A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Development of Knock-Out Muscle Cell Lines using Lentivirus-Mediated CRISPR/Cas9 Gene Editing
In This Article
Summary
The protocol describes how to generate knock-out myoblasts using CRISPR/Cas9, starting from the design of guide-RNAs to the cellular cloning and characterization of the knock-out clones.
Abstract
One important application of clustered regulatory interspaced short palindromic repeats (CRISPR)/Cas 9 is the development of knock-out cell lines, specifically to study the function of new genes/proteins associated with a disease, identified during the genetic diagnosis. For the development of such cell lines, two major issues have to be untangled: insertion of the CRISPR tools (the Cas9 and the guide RNA) with high efficiency into the chosen cells, and restriction of the Cas9 activity to the specific deletion of the chosen gene. The protocol described here is dedicated to the insertion of the CRISPR tools in difficult to transfect cells, such as muscle cells. This protocol is based on the use of lentiviruses, produced with plasmids publicly available, for which all the cloning steps are described to target a gene of interest. The control of Cas9 activity has been performed using an adaptation of a previously described system called KamiCas9, in which the transduction of the cells with a lentivirus encoding a guide RNA targeting the Cas9 allows the progressive abolition of Cas9 expression. This protocol has been applied to the development of a RYR1-knock out human muscle cell line, which has been further characterized at the protein and functional level, to confirm the knockout of this important calcium channel involved in muscle intracellular calcium release and in excitation-contraction coupling. The procedure described here can easily be applied to other genes in muscle cells or in other difficult to transfect cells and produce valuable tools to study these genes in human cells.
Introduction
With the progress of gene sequencing and the identification of mutations in genes of unknown functions in a specific tissue, the development of relevant cellular models to understand the function of a new target gene and confirm its involvement in the related pathophysiological mechanisms constitutes an essential tool. In addition, these models are of major importance for future therapeutic developments1,2, and constitute an interesting alternative to the development of knock-out animal models in straight line with the international recommendations for reduction in the use of animals in experimentation. Gene editing using CRISPR/Cas9 is among the most powerful tools currently available, which has allowed the development of many knock-out/knock-in models, and targeted gene validation using CRISPR/Cas9 is among the most widely used application of CRISPR/Cas93. The success of gene editing relies on the ability to introduce the CRISPR tools (the guide RNAs and the nuclease Cas9) in the target cell model, which can be a challenge in many difficult-to-transfect cells, such as muscle cells4. This challenge can be overcome with the use of virus, usually lentivirus, which has the great advantage to transduce efficiently many cell types and deliver its transgene. But its major drawback is the integration of the transgene in the host cell genome, leading to potential alteration of genes localized at the integration site and to the permanent expression of the transgene, which in the case of the nuclease Cas9 would result in damaging consequences5. A smart solution has been proposed by Merienne and colleagues6, which consists of introduction into the cells of a guide-RNA targeting the Cas9 gene itself, leading to Cas9 inactivation. An adaptation of this strategy is presented here as a user friendly and versatile protocol allowing to knock-out virtually any gene in difficult-to-transfect cells.
The goal of the protocol presented here is to induce the inactivation of a gene of interest in immortalized muscle cells. It can be used to knock-out any gene of interest, in different types of immortalized cells. The protocol described here contains steps to design the guide RNAs and their cloning into lentiviral plasmids, to produce the CRISPR tools in lentiviral vectors, to transduce the cells with the different lentiviruses, and to clone the cells to produce a homogenous edited cell line.
Using this protocol, immortalized human skeletal muscle cells have been developed with deletion of the type 1 ryanodine receptor (RyR1), an essential calcium channel involved in intracellular calcium release and muscle contraction7. The knock-out (KO) of the gene has been confirmed at the protein level using Western blot, and at the functional level using calcium imaging.
Protocol
Muscle biopsies were obtained from the Bank of Tissues for Research (Myobank, a partner in the EU network EuroBioBank, Paris, France) in accordance with European recommendations and French legislation. Written informed consent was obtained from all individuals. Immortalized myoblasts were kindly produced by Dr. V. Mouly (Myology Institute, Paris, France), and the protocols were approved by Myology Institute ethic committee (MESRI, n AC-2019-3502).
1. CRISPR guide design
- Identify the region of the gene to be deleted. Search for its genomic sequence using genome browser tools such as ensembl.org or genome.ucsc.edu and determine the chromosomic coordinates of the two regions to look for the guide RNA (gRNA), on both sides of the region to be deleted.
- For the gene used here, obtain the FASTA sequence of the RYR1 gene and the sequence of the exon 101 as follows. In ensembl, search RYR1 in the most recent version of the human genome, select the first entry, and click on the transcript of the protein coding sequence. Then, click on Exons to redirect to the list of exons of the gene.
- Click on Download Sequence and select Only Genomic Sequence to download the full consensus sequence of the whole gene. Scroll down the list of the exons and introns of the gene and select the targeted one(s).
- Find the corresponding nucleotide sequence in the gene. Select the nucleotide sequences of the introns immediately upstream and downstream of the exon to be deleted, which will be used to search for the gRNAs.
- Design the two gRNA, called guide 1 and guide 2 here, in the regions identified in step 1.1 (introns upstream and downstream the region to be deleted) using online tools such as Crispor.tefor.net8. Choose the two gRNAs separated by a few hundred base-pairs (bp), the sequence of each gRNA being exactly 20 nucleotides long without the protospacer adjacent motif (PAM). Choose the best guides available in order to limit off-target. See Figure 1 for an example of guide design.
- The sequence between the two guides is expected to be deleted, and to result in the knock-out of the gene of interest, thus choose the position of the two guides such that it deletes an essential sequence or an essential exon in the gene of interest. Make sure that the deleted sequence/exon is not present exclusively in an alternative transcript of the gene and/or that it encodes an important part of the protein, thus its deletion will result in a functional knock-out.
NOTE: Although the Cas9 cleavage site is predicted to occur 3 bp upstream of the protospacer adjacent motif (PAM), cleavage at a larger distance can also occur, thus a good solution is not to depend on the precise localization of the cleavage site, such as cleavage in intron.
- The sequence between the two guides is expected to be deleted, and to result in the knock-out of the gene of interest, thus choose the position of the two guides such that it deletes an essential sequence or an essential exon in the gene of interest. Make sure that the deleted sequence/exon is not present exclusively in an alternative transcript of the gene and/or that it encodes an important part of the protein, thus its deletion will result in a functional knock-out.
- Determine the reverse complement (RC) sequence for each gRNA, without the PAM, in order to have the following sequences: Guide 1 and Guide 1-RC, Guide 2, and Guide 2-RC.
- Order the primers presented in Table 1 to perform cloning of the plasmids. Throughout the protocol, use primers at a concentration of 10 nM in sterile H2O.
NOTE: The sequences added to the gRNAs in these primers (bolded and underlined) correspond to the sequence of the plasmid before and after the guide, promotor, and trans-activating-Crispr RNA (tracrRNA), respectively and should not be modified in order to ensure a good overlap between the primers and the plasmid.
2. Plasmid cloning
NOTE: In this step, the gRNAs will be inserted in the plasmid backbone for lentivirus production. A cassette encoding the two gRNAs is first produced by successive polymerase chain reactions (PCR), using the overlapping primers. The new cassette is then inserted into the lentiviral backbone plasmid #87919.
- Obtain the following plasmids: plasmid #87919, coding for CRISPR guide RNAs in a lentiviral vector and plasmid #87904 coding for the SpCas9 sequence in a lentiviral vector.
- Cassette construction
NOTE: The cloning protocol is summarized in Figure 2.- Run a PCR (A) reaction, with 2 µL of plasmid #87919, 2 µL of primer_XmaIF, 2 µL of primer_Guide1R, 25 µL of polymerase mix, and 19 µL of H2O. Run the following PCR program (program 1): initial denaturation 5 min at 98 °C, followed by 30 cycles of: 30 s at 98 °C, 30 s at 60 °C, 1 min 45 s at 72 °C, and a final elongation of 7 min at 72 °C. The Tm of the primers described at step 1.4 is 60 °C.
NOTE: Although the elongation time in program 1 appears quite long, this elongation time has been chosen to ensure the production of enough material of the right size. Indeed, because of the repeated sequences in the plasmid (the sequence of tracrRNA after RNA guides is repeated three times in the plasmid #87919) the PCR amplification of the expected DNA is difficult, and additional smaller bands are produced in the successive PCRs. Thus, because of the competition between the different PCR products, either the elongation time has been increased (to favor the longest one and to have enough purified material at the end), or a touch down PCR (program 2) has been used for the long fragment (such as described for the PCR (final) in step 2.2.6). - Separate the PCR products on a 1% agarose gel in Tris-Borate-EDTA buffer (TBE), excise and purify the 300 bp fragment. Perform purification using a dedicated kit following the manufacturer's instruction, with elution in a final volume of 20 µL. Either use the purified fragment directly or store at -20 °C for step 2.2.5.
- Run a PCR (B) reaction, with 2 µL of plasmid #87919, 2 µL of primer_Guide1F, 2 µL of primer_Guide2R, 25 µL of polymerase mix, and 19 µL of H2O using the PCR program 1. Separate the PCR products on a 1% agarose gel in TBE, excise and purify the 400 bp fragment in 20 µL of elution buffer. Either use the purified fragment directly or store at -20 °C for step 2.2.5.
- Run a PCR (C) reaction, with 2 µL of plasmid #87919, 2 µL of primer_Guide2F, 2 µL of primer_BlpIR, 25 µL of polymerase mix, and 19 µL of H2O using PCR program 1. Separate the PCR products on a 1% agarose gel in TBE buffer. Excise and purify the 600 bp fragment in 20 µL of elution buffer. Either use the purified fragment directly or store at -20 °C for step 2.2.6.
NOTE: Another fragment at 900 bp may be visible, due to hybridization of the primer on the repeated regions in the plasmid, as described in the NOTE of step 2.2.1. If present, this band should be discarded. - Run a PCR (D) reaction, with 2 µL of elution PCR A (from step 2.2.2), 2 µL of elution PCR B (from step 2.2.3), 2 µL of primer_XmaIF, 2 µL of primer_Guide2R, 25 µL of polymerase mix, and 19 µL of H2O, with PCR program 1. Separate the PCR products on a 1% agarose gel in TBE buffer; excise and purify the 700 bp fragment in 20 µL of elution buffer. Either use the purified fragment directly or store at -20 °C for step 2.2.6.
NOTE: Other bands may be visible at >1,000 bp, 400 bp, and 300 bp, due to hybridization of the primers on the repeated regions and should be discarded. - Run a PCR (final) reaction, with 4 µL of elution PCR C (from step 2.2.4), 4 µL of elution PCR D (from step 2.2.5), 4 µL of primer_XmaIF, 4 µL of primer_BlpIR, 50 µL of polymerase mix, and 34 µL of H2O. The program used is the following (PCR program 2): initial denaturation for 5 min at 98 °C; six cycles of: 30 s at 98 °C, 30 s at 66 °C (reduction in hybridization temperature of 1 °C per cycle), 1 min 45 at 72 °C; 35 cycles of: 30 s at 98 °C, 30 s at 60 °C, 1 min 45 at 72 °C, and a final elongation of 5 min at 72 °C.
NOTE: The PCR program for this final amplification is a touch down PCR, different from the previous one, because of the large size of the final amplification which contains two repeated tracrRNA sequences just after each guide. - Separate the PCR products on a 1% agarose gel in TBE buffer; excise and purify the final cassette, which migrates at about 1,300 bp in 20 µL of elution buffer. Quantify the eluted product. Either use the purified fragment directly or store at -20 °C for step 2.3.2.1. This is the final product that will be inserted in the lentiviral plasmid.
NOTE: Other fragments may be visible at >1000 bp, 400 bp and 300 bp, corresponding to incomplete PCR fragments which should be discarded.
- Run a PCR (A) reaction, with 2 µL of plasmid #87919, 2 µL of primer_XmaIF, 2 µL of primer_Guide1R, 25 µL of polymerase mix, and 19 µL of H2O. Run the following PCR program (program 1): initial denaturation 5 min at 98 °C, followed by 30 cycles of: 30 s at 98 °C, 30 s at 60 °C, 1 min 45 s at 72 °C, and a final elongation of 7 min at 72 °C. The Tm of the primers described at step 1.4 is 60 °C.
- Insertion of the gRNAs cassette in the lentiviral plasmid backbone.
- Linearize the plasmid by double digestion of the plasmid #87919 with the restriction enzymes XmaI and BlpI.
- Prepare the reaction with 15 µL of recommended buffer, 15 µL of plasmid (1µg/µL), 7.5 µL of BlpI enzyme (at 10 U/µL), 7.5 µL of XmaI enzyme (at 10 U/µL), and 112.5 µL of H2O. Incubate for 1 h at 37 °C, and then for 20 min at 65 °C. Load the total amount on a 1% agarose gel, cut out and purify the ~10 kb plasmid with an appropriate kit. Elution is performed in 20 µL of buffer, and the eluted product is quantified using optical density measurement.
NOTE: Use an appropriate purification protocol for large DNA fragment, such as the protocol described by Sun and coll9.
- Prepare the reaction with 15 µL of recommended buffer, 15 µL of plasmid (1µg/µL), 7.5 µL of BlpI enzyme (at 10 U/µL), 7.5 µL of XmaI enzyme (at 10 U/µL), and 112.5 µL of H2O. Incubate for 1 h at 37 °C, and then for 20 min at 65 °C. Load the total amount on a 1% agarose gel, cut out and purify the ~10 kb plasmid with an appropriate kit. Elution is performed in 20 µL of buffer, and the eluted product is quantified using optical density measurement.
- Ligate the gRNA cassette and the plasmid. Prepare the reaction mix with gRNA cassette from step 2.2.7 and the linearized plasmid from step 2.3.1.1, add 2 µL of enzyme and H2O to have a final volume of 10 µL. Incubate for 15 min at 50 °C to produce the final plasmid called p_guides.
NOTE: The DNA cassette amount should be between 50-100 ng and plasmid amount between 100-200 ng, with a 2:1 molar ratio.
- Linearize the plasmid by double digestion of the plasmid #87919 with the restriction enzymes XmaI and BlpI.
- Use 2 µL of the freshly prepared plasmid to transform chemically competent E.Coli such as Stbl3 (50 µL) or XL10-Gold and spread on LB agar plate with 100 µg/mL ampicillin after 1 h growth at 37 °C without antibiotic. Incubate at 37 °C overnight. Pick a few colonies and perform a mini prep using a commercial kit following the manufacturer's instructions.
- Perform a PCR amplification on the miniprep DNA with Primer_XmaI and Primer_BlpR using PCR program 1 (see Figure 2). Separate the PCR products on a 1% agarose gel in TBE buffer. Select a few colonies (~5) with a band at the expected size of about 1300 bp.
- Perform DNA sequencing of the selected colonies, using the Primer_XmaI or Primer_BlpR, to confirm the correct insertion of the gRNA cassette.
NOTE: One of the sequence-verified colony is further used in the study, and the plasmid is called the p_guides. - Repeat from step 2.2 (cassette construction) with the Primers-Killer F and R, and the Primers-mCherry F and R. Use one sequence-verified colony for further analysis. The plasmid is called p_Killer.
3. Lentivirus production
- Produce and purify a large amount of all the required plasmids using an endotoxin free maxi-prep kit following the manufacturer's instructions. Prepare aliquots at 2 µg/µL. Store at -20 °C
- Preparation of cells (Day 1)
- Prepare 18 plates of 145 cm seeded with 1 x 106 HEK293 cells per plate in 16 mL of medium composed of Dulbecco's modified eagle medium (DMEM) high glucose pyruvate, supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Amplify the cells at 37 °C, in a 5% CO2 incubator for 3 days.
NOTE: Production of lentivirus must be performed with caution in a Biosafety Level 2 laboratory, using adapted protection gears, including disposable protective suit, protective cap, and gloves. All the experiments must be done under a laminar flow hood (BSLII safety cabinet) with filter tips. All the solution containing lentiviruses and all used plastic/glass waste must be inactivated with ethanol 70% or other virus inactivator.
- Prepare 18 plates of 145 cm seeded with 1 x 106 HEK293 cells per plate in 16 mL of medium composed of Dulbecco's modified eagle medium (DMEM) high glucose pyruvate, supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Amplify the cells at 37 °C, in a 5% CO2 incubator for 3 days.
- Transfection of cells (Day 4)
- Check the confluence of the cells, to be sure they have reached 60%-65% confluence.
- Prepare the transfection solution containing for each plate: 20.8 µg of the plasmid of interest (p-guides, or p-Killer, or pCas9 #87904), 4.8 µg of the plasmid encoding the envelope (VSV-G, #8454), 20.8 µg of the plasmid psPAX2 (#12260) for lentiviral packaging, 136 µL of calcium phosphate and adjust with H2O to a final volume of 1,000 µL. Add this solution dropwise and under agitation to 1 mL of 2x HEPES-buffered saline (HBS).
NOTE: Do not prepare a mix for all the plates at the same time to ensure an optimal preparation of reagents; prepare a mix for six plates at the same time, e.g., for 18 plates, prepare three times a mix for six plates. - Incubate at room temperature (RT) for at least 10 min and add 2 mL of solution dropwise to the cells. Homogenize the transfection reagent with gentle agitation of the plate backward, forward, up, and down, incubate at 37 °C, 5% CO2 for at least 5 h.
NOTE: From this point and until the end of the production of lentivirus, wear additional protection gear, including a second pair of gloves, protection sleeves, and a disposable plastron. - Five hours after the transfection, remove the medium from the plates and rinse with PBS to get rid of transfection reagents. Add 12 mL of fresh medium and incubate 48 h at 37 °C, 5% CO2.
- Collection of the viral particles (Day 6)
- Collect and pool the medium from all the plates. Centrifuge at 800 x g for 5 min at 4 °C, to pellet cellular debris. Filter the supernatant using a 0.45 µm filter (multiple filters required).
- Centrifuge at 68,300 x g for 2 h at 4 °C in a swinging bucket rotor. Remove the supernatant and let the tubes upside down under the safety cabinet on a paper for 5-10 min to remove as much liquid as possible, and then add 100 µL of HEK proliferation medium per pellet. After at least 2 h at 4 °C, resuspend the pellets by pipetting up and down. Pool all the resuspended pellets.
NOTE: Pellets can be left in the medium overnight at 4 °C before aliquoting. - Aliquot lentivirus in 10 µL or 25 µL sample size (depending on the use) and snap-freeze with liquid nitrogen. Store at -80 °C. Do not freeze an aliquot that has been thawed.
- Repeat steps 3.2 to 3.4 with other plasmids of interest in order to produce LV-guides, LV-Killer and LV-Cas9.
NOTE: As an alternative, the lentiviruses can be purchased from a company or a virus facility.
4. Lentivirus titration
NOTE: The virus titration is performed on HEK293 cells. The titration is important to incorporate in the subsequent steps a precise number of lentiviruses per cell (whatever the batch of lentivirus), for the cells of interest. The number of viral particles that efficiently transduce a cell is called the multiplicity of infection (MOI): MOI 10 thus correspond to the introduction of 10 viral particles per cell. As the freezing/thawing cycle affects the viability of the lentivirus, the titration is performed with a frozen lentivirus aliquot, and each subsequent experiment will be performed with a new aliquot of the same pool. One titration method is described here, but other methods can be used.
- On day 1, seed 1 x 105 cells per plate in five 35 mm plates with glass coverslips on the bottom and two 35 mm plates without coverslips.
- On day 2, from the two plates without coverslips, collect and count the number of cells after trypsinization, and determine the average amount of cells per plate (N).
- Prepare 100 µL of lentivirus diluted at 1/10 in proliferation medium. Transduce the five cultured plates with different volumes of diluted virus, from 1 to 50 µL of diluted virus. For this protocol, use the following volumes to transduce the five plates: 1 µL, 5 µL, 10 µL, 20 µL, 50 µL. Incubate for 48 h at 37 °C, in a 5% CO2.
- On day 4, fix the cells by incubation of the coverslips at RT for 20 to 30 min in 4% paraformaldehyde. For the LV-Cas9, permeabilize the cells with 0.1% Triton X100 in Phosphate Buffer Saline (PBS) for 10 min at RT, saturate in PBS-0.1% Triton X100-2% Goat Serum- 0.5% Bovine Serum Albumin (BSA) for 20 min at RT, and label for 45 min at RT with primary antibody anti-V5 (dilution 1/400) followed by 30 min incubation at RT with fluorescent secondary antibody. Label the nuclei with Hoescht (10 µg/mL in PBS) for 10 min at RT.
- Mount the coverslips on a slide and observe using a fluorescent microscope equipped with a 20x objective. For LV-guides and LV-Killer, after fixation proceed directly to the nuclei labeling and mount the coverslips. For each coverslip, count the total number of nuclei (total number of cells) in the field of view and the number of cells labeled (either with V5 or mCherry), and determine the ratio of cells labeled for each coverslip.
- Select a coverslip in which the ratio of transduced cells is at least 10% and not exceeding 50%. Determine the transduction efficiency (X) for this coverslip, and note the volume of diluted virus (V, in µL) used to obtain this transduction efficiency.
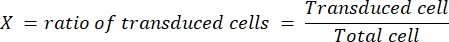
- Determine the titer (in infectious particles, represented as ip/mL) of the virus according to the following formula:

The dilution factor is the dilution of the lentivirus performed in step 4.3. We routinely obtained a final titer of 1 x 109 ip/mL for LV-Cas9 and 1 x 1010 ip/mL for LV-guide, determined in HEK cells.
5. Myoblast transduction
NOTE: The immortalized myoblasts are successively transduced with the three lentiviruses previously produced. They are maintained at a density below 50% in a proliferation medium composed of Ham's F10 supplemented with 20% FBS, 2% penicillin/streptomycin, 2% Ultroser G, and cultured at 37 °C, 5% CO2.
- Determine the volume of lentivirus needed to treat the chosen number of cells with MOI 10 for LV-Cas9 and LV-guides and MOI 20 for LV-killer, according to the following formula:
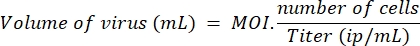
NOTE: In a parallel experiment, the transduction efficiency has been compared on myoblasts and HEK, using a control lentivirus (lenti-GFP), and we have determined that five times more lentiviruses are needed to transduce a myoblast efficiently compared to a HEK cell. Thus, MOI 10 measured on HEK corresponds to two viral particles per myoblast. The MOI used here are calculated on HEK cells. - On day 1, seed 96-well plates with 10,000 cells in 100 µL of proliferation medium per well. On day 2, transduce the cells under the safety cabinet by adding the appropriate volume of LV-guides and LV-Cas9 calculated in step 5.1. Return the cells to the incubator until day 7.
NOTE: The use of MOI higher than 25, especially for LV-Cas9, may not improve the number of edited cells due to higher cell death at a high concentration of lentivirus. - On day 7, perform trypsinization and count the cells. Seed the cells at a confluency of 40% to 50% in a new plate and return the cells to the incubator. Five hours later, transduce the cells with LV-Killer at a MOI of 20 (volume calculated in step 5.1). Amplify the cells for 5 to 10 days after transfection, for at least two passages, and always maintain them at a low confluency of <50%. The cells are ready for the next step, cellular cloning, when their growth is returned to normal (estimated by the division time).
