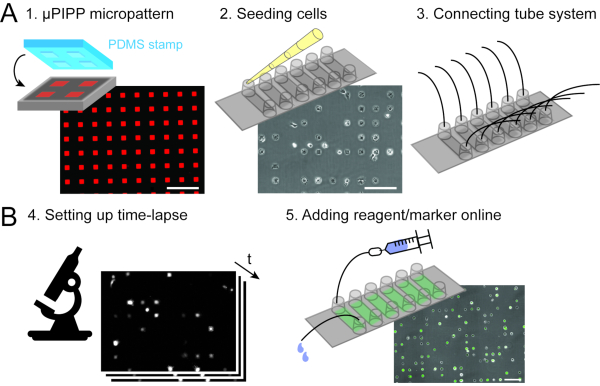
Figure 2: Data acquisition combining single-cell microarrays (A) with scanning time-lapse microscopy (B). As preparation of the time-lapse experiment, a single-cell array with a 2D micropattern of adhesion squares is prepared (1), followed by cell seeding and the alignment of the cells on the micropattern (2) as well as the connection of a perfusion system to the six-channel slide, which enables liquid handling during the time-lapse measurement (3). A scanning time-lapse experiment is set up ....
Results
The LISCA approach enables to efficiently collect fluorescence time courses from single cells. As a representative example we outline how the LISCA method is applied to measure single-cell eGFP expression after transfection. The data of the LISCA experiment is used to assess mRNA delivery kinetics, which is important for the development of efficient mRNA drugs.
In particular we demonstrate the different impact of two lipid-based mRNA delivery systems with respect to the time point of translati.......
Discussion
Here we described LISCA as a versatile technique to follow cellular kinetics of intracellular fluorescent labels at the single-cell level. In order to perform a successful LISCA experiment, each of the described steps of the protocol section must be established individually and then all steps must be combined. Each of the three major aspects of LISCA feature crucial steps.
Single-cell microarray fabrication
The quality of the microarray is crucial as the cellular alignment on th.......
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgements
This work was supported by grants from the German Science Foundation (DFG) to Collaborative Research Center (SFB) 1032. Support by the German Federal Ministry of Education, Research and Technology (BMBF) under the cooperative project 05K2018-2017-06716 Medisoft as well as a grant from the Bayerische Forschungsstiftung are gratefully acknowledged. Anita Reiser was supported by a DFG Fellowship through the Graduate School of Quantitative Biosciences Munich (QBM).
....Materials
| Name | Company | Catalog Number | Comments |
| Adtech Polymer Engineering PTFE Microtubing | Fisher Scientific | 10178071 | |
| baking oven | Binder | 9010-0190 | |
| CFI Plan Fluor DL 10x | Nikon | MRH20100 | |
| Desiccator | Roth | NX07.1 | |
| Eclipse Ti-E | Nikon | ||
| eGFP mRNA | Trilink | L-7601 | |
| Female Luer to Tube Connector | MEDNET | FTL210-6005 | |
| Fetal bovine serum | Thermo Fisher | 10270106 | |
| Fibronectin | Yo Proteins | 663 | |
| Filter set eGFP | AHF | F46-002 | |
| Fisherbrand Translucent Platinum-Cured Silicone Tubing | Fisher Scientific | 11768088 | |
| HEPES (1 M) | Thermo Fisher | 15630080 | |
| Incubation Box | Okolab | OKO-H201 | |
| incubator | Binder | 9040-0012 | |
| L-15 without phenol red | Thermo Fisher | 21083027 | |
| Lipofectamine 2000 | Thermo Fisher | 11668027 | |
| Male Luer | in-house fabricated consisting of teflon | ||
| Male Luer to Tube Connector | MEDNET | MTLS210-6005 | alternative to in-house fabricated male luers |
| NaCl (5 M) | Thermo Fisher | AM9760G | |
| Needleless Valve to Male Luer Connector | MEDNET | NVFMLLPC | |
| NIS Elements | Nikon | Imaging software Version 5.02.00 | |
| NOA81 | Thorlabs | NOA81 | Fast Curing Optical Adhesive for tube system assembly |
| Opti-MEM | Thermo Fisher | 31985062 | |
| PCO edge 4.2 M-USB-HQ-PCO | pco | ||
| Phosphate buffered saline (PBS) | in-house prepared | ||
| Plasma Cleaner | Diener Femto | Pico-BRS | |
| PLL(20 kDa)-g[3.5]-PEG(2 kDa) | SuSoS AG | ||
| silicon wafer mit mircorstructures | in-house fabricated | ||
| Sola Light Engine | Lumencor | ||
| sticky slide VI 0.4 | ibidi | 80608 | |
| Sylgard 184 Silicone Elastomer Kit | Dow Corning | 1673921 | |
| Tango 2 | Märzhäuser | 00-24-626-0000 | |
| Ultrapure water | in-house prepared | ||
| uncoated coverslips | ibidi | 10813 | |
| Injekt-F Solo, 1 mL | Omilab | 9166017V | with replacement sporn |
References
- Altschuler, S. J., Wu, L. F. Cellular heterogeneity: do differences make a difference. Cell. 141 (4), 559-563 (2010).
- Locke, J. C., Elowitz, M. B. Using movies to analyse gene circuit dynamics in single cells.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved