Simple and Fast Rolling Circle Amplification-Based Detection of Topoisomerase 1 Activity in Crude Biological Samples
In This Article
Summary
A protocol for the sensitive and quantitative detection of topoisomerase 1 activity using the rolling circle enhanced enzyme activity detection assay is described. The method allows detection of topoisomerase 1 activity from purified components or cell/tissue extracts. This protocol has wide-ranging applications in any field involving detection of enzymatic activity.
Abstract
Isothermal amplification-based techniques such as the rolling circle amplification have been successfully employed for the detection of nucleic acids, protein amounts, or other relevant molecules. These methods have shown to be substantial alternatives to PCR or ELISA for clinical and research applications. Moreover, the detection of protein amount (by Western blot or immunohistochemistry) is often insufficient to provide information for cancer diagnosis, whereas the measurement of enzyme activity represents a valuable biomarker. Measurement of enzyme activity also allows for the diagnosis and potential treatment of pathogen-borne diseases. In all eukaryotes, topoisomerases are the key DNA-binding enzymes involved in the control of the DNA topological state during important cellular processes and are among the important biomarkers for cancer prognosis and treatment.
Over the years, topoisomerases have been substantially investigated as a potential target of antiparasitic and anticancer drugs with libraries of natural and synthetic small-molecule compounds that are investigated every year. Here, the rolling circle amplification method, termed rolling circle enhanced enzyme activity detection (REEAD) assay that allows for the quantitative measurement of topoisomerase 1 (TOP1) activity in a simple, fast, and gel-free manner is presented.By cleaving and ligating a specially designed DNA substrate, TOP1 converts a DNA oligonucleotide into a closed circle, which becomes the template for rolling circle amplification, yielding ~103 tandem repeat rolling circle products. Depending on the nucleotide incorporation during the amplification, there is the possibility of different readout methods, from fluorescence to chemiluminescence to colorimetric. As each TOP1-mediated cleavage-ligation generates one closed DNA circle, the assay is highly sensitive and directly quantitative.
Introduction
Topoisomerases belong to the class of DNA-modifying enzymes and many of these have proven useful as biomarkers for human diseases1,2,3,4,5,6. TOP1 is involved in resolving the topological stress associated with cellular processes such as DNA replication, gene transcription, recombination, and chromosomal segregation7. TOP1 can resolve both negative and positive supercoils by a mechanism that involves the formation of a transient single-stranded break in the DNA8,9. After binding to DNA, TOP1 positions the active site tyrosine (Tyr723) to perform a nucleophilic attack on the phosphodiester backbone. A TOP1-DNA cleavage complex is then generated with the enzyme covalently attached to the 3'-end of the broken DNA strand. This releases torsional stress by allowing the 5'-end of the cleaved strand to revolve around the intact strand. Finally, the hydroxyl group of the 5'-end performs a nucleophilic attack on the 3'-phosphotyrosyl bond. As a result, TOP1 is released, and the DNA backbone is restored8.
Several assays have been developed to investigate the steps of the TOP1 catalytic cycle, including the relaxation assay10, the electrophoretic mobility shift assay (EMSA)11,12, the DNA suicide cleavage-ligation assays13,14, and the in vivo complex of enzymes (ICE) assay15. However, these assays have several limitations as they are dependent on gel electrophoresis, which requires DNA intercalating agents, or they require highly specialized training and equipment. Moreover, the assays require large amounts of purified TOP1 enzyme (ranging from 1 to 5 ng for the relaxation assay and 50 to 200 ng for the EMSA and the cleavage-ligation assay) or extracts from at least 106 cells to perform optimally. Therefore, a highly sensitive method that enables the specific detection of TOP1 activity in crude biological samples and at the single catalytic event level, called the REEAD assay, has been developed16.
Here, a protocol for the detection of TOP1 activity using the REEAD assay16 is presented. A schematic representation of the assay is depicted in Figure 1. A custom-designed silicone grid is attached to a functionalized slide to create a glass-slide multiwell setup, called wellmaker in the following (Figure 1A). This is followed by coupling of a 5'-amino modified primer to the functional NHS groups in the wells of the slide (Figure 1B). TOP1-mediated cleavage and ligation reaction converts a specific DNA substrate to a closed circle. The TOP1-specific substrate (Figure 1C) spontaneously folds into a dumbbell-shape containing a double-stranded stem and two single-stranded loops. One of the loops is complementary to the surface-anchored primer. The stem region contains a favored TOP1 cleavage site three bases upstream from the 3'-end and a 5'-hydroxyl overhang. Circularization of the substrate can be achieved using either cell/tissue extract (Figure 1C) or recombinant, purified TOP1 (Figure 1D).
When the substrate is cleaved by TOP1, the enzyme becomes transiently bound to the 3'-end and the three-base fragment diffuses, allowing the 5'-overhang to anneal to the substrate and facilitating TOP1-mediated ligation. The ligation results in circularization of the substrate and subsequently the dissociation of TOP1. The closed circle is hybridized to the surface-anchored primer (Figure 1E) and is used as a template for isothermal rolling circle amplification (RCA) mediated by the phi29 polymerase, which can perform RCA with strand displacement, yielding 103 tandem repeat products. During the RCA step, fluorescently labeled (Figure 1F) or biotin-coupled (Figure 1G) nucleotides can be incorporated17. Incorporation of fluorescently labeled nucleotides allows for detection of the RCPs either using a fluorescence microscope or a fluorescence scanner. Alternatively, a horseradish peroxidase (HRP)-coupled anti-biotin antibody (Figure 1H) can bind the biotinylated nucleotides, thus allowing for detection of the rolled circle products (RCPs) using either enhanced chemiluminescence (ECL) or by the conversion of 3,3',5,5'-tetramethylbenzidine (TMB) into a detectable color. The RCA follows a linear reaction kinetic, making the REEAD assay directly quantitative, as one RCP represents a single TOP1-mediated cleavage-ligation reaction. Here it is shown that this assay can be used to detect TOP1 activity as a purified recombinant enzyme or extracted from crude samples and as an anti-TOP1 drugs screening tool.
Protocol
NOTE: Find a list of buffer compositions, equipment, and other materials required in Table of Materials.
1. Cell culture
- Culture the preferred cells in suitable medium and grow them according to instructions. As an example, grow Caco2, colorectal adenocarcinoma-derived cells, in Minimal Essential Medium (MEM) supplemented with 20% fetal bovine serum (FBS), 1% non-essential amino acids (NEAA), 100 units/mL penicillin, and 100 mg/mL streptomycin. Maintain the cell cultures in a humidified incubator (5% CO2/95% air atmosphere at 37 °C). Plate the cells into tissue culture flasks and split every 3 days to maintain the cells at 70% confluency.
- Harvest the cells from an 70% confluent flask by trypsin treatment (0.25% trypsin, 0.02% EDTA solution) and wash with phosphate buffered saline (PBS).
- Resuspend the cell pellet in PBS to adjust the cell concentration to 0.5 × 106 cells/tube. Spin at 200 × g for 5 min and carefully aspirate the supernatant.
NOTE: Cells used for REEAD must be used fresh and kept on ice. The results obtained with Caco2 cells has been recently published17. The assay has been tested with a variety of cell lines, including colon, breast, lung, and cervical cancer derived cells18,19,20,21,22,23, but it can be used with all crude biological samples containing TOP1, such as tissues, blood, and saliva.
2. Preparation of functionalized slides
- Attach the custom-designed silicone isolator grid to the functionalized slide, thereby making the wellmaker. Press the silicone to avoid the formation of air bubbles (see Figure 1A).
- Prepare a mixture of 5 µM 5'-amino primer in 1x print buffer. Add 4 µL of the mixture to each well and put the wellamker in a hybridization chamber with saturated NaCl at temperatures between 15 °C and 25 °C, protected from light.
NOTE: A hybridization chamber can easily be made by using a pipette tip box filled with saturated NaCl. The hybridization takes a minimum of 16 h and a maximum of 72 h. See the sequence of the 5'-amino primer in Figure 1B. The slides are functionalized with NHS groups to allow for the binding of amino-modified oligonucleotides.
3. Generation of closed circular substrates
- Preparation of closed circular substrates with recombinant TOP1 or cell extract.
- Lyse a cell pellet of 0.5 × 106 cells in 500 µL of Lysis Buffer to reach a cell density of 1,000 cells/µL. Incubate for 10 min on ice.
- Prepare a circle mixture of 2 µL of 5 µM TOP1-specific substrate (the sequence of the substrate is as following: 5'-AGA AAA ATT TTT AAA AAA ACT GTG AAG ATC GCT TAT TTT TTT AAA AAT AAA TCT AAG TCT TTT AGA TCC CTC AAT GCA CAT GTT TGG CTC CGA TCT AAA AGA CTT AGA-3') and 2 µL of recombinant TOP1 enzyme or 2 µL of cell extract (see step 3.1.1) in 16 µL of 1x TOP1 Reaction Buffer.
NOTE: The concentration of recombinant TOP1 used depends on the readout method chosen (see Figure 2, Figure 3, Figure 4, and Figure 5). See the sequence of the TOP1-specific substrate in Figure 1C. - Incubate for 30 min at 37 °C (see Figure 1C,D).
- Stop the reaction by adding 2 µL of 1% SDS.
- Preparation of closed circular substrates in the presence of drugs
- Prepare a circle mixture of 2 µL of 5 µM TOP1-specific substrate and 1 µL of 100% DMSO or 1 µL of 1.6 mM camptothecin (CPT) in 14 µL of 1x TOP1 Reaction Buffer. Add 2 µL of 5 ng/µL recombinant TOP1 enzyme.
NOTE: Other small molecule compounds or drugs can be used. In that case, a titration of the compound should be made. If the compound is dissolved in a solvent other than DMSO, this should be used as a control instead of DMSO. - Incubate for 1 min at 37 °C.
- Stop the reaction by adding 2 µL of 1% SDS.
- Prepare a circle mixture of 2 µL of 5 µM TOP1-specific substrate and 1 µL of 100% DMSO or 1 µL of 1.6 mM camptothecin (CPT) in 14 µL of 1x TOP1 Reaction Buffer. Add 2 µL of 5 ng/µL recombinant TOP1 enzyme.
NOTE: Step 3 can be started in parallel with step 2, and the circles can be stored at 4 °C overnight. Alternatively, the DNA circles can be prepared at the same time as step 4 and used immediately. See the sequence of the TOP1-specific substrate in Figure 1C. The substrate folds into a dumbbell shape with a double stem region containing a preferred TOP1 cleavage site and two single-stranded loops. The open dumbbell substrate is converted into a closed circular substrate by TOP1-mediated cleavage and ligation and is hereafter referred to as a circle. As there is an excess of the 5'-amino primer, there will be no competition between open and closed TOP1-specific substrates.
4. Blocking of the wellmaker
- Immerse the wellmaker in a 5 cm x 5 cm tray filled with Buffer 1 that was preheated to 50 °C. By using a pipette, push the liquid inside the wells to make sure that there are no air bubbles. Incubate for 30 min at 50 °C.
- Remove Buffer 1 and wash 2 x 1 min with dH2O. Shake vigorously by hand.
- Add Buffer 2 that has been preheated to 50 °C. Make sure that there are no air bubbles in the wells. Incubate for 30 min at 50 °C.
- Wash 2 x 1 min with dH2O. Shake vigorously by hand.
- Wash with 70% EtOH for 1 min. Shake vigorously by hand.
- Let the wellmaker setup air-dry.
NOTE: Use compressed air to dry the wells. Alternatively, it is possible to blow air using a Pasteur pipette. Ensure that the wells are dry before proceeding.
5. Hybridization of the circles to the wellmaker
- Add 4 µL of the circles (made in step 3) to each corresponding well.
- Place the wellmaker in a humidity chamber for 1 h with dH2O at 37 °C.
NOTE: A humidity chamber can be made by using a box for pipette tips filled with dH2O. Alternatively, the hybridization can be done overnight at 25 °C in the humidity chamber.
6. Washing
- Wash the wellmaker with Buffer 3.
- Remove Buffer 3 and replace with Buffer 4.