NOTE: The proliferation may be a bit slower after the transduction. The normal cell growth can be determined before this experimental procedure by estimation of the division time of the cells.
6. Cellular cloning
NOTE: As myoblast transduction is difficult and never reaches 100% efficiency, even when using lentivirus, cellular cloning is required in order to get a fully corrected cell line. This is only possible with immortalized cells, or cells that can be cultured and amplified during few weeks/months.
- Trypsinize and count the cells. Dilute the cells in proliferation medium at 10 cells/mL and seed the cells at 1 cell/well in 96-well plates containing 100 µL/well of medium.
NOTE: The number of plates to be seeded depends on the probability of the expected gene editing, 2 to 10 plates are routinely used. - Monitor the cells for growth and progressively amplify each well in a larger plate until reaching at least a 35 mm plate, while maintaining the confluency of the myoblasts below 50%. This step can last 2-6 weeks, depending on the cells used and their ability to grow once isolated in a well.
7. Clone selection
NOTE: This step is performed to identify which of the growing clones have been appropriately modified.
- Design a set of primers composed of a primer located before the first guide (Primer_BeforeGuide1F) and another primer located after the second guide (Primer_AfterGuide2R) in order to amplify the region enclosing the putatively modified sequence. See Table 1 for the primers used here.
- Collect cells from each clone, spare at least 300,000 cells for future amplification, and extract genomic DNA using any standard protocol on the remaining cells.
- If a large number of clones are growing, to discard the non-edited ones, perform a rapid test by pooling cells from five clones in the same tube to extract the DNA from this pool and test with PCR. Repeat with as many clones as necessary. Then, further separate the pools which contained edited cells to perform individual analysis.
- Control the editing by PCR as follows. Prepare the PCR reaction with 1 µL of Primer_BeforeGuide1F, 1 µL of Primer_AfterGuide2R, 12.5 µL of polymerase mix, 3 µL of genomic DNA, and 7.5 µL of H2O. Amplify in a thermocycler according to the manufacturer's instructions and primers parameters. Run on a 1% agarose gel to identify the edited clones.
- Control the editing by sequencing as follows. Perform Sanger sequencing of the selected clones to confirm the deletion and to identify how the editing has been performed in each clone. Keep more than one edited clone to make sure that only the targeted gene has been modified and is responsible for the physiological effect observed and keep a non-edited clone that will be used as a control clone (CTRL) in the subsequent experiments.
- Expand the selected clones. Once the confluency of each clone has reached about 50%, trypsinize the cells and plate the cells in a larger dish, until enough cells have been produced to perform the biochemical and functional characterizations (usually more than 1 x 106 per clone), and store frozen aliquots of each clone for future use.
8. Characterization of edited clones
NOTE: Once a few clones have been picked and confirmed by DNA sequencing, the deletion of the targeted gene can be confirmed at the protein level using Western blot, and at the functional level if a functional cellular assay is available for this gene. In the case of RYR1-KO, as RyR1 is a calcium channel, the functional characterization has been performed using calcium imaging on cultured cells.
- Protein expression in edited clones
NOTE: RyR1 is expressed only in differentiated myotubes10. Its expression has been evaluated in myotubes using Western blot, to confirm the deletion at the protein level of RyR1, as well as the deletion of the Cas9 protein.- Plate 200,000 cells in proliferation medium (described above, step 5) on a surface of about 1.76 cm2 in a 35 mm plate coated with laminin (surface corresponding to a 200 µL drop of laminin at 10 mg/mL in PBS with calcium). Once the cells are stuck to the plate after being incubated for 2-3 h at 37 °C, 5% CO2, shift the culture medium to a differentiation medium composed of DMEM low glucose + 10% Horse serum + 1% penicillin/streptomycin, and return the cells to the incubator for 6 days.
- After 6 days of differentiation, collect and lyse the cells with 200 µL of RIPA supplemented with protease inhibitors. Determine protein concentration using the Folin Lowry method11.
- Load 15 µg of protein, after denaturation for 30 min at RT in Laemmli denaturation buffer, on a 5%-15% gradient acrylamide gel. After electrophoretic separation, transfer the proteins on Immobilon P at 0.8 V for 4 h11.
- After saturation of the membrane for 30 min at RT in PBS containing 0.1% Tween 20 and 5% Non-fat dry milk, incubate the membrane with the primary antibodies diluted in the same buffer for 2 h at RT or overnight at 4 °C, wash the membrane 5x for 5 min with PBS-0.1% Tween 20 and incubate the membrane with the secondary antibodies for 1 h at RT. The primary antibodies used are: antibodies against V5-tag (dilution: 1/5000) to detect Cas9, anti-GAPDH (dilution: 1/1000) as a loading control, anti-RyR1 antibody12,13 (dilution: 1/10.000), antibody against the alpha 1 subunit of DHPR (dilution: 1/1000) and antibody against the myosin heavy chain MF20 (dilution: 1/1000).
- Wash the membrane 5x for 5 min with PBS-0.1% Tween 20, dry the excess of liquid and add the chemiluminescent substrate. Proceed as recommended by the substrate provider to detect the chemiluminescent signal.
- Functional characterization of edited clones
NOTE: The function of RyR1 was assessed using calcium imaging in differentiated myotubes, produced from CTRL or KO clones14.- Plate 50,000 cells on a 0.2 cm2 surface in the center of 35 mm dishes coated with laminin (surface covered by a 50 µL laminin drop, at 10 mg/mL in PBS with calcium) and induce differentiation for 6 days as described in step 8.1.1. Prepare three plates for each stimulation, to have a biological triplicate.
- Load the myotubes with 50 µL of fluo 4-direct, diluted 1:1 in differentiation medium and incubated for 30 min at 37 °C. Rinse the cells twice with KREBS buffer supplemented with glucose at 1 mg/mL.
- Measure the fluorescence variations with an inverted fluorescent microscope or a confocal microscope using a 10x objective. Install the plate on the stage of the microscope and start the acquisition at 1 frame per second for 90 s.
- Remove the remaining KREBS and stimulate the cells at frame 25 by addition of 2 mL of KCl for membrane depolarization (140 mM final concentration) or 2 mL of 4 CmC (500 µM final concentration) for RyR1 direct stimulation. Ensure that at least 10 myotubes are present in the field recorded.
- Quantify the fluorescence variation in each myotube, using a dedicated software. Select for analysis at least 10 myotubes per dish (ideally 20-30 myotubes per dish), draw a line (or a Region of Interest (ROI)) on the long axis of each myotube and collect the fluorescence F along this line for all the frames.
- Determine the initial fluorescent value, F0, corresponding to frames 1 to 24. Plot the fluorescent variation (F-F0)/F0 as a function of time from 0 to 90 s. Repeat the experiment three times to obtain the fluorescence variation from at least 90 myotubes from three different cultures. Pool all the results for the 90 myotubes and calculate the mean ± SEM of (F-F0)/F0 at each time frame. Quantify the peak amplitude of calcium release for each stimulation and each clone.
Results
This protocol was applied to immortalized myoblasts from a healthy subject15 (so-called HM cells, for human myoblasts), in which the RyR1 has been previously characterized16, in order to knock out the RYR1 gene encoding the RyR1 protein. The design of the guides RNA was made to delete the sequence encompassing part of exon 101 and intron 101 of the gene. Deletion of part of exon 101 is foreseen to result in disruption of the reading frame. In addition, exon 101 enc...
Discussion
A major step on the way to the characterization of genes of unknown function involved in pathologies is the development of relevant cellular models to study the function of these genes. The use of gene editing using CRISPR/Cas9 is an exponentially growing field of research, and the development of knock-out models as presented here is among its most widely used applications. In this context, we propose here a versatile protocol to develop a human cell line knock-out in any gene of interest, allowing the characterization o...
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgements
This work was funded by grants from Association Française contre les myopathies (AFM-Téléthon) and from Auvergne-Rhône Alpes Région (AURA).