- Remove Buffer 4 and wash with 70% EtOH.
- Remove the EtOH and let the wellmaker dry.
NOTE: All the washing steps are performed for 1 min in a 5 cm x 5 cm tray by fully submerging the slide. Always make sure that there are no air bubbles inside the wells.
7. Rolling circle amplification
- Prepare an RCA mixture of 1x Phi29 Reaction Buffer supplemented with 0.2 µg/µL BSA, 1 unit of Phi29 polymerase, and either 0.25 mM dNTP and 0.0125 mM ATTO-488-dUTP for fluorescence readout, or 0.1 mM dATP, 0.1 mM DTTP, 0.1 mM dGTP, 0.09 mM dCTP, and 0.01 mM Biotin-dCTP for colorimetric/chemiluminescence readout. Add 4 µL to each well. See Figure 1E.
NOTE: Preparation of the RCA mixture must be done on ice. When incorporating fluorescent nucleotides in the RCA, avoid direct light from this step to protect the fluorophores. - Incubate for 2 h at 37 °C in the humidity chamber. In case of fluorescent nucleotides used, set the humidity chamber in the dark. See Figure 1F,G.
8. Washing
- For the chemiluminescence or colorimetric readout protocols, wash the wellmaker as in step 6, and then proceed to step 11.
- For the fluorescence readout protocol, remove the silicone grid using tweezers before washing as in the following steps.
- Wash the slide for 10 min in Buffer 3.
- Remove Buffer 3 and replace with Buffer 4. Wash for 5 min.
- Remove Buffer 4 and wash for 1 min in 70% EtOH.
- Remove the EtOH and let the wellmaker dry.
9. Visualization of fluorescent rolling circle products using a fluorescence scanner
- Scan the slide in a fluorescence scanner using the filters corresponding to the fluorophore used. Use the maximum photomultiplier possible that does not give saturation.
NOTE: The fluorescent scanner used for image acquisition in this protocol is equipped with a 473 nm laser and a FAM filter-excitation 490 nm, emission 520 nm. - Import the picture into ImageJ and change the image type to 8-bit (Image | Type | 8-bit). To measure the bands separately, limit the measured area by using the rectangle drawing tool from the toolbar. Draw the desired area and measure the intensity (Analyze | Measure). Then, move the originally drawn area to the next band and measure.
- Plot the data in the desired software. Representative images including quantification extracted from ImageJ are depicted in Figure 2.
10. Visualization of fluorescent rolling circle products using a fluorescence microscope
- With an EtOH-resistant pen, draw the position of the wells before removing the silicone grid and wash the slide as in step 8.2. After the washing steps, mount the slide with 1 µL of mounting medium without 4',6-diamidino-2-phenylindole (DAPI) and add a cover glass. To allow for visualization in the camera, glue the slide onto a 76 mm x 26 mm microscope slide.
- Analyze using a fluorescence microscope equipped with a 60x oil immersion objective and a camera. Take 12-15 images of each sample/well.
- Import the pictures to ImageJ and stack the images (Image | Stacks | Image to stack). Change the image type to 8-bit (Image | Type | 8-bit).
- Set the threshold (Image | Adjust | Threshold).
- Set the threshold as follows: set the lower bar and adjust the upper bar so that only the right signals are red and the background is black. Ensure that the threshold is as low as possible before the real signals start to disappear.
- Sweep through the individual images and make sure that they correspond to the images before setting the threshold. Note that the threshold may change from experiment to experiment.
- Count the signals (Analyze | Analyze particles). Ensure that the summarized field in the ImageJ settings is checked.
- Export the results to a spreadsheet or other software for further data processing. Representative images including quantification extracted from ImageJ are depicted in Figure 3.
11. Coupling of HRP-conjugated anti-biotin antibody to the biotin-labeled rolling circle products
- Add 4 µL of HRP-conjugated anti-biotin antibody diluted 1:300 in Buffer 5 supplemented with 5% non-fat skimmed milk and 5% BSA to each well.
- Incubate for 50 min at 15-25 °C in the humidity chamber (see Figure 1H).
- Wash 3 x 3 min with Buffer 5.
- Dry the wellmaker.
12. Visualization of the biotin-labeled rolling circle products
- Visualization with ECL
- Mix 40 µL of ECL Luminol and 40 µL of H2O2 immediately before use. Add 2 µL to each well.
- Visualize the slide using a CCD camera or on X-ray films.
- Visualization with TMB
- After step 11.4, remove the silicone grid. Then, add 400 µL of TMB on top of the entire slide and put the slide in a humidity chamber. Wait for ~5-10 min for color development. After the color development, wash the slide with 70% EtOH.
- Visualize the color development with the naked eye. Take a photograph of the slide with a photo camera or mobile phone.
- Import the picture to ImageJ and change the image type to 8-bit (Image | Type | 8-bit). To measure the bands separately, limit the measured area by using the rectangle drawing tool from the toolbar. Draw the desired area and measure the intensity (Analyze | Measure). Then, move the originally drawn area to the next band and measure.
- Plot the data in the desired software. Representative images including quantification extracted from ImageJ are depicted in Figure 4, Figure 5, and Figure 6.
Representative Results
Here, a protocol for the detection of TOP1 activity using the REEAD assay is presented16. The protocol was used to detect recombinant TOP1 activity using four different readout methods: fluorescence scanner, fluorescence microscope, chemiluminescence, and colorimetric. The protocol was also used to detect the activity of TOP1 extracted from Caco2 cells, as an example of crude extract, with chemiluminescence or TMB readout. Moreover, the protocol was used as a drug screening tool, to detect the inhibition of recombinant TOP1 activity by CPT, as an example of a TOP1-specific inhibitor, using the chemiluminescence readout method.
The results obtained when analyzing the TOP1 activity using the fluorescence readout are depicted in Figure 2 and Figure 3. A representative scan of the fluorescent slide and the resulting quantification are shown in Figure 2. As evident from the quantification, this readout has a detection limit of 12.5 ng of TOP1. Representative images and the resulting quantification obtained when using the fluorescent microscope readout are shown in Figure 3. Using this readout method, the detection limit is as little as 0.1 ng of TOP1. The results obtained when analyzing the TOP1 activity using the chemiluminescence readout is depicted in Figure 4, and the results from the colorimetric readout are depicted in Figure 5. The detection limit of these readout methods is at 6 ng of TOP1 or as TOP1 extracted from 312 Caco2 cells. Figure 6 shows the measurements of the TOP1 activity in the presence of 80 µM CPT or DMSO as an example of drug screening application. The representative image and the resulting quantification of three independent experiments show that CPT inhibits TOP1-mediated circularization of the substrate as expected24,25.
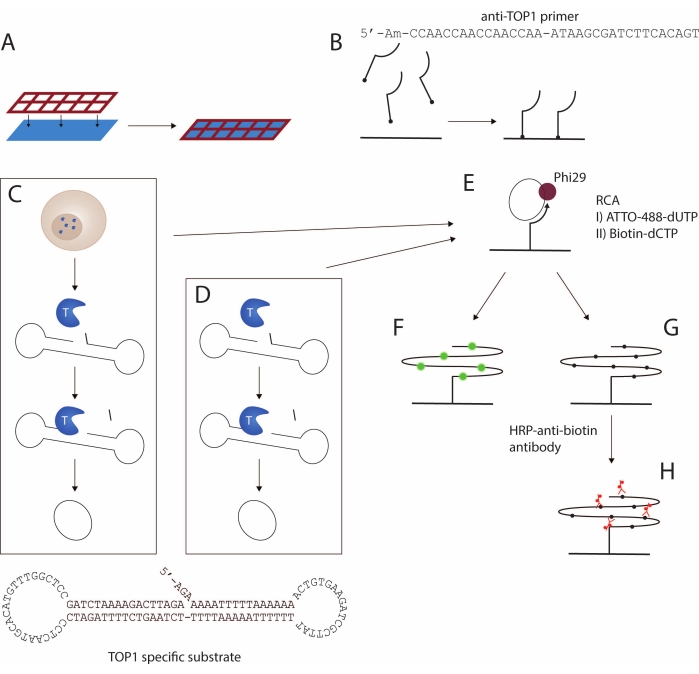
Figure 1: Schematic representation of the REEAD protocol. (A,B) Preparation of the functionalized slides. The silicone grid is attached to the functionalized slide. Then, the 5'-amino primer is coupled to the slide, thereby enabling amplification of the TOP1-specific substrate. (C,D) Generation of closed circular substrates. The TOP1-specific substrate folds into a dumbbell shape with a preferred TOP1 cleavage site in the double-stranded stem and a primer-binding sequence in one of the two single-stranded loops. (C) TOP1 is released upon lysis of the cells. Upon TOP1-mediated cleavage and ligation, the substrate is converted into a closed circle; (D) purified, recombinant TOP1 is used. (E) Rolling circle amplification. The closed circular substrates are hybridized to the surface-anchored primer and amplified by RCA using Phi29 polymerase. The RCA can be achieved either by incorporation of fluorescent nucleotides as in F or biotinylated nucleotides as in G. The fluorescent rolling circle products are visualized using either a fluorescence microscope or a fluorescence scanner. (H) Coupling of the HRP-conjugated anti-biotin antibody. The biotinylated rolling circle products are incubated with the HRP-conjugated anti-biotin antibody. The signal development is mediated by the HRP enzyme, and the signals are visualized either by ECL and detected using a CCD camera or by using TMB generating a colorimetric visualization. Abbreviations: REEAD = rolling circle enhanced enzyme activity detection; RCA = rolling circle amplification; TOP1 = topoisomerase 1; HRP = horseradish peroxidase; ECL = enhanced chemiluminescence; TMB = 3,3',5,5'-tetramethylbenzidine. Please click here to view a larger version of this figure.
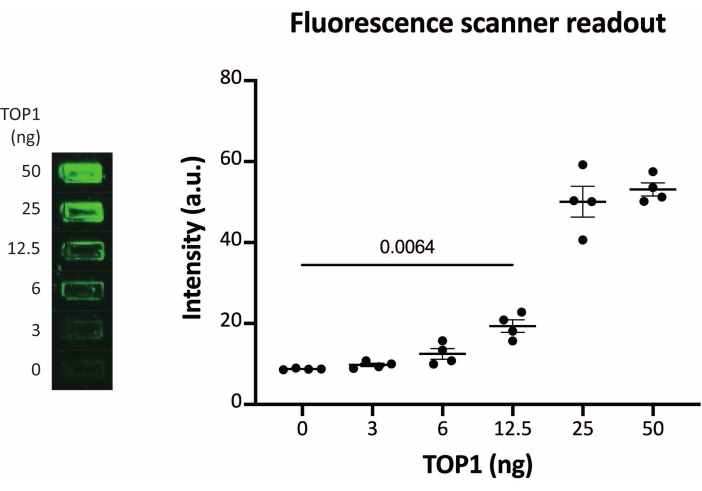
Figure 2: Measurements of TOP1 activity using the fluorescence scanner. Left panel shows a representative image of the slide when analyzing the activity of 3-50 ng of TOP1 using the fluorescence scanner readout protocol. Right panel shows the resulting quantification. t-test with Welch's correction, p = 0.0064, n = 4. Abbreviation: TOP1 = topoisomerase 1. Please click here to view a larger version of this figure.
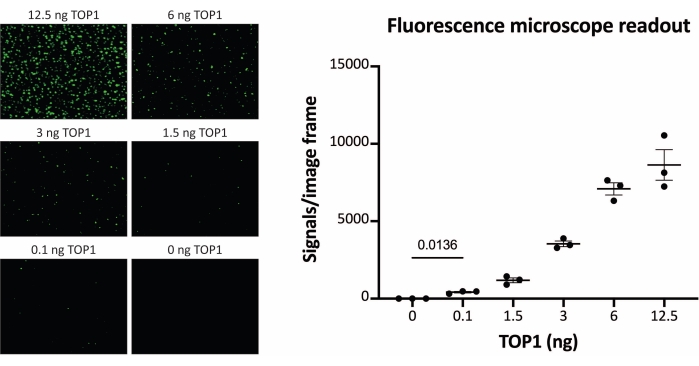
Figure 3: Measurements of TOP1 activity using the fluorescence microscope. Left panel shows representative images of the slide when analyzing the activity of 0.1-12.5 ng of TOP1 using the fluorescence microscope readout protocol. Note that the images are cropped versions to properly show the dots. For this reason, they do not resemble the number of the signals in the quantification shown in the right panel. t-test with Welch's correction, p = 0.0136, n = 3. Abbreviation: TOP1 = topoisomerase 1. Please click here to view a larger version of this figure.
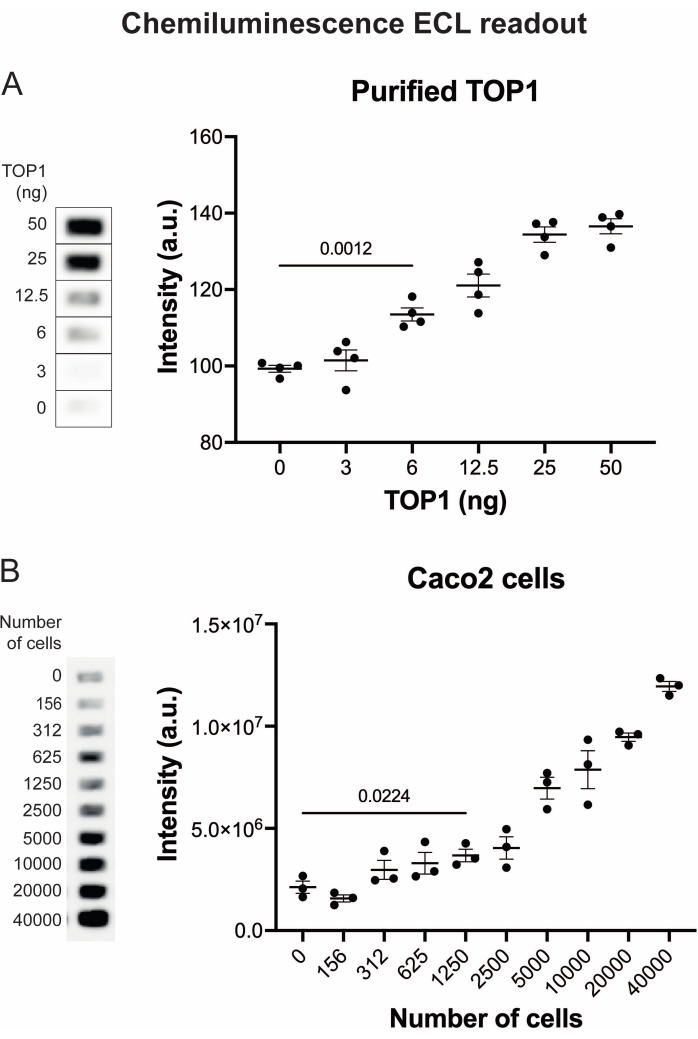
Figure 4: Measurements of TOP1 activity using the chemiluminescence readout. (A) Left panel shows a representative image of the slide when analyzing the activity of 3-50 ng of TOP1 using the chemiluminescence readout protocol. Right panel shows the resulting quantification. t-test with Welch's correction, p < 0.0001, n = 4. (B) Left panel shows a representative image of the slide when analyzing the activity of TOP1 extracted from 156-40,000 Caco2 cells using the chemiluminescence readout protocol. Right panel shows the resulting quantification. t-test with Welch's correction, p = 0.0224, n = 3. Abbreviations: TOP1 = topoisomerase 1. Please click here to view a larger version of this figure.
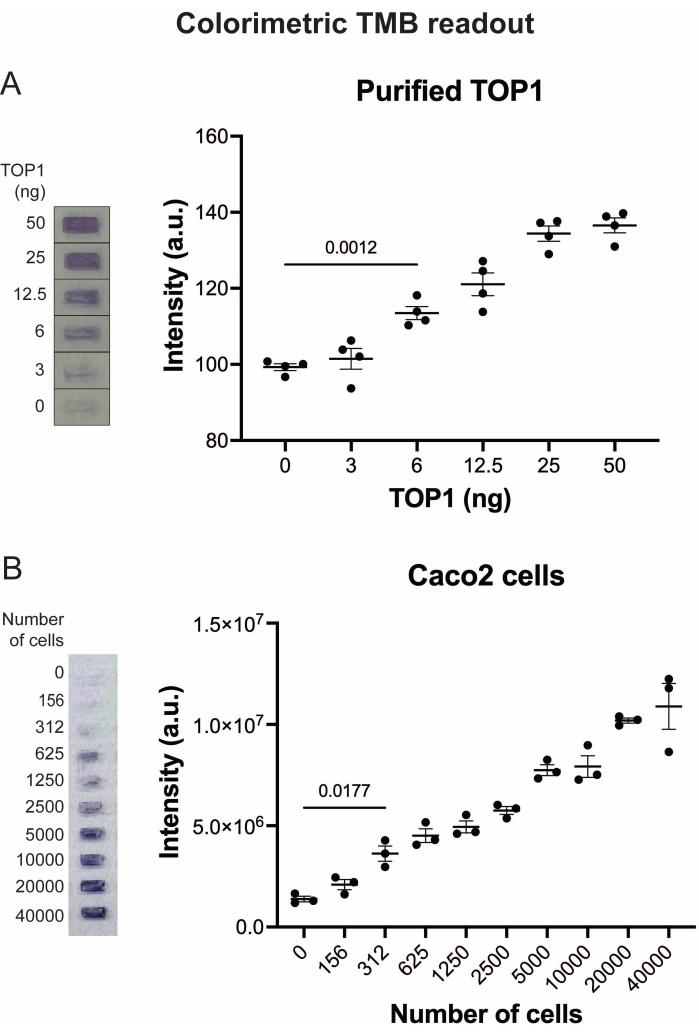
Figure 5: Measurements of TOP1 activity using the colorimetric readout. (A) Left panel shows a representative image of the slide when analyzing 3-50 ng of TOP1 using the colorimetric readout protocol. Right panel shows the resulting quantification. t-test with Welch's correction, p = 0.0012, n = 4. (B) Left panel shows a representative image of the slide when analyzing the activity of TOP1 extracted from 156-40,000 Caco2 cells using the colorimetric readout protocol. Right panel shows the resulting quantification. t-test with Welch's correction, p = 0.0117, n = 3. Abbreviations: TOP1 = topoisomerase 1; TMB = 3,3',5,5'-tetramethylbenzidine. Please click here to view a larger version of this figure.
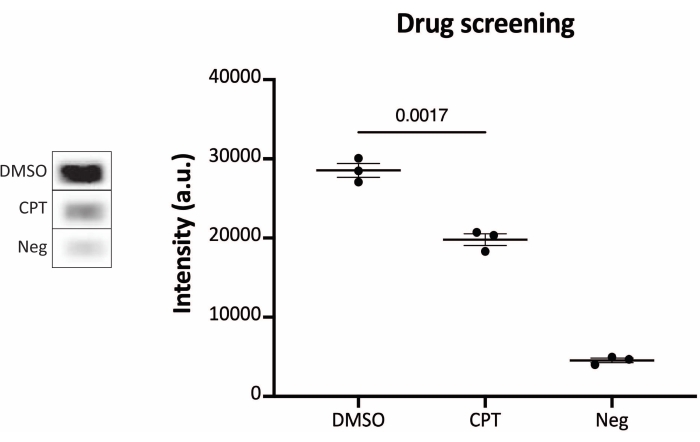
Figure 6: Measurements of TOP1 activity after drug treatment using the chemiluminescence readout protocols. Left panel shows a representative image of the slide when analyzing 10 ng of TOP1 in the presence of 5% DMSO or 80 µM CPT for 1 min. Right panel shows the resulting quantification. t-test with Welch's correction, p = 0.0017, n = 3. Abbreviations: TOP1 = topoisomerase 1; CPT = camptothecin; DMSO = dimethylsulfoxide; Neg = negative control. Please click here to view a larger version of this figure.
Discussion
Topoisomerases represent a class of DNA-modifying enzymes that attract high research and clinical interest, being the target of small-molecule compounds with potential effect in anticancer treatment or in the combat of infectious diseases. Moreover, the activity of human TOP1 has proven to be an effective biomarker of cancer prognosis and treatment18,19,23. Although it is the TOP1 activity that influences the efficacy of a chemotherapeutic inhibitor26, it is frequently the quantity of DNA-RNA or the amount of TOP1 that is assessed in clinical settings. This is due to the lack of fast, easy, and gel-based free tools that can provide accurate and precise quantification of topoisomerase activity in every laboratory settings.
Here, a protocol for the REEAD assay allowing the measurement of TOP1 activity in a gel-free manner is described. In this protocol, purified TOP1 or crude extract from cells is incubated with a specially designed DNA dumbbell-shaped substrate which, upon cleavage/ligation mediated by TOP1 is converted into a closed, circular molecule. The circles are then hybridized onto a glass surface and amplified by RCA. To allow for ease-of-use in the performing of reactions happening on the slide, a silicone grid is attached to the glass, making individual wells where the reactions take place. In this way, the protocol takes advantage of a multiwell system-a silicone grid-called wellmaker.
The protocol was used to detect the activity of recombinant TOP1 and TOP1 extracted from colorectal adenocarcinoma cells Caco2, used as an example. Moreover, as an example of REEAD to be used as a drug screening tool, the protocol was used to detect TOP1 activity inhibition by the well-known TOP1 inhibitor CPT24,25. The assay was coupled with different readout methods-fluorescence microscopy, which gives a high sensitivity, and a fluorescence scanner, chemiluminescence, or colorimetric, which require less specialized equipment and training. The highly sensitive fluorescence microscope readout has the limitations of the need for a good-quality fluorescence microscope setting, skilled personnel, and time-consuming image acquisition and analysis.
For these reasons, the fluorescence scanner readout that allows faster acquisition and analysis even if at the expense of the sensitivity is presented. In case of the lack of a fluorescence scanner, two excellent alternatives, chemiluminescence and colorimetric readout methods, can be considered. Both methods are fast and simple, requiring no expensive equipment or specialized training. In all the readout formats, REEAD has many advantages compared to the state-of-the-art assays, which are more time-consuming, gel-based (with the requirements of intercalating agents), and less directly quantitative. However, there are a few critical steps in the presented protocol. When handling the drug screening, the time points, the drug concentration, and the ratio of TOP1 amount/DNA substrate should be optimized compared to the described settings optimized for CPT. The drug can be investigated performing a preincubation with the DNA substrate or a preincubation with the TOP1 enzyme. This can give valuable information about the ability of the molecule to inhibit TOP1 and offer insight of the drug mechanism of action.
Moreover, if experiencing low or absent signals, this is most likely due to an inefficient lysis of the crude sample or a degradation of TOP1 in the extract due to improper use of protease inhibitors. In addition, this can also be due to inadequate storage of the important assay components, such as recombinant TOP1, phi29 polymerase, nucleotides, substrate, and primer. Finally, repeated freeze-thaw cycles of the oligonucleotides should be avoided, as this dramatically affects the assay performance. When crude extract is used, the extraction efficiency of the specific biological sample needs to be optimized and the amount and activity of TOP1 may vary from the example reported here. For this reason, every crude extract to be tested will require a titration for the identification of the assay detection limit and range of sensitivity when using that particular sample. The protocol has been optimized for the detection of human TOP1. The activity of other eukaryotic TOP1s can be measured, but the NaCl concentration and incubation time might need to be optimized according to the enzyme purification method and optimal enzyme activity. More circularized substrates will result from prolonged incubation, whereas fewer circularized substrates will result from shortened incubation.
In addition to the presented applications, REEAD allows for the measurement of the TOP1 activity in crude extracts from small biopsies from cancer patients18, the prediction of the cytotoxic anticancer effect of CPT in cancer cell lines20,21,22, and even the detection of the enzyme activity in single cells20,21. Moreover, the presented REEAD setting using the wellmaker enables multiwell-based drug screening of libraries of synthetic or natural compounds. In addition to this, a modified version of the REEAD assay, called REEAD C/L, has been developed. This setup enables separate investigations of the cleavage and ligation steps of the TOP1 reaction27. With the REEAD C/L, it is possible to determine the mechanism of action of small molecule inhibitors and characterize them as TOP1 catalytic inhibitors with potential antiparasitic effects28 or as TOP1 poisons to be used for anticancer treatment24,25. Finally, with specific redesign of the DNA substrates to match the different requirements (for DNA binding or cleavage-ligation) of other TOP1 enzymes, REEAD has also been used for the detection of infectious diseases (such as in the case of Plasmodium falciparum, causing malaria29), or Leishmania donovani and Monkeypox virus (data not shown). Or, it can be used to identify small molecule compounds with antipathogenic effects. The presented protocol provides scientists in the TOP1 field an easy method to detect enzyme activity with little to no optimization and with the possibility to be adapted to even wider applications in the future.
Disclosures
The authors C.T. and K.M. are employees of VPCIR biosciences ApS. C.T., B.R.K., and M.S. are shareholders and/or share option holders. C.T., B.R.K., and M.S. declare that they are named inventors of the patent EP2022/057172 filed in the name of VPCIR biosciences ApS. The other authors declare that they have no competing interests.
Acknowledgements
The authors would like to thank laboratory technician Noriko Y. Hansen, Department of Molecular Biology and Genetics, Aarhus University, for technical assistance in relation to Phi29 and TOP1 enzyme purifications.
Materials
| Name | Company | Catalog Number | Comments |
| Equipment | |||
| Camera for fluorescence image analysis | |||
| CCD camera | For chemiluninescence image analysis | ||
| Fluorescence microscope | Olympus | Equipped with 60x oil immersion objective and a GFP filter ( https://www.edmundoptics.com/p/gfp-filter-cube-set-olympus/21527/) | |
| Fluorescence scanner | Typhoon | FLA 9500 | Equipped with 473 laser and FAM filter |
| Humidity chamber with dH2O | |||
| Hybridization chamber with saturated NaCl | |||
| ImageJ software | Fiji | Download at: https://imagej.net/software/fiji/downloads | |
| Materials | |||
| Cell culture | |||
| Cell growth medium appropiate for the chosen cell line | |||
| Trypsin | Sigma | #T4174 | |
| Pen strep | Sigma | #P4300 | |
| Tissue culture flasks | ThermoFisher | #178983 | |
| FBS | Gibco | #10270106 | |
| NEEA | Sigma | #M7145 | |
| PBS | ThermoFisher | #10010023 | |
| Functionalized slides | |||
| 5'-amine anti-TOP1 primer | Sigma | 5’-/5AmMC6/CCA ACC AAC CAA CCA AAT AAG-3’ | |
| Activated Codelink HD slides | SurModics | #DHD1-0023 | |
| Silicon grid | Grace-biolabs | Custom made | |
| Pertex glue | Histolabs | #00801 | |
| Microscope slide 76 x 26 mm | Hounisen | #2510 1201BL | Other microscope slide of same dimension can be used |
| DNA circles and rolling circle amplification | |||
| TOP1 specific substrate | Sigma | 5’-AGA AAA ATT TTT AAA AAA ACT GTG AAG ATC GCT TAT TTT TTT AAA AAT AAA TCT AAG TCT TTT AGA TCC CTC AAT GCA CAT GTT TGG CTC CGA TCT AAA AGA CTT AGA-3’ | |
| ATTO-488-dUTP | Jena Biosciences | #95387 | |
| Biotin-dCTP | Jena Biosciences | #NU-809-BIO16L | |
| dNTP | ThermoFisher | #R0181 | |
| Phi29 polymerase | VPCIR Biosciences | #10010041 | |
| Recombinant TOP1 | VPCIR Biosciences | #10010001 | |
| Detection of rolling circle products | |||
| BioFX TMB | SurModics | #ESPM-0100-01 | |
| Cover glass | |||
| ECL mixture | Cytiva | #RPN2236 | |
| HRP conjugated anti-biotin antibody | Merck | #A4541 | |
| Mounting medium (vectachield) | Vector laboratories | #H-1000 | |
| Buffer list | |||
| 1x PE Buffer | 1 mM EDTA, 8.12 mM Na2HPO4·2H2O, 1.88 mM NaH2PO4·H2O | ||
| 6x Print Buffer | 300 mM Na3PO4, pH 8.5 | ||
| Blocking solution | Buffer 5 supplemented with 5% non-fat milk and 5% BSA pH 9 | ||
| Lysis Buffer | 10 mM Tris-HCl pH 7.5, 1 mM EDTA + protease inhibitors | ||
| 10x TOP1 Reaction Buffer | 50 mM CaCl2, 50 mM MgCl2, 100 mM Tris-HCl pH 7.5 | ||
| 10x Phi29 Reaction Buffer | 330 mM Tris-acetate pH 7.5, 100 mM Mg-acetate, 660 mM K-acetate, 1% Tween20, 200 mM DTT | ||
| Buffer 1 | 50 mM Tris, 50 mM Tris-HCl, 32 mM ethanolamine. NB: stored at 50 °C. | ||
| Buffer 2 | 4x SSC, 0.1% SDS. NB: stored at 50 °C. | ||
| Buffer 3 | 100 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.3% SDS | ||
| Buffer 4 | 100 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% Tween20 | ||
| Buffer 5 | 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% Tween20 | ||
| Chemicals for buffers | |||
| Tris | Sigma | #T1506 | |
| HCl | Sigma | #1.00317.2011 | |
| EDTA | Sigma | #1.08418.1000 | |
| PMSF | Sigma | #05056489001 | |
| SDS | Applichem | #436143 | |
| Na2HPO4 | Sigma | #1.06580.1000 | |
| NaH2PO4 | Sigma | #1.06346.1000 | |
| Ethanolamine | Sigma | #411000 | |
| SSC | Invitrogen | #15557-036 | |
| Tris-acetate | Sigma | #T1258 | |
| Mg-acetate | Sigma | #M0631 | |
| K-acetate | Sigma | #1.04820.1000 | |
| Tween20 | Sigma | #8.22184.0500 | |
| DTT | Sigma | #D0632 | |
| CaCl2 | Sigma | #1.02382.1000 | |
| MgCl2 | Sigma | #M2670 | |
| NaCl | Sigma | #1.06404.5000 | |
| Skimmed milk powder | Sigma | #70166 | |
| BSA | Sigma | #A4503 |
References
- Baldwin, E. L., Osheroff, N. Etoposide, topoisomerase II and cancer. Current Medicinal Chemistry-Anti-Cancer Agents. 5 (4), 363-372 (2005).
- Gilbert, D. C., Chalmers, A. J., El-Khamisy, S. F. Topoisomerase I inhibition in colorectal cancer: Biomarkers and therapeutic targets. British Journal of Cancer. 106 (1), 18-24 (2012).
- Ikeguchi, M., et al. TopoisomeraseI expression in tumors as a biological marker for CPT-11 chemosensitivity in patients with colorectal cancer. Surgery Today. 41 (9), 1196-1199 (2011).
- Meisenberg, C., et al. Clinical and cellular roles for TDP1 and TOP1 in modulating colorectal cancer response to irinotecan. Molecular Cancer Therapeutics. 14 (2), 575-585 (2015).
- Palshof, J. A., et al. Topoisomerase I copy number alterations as biomarker for irinotecan efficacy in metastatic colorectal cancer. BMC Cancer. 17 (1), 1-10 (2017).
- Proszek, J., et al. Topoisomerase I as a biomarker: Detection of activity at the single molecule level. Sensors. 14 (1), 1195-1207 (2014).
- Leppard, J. B., Champoux, J. J. Human DNA topoisomerase I: Relaxation, roles, and damage control. Chromosoma. 114 (2), 75-85 (2005).
- Champoux, J. J. DNA topoisomerases: structure, function, and mechanism. Annual Review of Biochemistry. 70 (1), 369-413 (2001).
- Redinbo, M. R., et al. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 279 (5356), 1504-1513 (1998).
- Nitiss, J. L., Soans, E., Rogojina, A., Seth, A., Mishina, M. Topoisomerase assays. Current Protocols in Pharmacology. 57 (1), 3 (2012).
- Tesauro, C., et al. Erybraedin C, a natural compound from the plant Bituminaria bituminosa, inhibits both the cleavage and religation activities of human topoisomerase I. Biochemical Journal. 425 (3), 531-539 (2010).
- Keller, J. G., et al. Topoisomerase 1 inhibits MYC promoter activity by inducing G-quadruplex formation. Nucleic Acids Research. 50 (11), 6332-6342 (2022).
- Christiansen, K., Westergaard, O. Characterization of intra- and intermolecular DNA ligation mediated by eukaryotic topoisomerase I. Role of bipartite DNA interaction in the ligation process. Journal of Biological Chemistry. 269 (1), 721-729 (1994).
- Svejstrup, J. Q., Christiansen, K., Andersen, A. H., Lund, K., Westergaard, O. Minimal DNA duplex requirements for topoisomerase I-mediated cleavage in vitro. Journal of Biological Chemistry. 265 (1), 12529-12535 (1990).
- Anand, J., Sun, Y., Zhao, Y., Nitiss, K. C., Nitiss, J. L. Detection of topoisomerase covalent complexes in eukaryotic cells. Methods in Molecular Biology. , 283-299 (2018).
- Stougaard, M., et al. Single-molecule detection of human topoisomerase I cleavage-ligation activity. ACS Nano. 3 (1), 223-233 (2009).
- Keller, J. G., Petersen, K. V., Knudsen, B. R., Tesauro, C. Simple and fast DNA-based tool to investigate topoisomerase 1 activity, a biomarker for drug susceptibility in colorectal cancer. Recent Underst. Colorectal Cancer Treatment. , (2022).
- Jakobsen, A. K., et al. Correlation between topoisomerase I and tyrosyl-DNA phosphodiesterase 1 activities in non-small cell lung cancer tissue. Experimental and Molecular Pathology. 99 (1), 56-64 (2015).
- Jakobsen, A. K., et al. TDP1 and TOP1 as targets in anticancer treatment of NSCLC: Activity and protein level in normal and tumor tissue from 150 NSCLC patients correlated to clinical data. Lung Cancer. 164, 23-32 (2022).
- Keller, J. G., et al. On-slide detection of enzymatic activities in selected single cells. Nanoscale. 9 (36), 13546-13553 (2017).
- Keller, J. G., Stougaard, M., Knudsen, B. R. Enzymatic activity in single cells. Trends in Biotechnology. 29 (5), 222-230 (2019).
- Tesauro, C., et al. Different camptothecin sensitivities in subpopulations of colon cancer cells correlate with expression of different phospho-isoforms of topoisomerase i with different activities. Cancers. 12 (5), 1240 (2020).
- Tesauro, C., et al. Topoisomerase i activity and sensitivity to camptothecin in breast cancer-derived cells: A comparative study. BMC Cancer. 19 (1), 1-15 (2019).
- Pommier, Y. DNA topoisomerase I Inhibitors: Chemistry, biology, and interfacial inhibition. Chemical Reviews. 109 (7), 2894-2902 (2009).
- Pommier, Y. Drugging topoisomerases: lessons and challenges. ACS Chemical Biology. 8 (1), 82-95 (2013).
- Bailly, C. Irinotecan: 25 years of cancer treatment. Pharmacological Research. 148, 104398 (2019).
- Petersen, K. V., et al. Simple and fast dna based sensor system for screening of small-molecule compounds targeting eukaryotic topoisomerase 1. Pharmaceutics. 13 (8), 1255 (2021).
- García-Estrada, C., Prada, C. F., Fernández-Rubio, C., Rojo-Vázquez, F., Balaña-Fouce, R. DNA topoisomerases in apicomplexan parasites: promising targets for drug discovery. Proceedings of the Royal Society B: Biological Sciences. 277 (1689), 1777-1787 (2010).
- Hede, M. S., et al. Detection of the malaria causing Plasmodium parasite in saliva from infected patients using topoisomerase I activity as a biomarker. Scientific Reports. 8 (1), 1-12 (2018).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2025 MyJoVE Corporation. All rights reserved