Materials
| Name | Company | Catalog Number | Comments |
| Anti-CACNA1S antibody | Sigma-Aldrich | HPA048892 | Primary antibody |
| Blp I | NE BioLabs | R0585S | Restriction enzyme |
| CalPhos Mammalian Transfection Kit | Takara | 631312 | Transfection kit |
| Easy blot anti Mouse IgG | GeneTex | GTX221667-01 | HRP secondary antibody |
| Easy blot anti Rabbit IgG | GeneTex | GTX221666 | HRP secondary antibody |
| Fluo-4 direct | Molecular Probes | F10472 | Calcium imaging |
| GAPDH(14C10) Rabbit mAb | Cell Signaling Technology | #2118 | Primary antibody |
| HindIII | Fermentas | ER0501 | Restriction enzyme |
| InFusion HD Precision Plus | Takara | 638920 | Ligation kit |
| MasterMix Phusion High Fidelity with GC | ThermoFisher Scientific | F532L | Mix for PCR reaction with High fidelity Taq polymerase and dNTPs |
| Myosin Heavy Chain antibody | DHSB | MF20 | Primary antibody |
| NucleoBond Xtra Maxi EF | Macherey-Nagel | REF 740424 | Maxipreparation kit for purification of plasmids |
| NucleoSpin Gel and PCR Clean-up | Macherey-Nagel | 740609 | DNA purification |
| NucleoSpin Tissue | Macherey-Nagel | 740952 | Kit for DNA extraction from cell |
| One Shot Stbl3 Chemically Competent E. coli | ThermoFisher Scientific | C737303 | Chemically competent cells |
| Plasmid #87904 | Addgene | 87904 | Lentiviral plasmid encoding the SpCas9 (for LV-Cas9) |
| Plasmid #87919 | Addgene | 87919 | Lentiviral backbone for insertion of cassette with guides (for LV-guide-target) |
| Plasmid #12260 | Addgene | 12260 | Lentiviral plasmid encoding lentiviral packaging GAG POL |
| Plasmid #8454 | Addgene | 8454 | Lentiviral plasmid encoding envelope protein for producing lentiviral and MuLV retroviral particles |
| V5 Tag Monoclonal Antibody | Invitrogene | R96025 | Primary antibody |
| XL10-Gold Ultracompetent Cells | Agilent | 200317 | Chemically competent cells |
| Xma I | NE BioLabs | R0180S | Restriction enzyme |
References
- Claussnitzer, M., Susztak, K. Gaining insight into metabolic diseases from human genetic discoveries. Trends in Genetics. 37 (12), 1081-1094 (2021).
- Fuster-García, C., García-Bohórquez, B., Rodríguez-Muñoz, A., Millán, J. M., García-García, G. Application of CRISPR tools for variant interpretation and disease modeling in inherited retinal dystrophies. Genes. 11 (5), 473 (2020).
- Modell, A. E., Lim, D., Nguyen, T. M., Sreekanth, V., Choudhary, A. CRISPR-based therapeutics: current challenges and future applications. Trends in Pharmacological Sciences. 43 (2), 151-161 (2022).
- Olson, E. N. Toward the correction of muscular dystrophy by gene editing. Proceedings of the National Academy of Sciences of the United States of America. 118 (22), (2021).
- Wu, X., Kriz, A. J., Sharp, P. A. Target specificity of the CRISPR-Cas9 system. Quantitative Biology. 2 (2), 59-70 (2014).
- Merienne, N., et al. The self-inactivating KamiCas9 system for the editing of CNS disease genes. Cell Reports. 20 (12), 2980-2991 (2017).
- Marty, I., Fauré, J. Excitation-contraction coupling alterations in myopathies. Journal of Neuromuscular Diseases. 3 (4), 443-453 (2016).
- Concordet, J. P., Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Research. 46, 242-245 (2018).
- Sun, Y., Sriramajayam, K., Luo, D., Liao, D. J. A quick, cost-free method of purification of DNA fragments from agarose gel. Journal of Cancer. 3, 93-95 (2012).
- Flucher, B. E., Conti, A., Takeshima, H., Sorrentino, V. Type 3 and type 1 ryanodine receptors are localized in triads of the same mammalian skeletal muscle fibers. The Journal of Cell Biology. 146 (3), 621-630 (1999).
- Hess, H. H., Lees, M. B., Derr, J. E. A linear Lowry--Folin assay for both water-soluble and sodium dodecyl sulfate-solubilized proteins. Analytical Biochemistry. 85 (1), 295-300 (1978).
- Garibaldi, M., et al. Dusty core disease' (DuCD): expanding morphological spectrum of RYR1 recessive myopathies. Acta Neuropathologica Communications. 7 (1), 3 (2019).
- Marty, I., et al. Biochemical evidence for a complex involving Dihydropyridine receptor and Ryanodine receptor in triad junctions of skeletal muscle. Proceedings of the National Academy of Sciences of the United States of America. 91 (6), 2270-2274 (1994).
- Oddoux, S., et al. Triadin deletion induces impaired skeletal muscle function. Journal of Biological Chemistry. 284 (50), 34918-34929 (2009).
- Mamchaoui, K., et al. Immortalized pathological human myoblasts: towards a universal tool for the study of neuromuscular disorders. Skeletal Muscle. 1, 34 (2011).
- Cacheux, M., et al. Functional characterization of a central core disease RyR1 mutation (p.Y4864H) associated with quantitative defect in RyR1 protein. Journal of Neuromuscular Diseases. 2 (4), 421-432 (2015).
- Luis, A. The old and the new: Prospects for non-integrating lentiviral vector technology. Viruses. 12 (10), 1103 (2020).
- Leenay, R. T., Beisel, C. L. Deciphering, communicating, and engineering the CRISPR PAM. Journal of Molecular Biology. 429 (2), 177-191 (2017).
- Salmon, P., Trono, D. Production and titration of lentiviral vectors. Current Protocols in Neurosciences. , (2006).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved