A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Infection of Primary Nasal Epithelial Cells Grown at an Air-Liquid Interface to Characterize Human Coronavirus-Host Interactions
In This Article
Summary
The nasal epithelium is the primary barrier site encountered by all respiratory pathogens. Here, we outline methods to use primary nasal epithelial cells grown as air-liquid interface (ALI) cultures to characterize human coronavirus-host interactions in a physiologically relevant system.
Abstract
Three highly pathogenic human coronaviruses (HCoVs) - SARS-CoV (2002), MERS-CoV (2012), and SARS-CoV-2 (2019) - have emerged and caused significant public health crises in the past 20 years. Four additional HCoVs cause a significant portion of common cold cases each year (HCoV-NL63, -229E, -OC43, and -HKU1), highlighting the importance of studying these viruses in physiologically relevant systems. HCoVs enter the respiratory tract and establish infection in the nasal epithelium, the primary site encountered by all respiratory pathogens. We use a primary nasal epithelial culture system in which patient-derived nasal samples are grown at an air-liquid interface (ALI) to study host-pathogen interactions at this important sentinel site. These cultures recapitulate many features of the in vivo airway, including the cell types present, ciliary function, and mucus production. We describe methods to characterize viral replication, host cell tropism, virus-induced cytotoxicity, and innate immune induction in nasal ALI cultures following HCoV infection, using recent work comparing lethal and seasonal HCoVs as an example1. An increased understanding of host-pathogen interactions in the nose has the potential to provide novel targets for antiviral therapeutics against HCoVs and other respiratory viruses that will likely emerge in the future.
Introduction
Seven human coronaviruses (HCoVs) have been identified to date and cause a range of respiratory diseases2. The common or seasonal HCoVs (HCoV-NL63, -229E, -OC43, and -HKU1) are typically associated with upper respiratory tract pathology and cause an estimated 10%-30% of common cold cases annually. Though this is the typical clinical phenotype associated with the common HCoVs, these viruses can cause more significant lower respiratory tract disease in at-risk populations, including children, older adults, and immunocompromised individuals3,4. Three pathogenic HCoVs have emerged and caused significant public health emergencies in the last 20 years, including severe acute respiratory syndrome (SARS)-CoV, Middle East respiratory syndrome (MERS)-CoV, and SARS-CoV-2. Lethal HCoVs are associated with more severe respiratory tract pathology, which is clearly illustrated by the >34% case-fatality rate associated with MERS-CoV cases (894 deaths from over 2,500 cases since its emergence in 2012)5,6. It is important to note that the lethal HCoVs also cause a range of respiratory tract diseases, from asymptomatic infections to lethal pneumonia, as seen with the ongoing COVID-19 pandemic7.
HCoVs, like other respiratory pathogens, enter the respiratory tract and establish a productive infection in the nasal epithelium8. Spread to the lower airway is thought to be associated with aspiration from the oral/nasal cavity to the lung, where HCoVs cause more significant lower respiratory tract pathology9,10,11. Thus, the nose serves as the initial portal for viral entry and is the primary barrier to infection with its robust mucociliary clearance machinery and unique innate immune mechanisms aimed at preventing further viral spread to the lower airway12,13. For example, nasal epithelial cells have been reported to express higher than average basal levels of antiviral interferons and interferon-stimulated genes, indicating that nasal cells may be primed for early responses to respiratory viruses14,15,16.
We have previously utilized patient-derived primary nasal epithelial cells grown at an air-liquid interface (ALI) to model HCoV-host interactions in the nose, where HCoV infections begin. Nasal ALI cultures are permissive to both pathogenic (SARS-CoV-2 and MERS-CoV) and common HCoVs (HCoV-NL63 and HCoV-229E) and offer various advantages over traditional airway epithelial cell lines such as A549 (a lung adenocarcinoma cell line)16,17. After differentiation, nasal ALI cultures contain a heterogeneous cellular population and exhibit many of the functions expected of the in vivo nasal epithelium, such as mucociliary clearance machinery18. Nasal cells also offer advantages over lower airway culture systems (such as human bronchial epithelial cells, HBECs), as the acquisition of nasal epithelial cells via cytologic brushing is significantly less invasive compared with using techniques such as bronchoscopy for attaining HBECs19,20,21.
This paper describes methods for utilizing this nasal ALI culture system to characterize HCoV-host interactions in the nasal epithelium. We have applied these methods in recently published works to compare SARS-CoV-2, MERS-CoV, HCoV-NL63, and HCoV-229E1,16,17. Though these methods and representative results emphasize the study of HCoVs in this nasal cell model, the system is highly adaptable to other HCoVs, as well as other respiratory pathogens. Further, these methods can be applied more broadly to other ALI culture systems in order to investigate viral replication and cellular tropism, as well as cytotoxicity and innate immune induction following infection.
Access restricted. Please log in or start a trial to view this content.
Protocol
The use of nasal specimens was approved by the University of Pennsylvania Institutional Review Board (protocol # 800614) and the Philadelphia VA Institutional Review Board (protocol # 00781).
1. Infection of nasal ALI cultures
NOTE: Acquisition of clinical specimens, as well as growth and differentiation of nasal ALI cultures, is outside the scope of this paper. Specific methods for culturing primary nasal epithelial cells can be found in recently published works utilizing these cultures18,22,23. The below protocols can additionally be applied to commercially available nasal epithelial ALI cultures if desired. Protocols and volumes detailed below are applicable to 24-well plate transwell inserts (6.5 mm diameter, 0.33 cm2 membrane surface area). If using ALI cultures grown on larger transwells (i.e., 12-well plates, 12 mm diameter, 1.12 cm2 surface area), adjust the volumes proportionally to reflect the transwell size.
- Day before infection:
- Wash ALI cultures 3x with phosphate-buffered saline (PBS) apically (add ~200 μL of warmed PBS, place in 37 °C incubator for 5 min, aspirate PBS, and repeat).
- Replace basal medium (500 μL).
- Allow cultures to equilibrate at the temperature at which infections will be conducted overnight (i.e., if infecting at 33 °C, place cultures in a 33 °C incubator after PBS washes).
NOTE: HCoVs associated with the common cold such as HCoV-229E and HCoV-NL63 are reported to replicate more efficiently at 33 °C. Additionally, the temperature of the in vivo nasal epithelium is 33 °C (this differs from the temperature of the lung, which is 37 °C).
- Dilute virus as needed in serum-free Dulbecco's Modified Eagle Medium (DMEM) to achieve the desired multiplicity of infection (MOI) in a total inoculum volume of 50 μL.
NOTE: Infections have typically been conducted at MOI = 5 (high MOI); however, infections at MOI = 0.5 (low MOI) have also been used for SARS-CoV-2 and common cold-associated HCoVs, and these result in comparable peak viral titers, but with variable kinetics (either MOI is acceptable). - Add inoculum apically, and place cultures back into the incubator for 1 h.
- Rock plates gently every 15 min during infection (holding plate firmly in both hands, rock forward and backward and side to side to ensure uniform adsorption of viral inoculum).
- After 1 h of incubation, aspirate viral inoculum, and wash each infected culture 3x with PBS to ensure removal of viral inoculum (for each wash, add 200 μL of PBS, incubate for 5 min, and aspirate or remove with a pipette).
- If desired, collect the third PBS wash to confirm adequate removal of input virus.
- Replace the basal medium with fresh medium on infected ALIs every 72 h during infection.
2. Collection of apical surface liquid (ASL) and titration of shed virus
- At predetermined time points following infection, add 200 μL of PBS to apical chamber of each infected transwell.
NOTE: Relevant time points vary depending on HCoV of interest and range from 24 h to 192 h post infection (see the representative results section for viral replication data for various HCoVs). - Pipette PBS up and down 5x to ensure maximal collection of apically shed virus, and collect entire volume into a microcentrifuge tube (this is the ASL sample).
NOTE: ASL includes shed viral particles in addition to mucus and other products secreted apically from ALI cultures. - Quantify infectious virus in ASL via standard viral plaque assay (serially dilute virus-containing samples in order to quantify the concentration of viral particles).
NOTE: Cell types and incubation period used for plaque assay will depend on the virus used: SARS-CoV-2 (VeroE6 cells); MERS-CoV (VeroCCL81 cells); HCoV-NL63 (LLC-MK2 cells); HCoV-229E (Huh7 cells). Specifics on how to conduct viral plaque assays are beyond the scope of this manuscript but have been detailed previously in a Journal of Visualized Experiments (JoVE) publication24. - If desired, collect the basal medium at various times post-infection to confirm the absence of basally released virus. HCoVs are typically released apically from nasal epithelial cells, but confirm this via plaque assay of undiluted basal medium.
- Store ASL samples at −80 °C if quantification by plaque assay will not occur on the day of collection.
3. Quantification of intracellular virus
- After collecting ASL sample, move each transwell into a clean 24-well plate preloaded with 500 μL of DMEM containing 2% fetal bovine serum (FBS) basally.
NOTE: DMEM with 2% FBS is used to stabilize the virus during subsequent freeze-thaw cycles. - Wash each transwell 3x apically with PBS to ensure complete removal of apically shed virus.
- After aspirating to remove the final PBS wash, add 100 μL of DMEM with 2% FBS to the apical compartment.
- Move the plate containing transwells with both apical and basal media into a −80 °C freezer, and complete three consecutive freeze-thaw cycles to lyse the cells.
- After the final freeze-thaw cycle, pool apical (100 μL) and basal (500 μL) media into a clean tube.
- Centrifuge at 500 × g for 10 min at 4 °C to pellet any cellular debris.
- Collect the supernatant. This is the intracellular virus sample for titration via standard plaque assay.
NOTE: Three-fold dilution occurs during the collection process relative to ASL collection; ASL samples are collected in 200 μL of PBS, while intracellular virus sample is collected into a total volume of 600 μL.
4. Transepithelial electrical resistance (TEER) measurement
NOTE: For TEER measurement, PBS supplemented with calcium and magnesium (PBS + Ca2+/Mg2+) should be used. An epithelial volt/ohmmeter set to read in ohms is used (see the Table of Materials).
- Clean, equilibrate, and blank the EVOM instrument according to the manufacturer's instructions; use an "empty" transwell with no nasal cells added for blanking. Record blank TEER measurement.
NOTE: If using multiple viruses, the EVOM instrument must be cleaned stringently between conditions to avoid cross-contamination (washes with 70% ethanol followed by deionized water are sufficient). - Move each infected transwell into a prelabeled, clean 24-well plate with 500 μL of PBS + Ca2+/Mg2+ basally to wash residual basal medium from the transwells.
- Add 200 μL of PBS + Ca2+/Mg2+ to the apical compartment of each transwell.
- Add 1 mL of PBS + Ca2+/Mg2+ to the Endohm-6 measurement chamber.
- Move each transwell into the Endohm-6 measurement chamber, and replace the lid of the chamber so that the apical electrode rests in the 200 μL of PBS in the apical compartment; the basal electrode is built into the bottom of the Endohm-6 chamber.
- Allow the EVOM reading to stabilize, and record raw TEER measurement.
- Collect ASL sample as described above after taking TEER measurement if titering is desired (collect in the 200 μL of PBS + Ca2+/Mg2+ that was added for TEER measurement).
NOTE: ASL sample collection can induce micro-tears in the epithelial barrier that may confound TEER readings, so ASL must be collected after TEER measurement. Further, basal medium must not be changed immediately before TEER readings as this may also impact TEER values. - To convert raw TEER readings to final measurements in Ohms/cm2, subtract blank TEER value, and multiply this value by the surface area of the transwell membrane using equation (1):
TEER = [TEER reading - blank TEER value] × (surface area of transwell) (1) - For HCoVs, evaluate TEER every 24 h or 48 h following infection; the kinetics of TEER changes often vary among viruses and necessitate troubleshooting at various time points.
- When measuring TEER, always include mock-infected cultures and evaluate at each time point (negative control).
NOTE: For mock cultures, TEER measurements should remain stable or may slightly increase from baseline after differentiation of the cultures is completed. Surface area of membrane supports will vary depending on size and manufacturer of transwells; 24-well transwells typically have a surface area of 0.33 cm2.
5. Measurement of cytotoxicity during infection via lactate dehydrogenase (LDH) assay
NOTE: In this work, LDH content in ASL samples was quantified using a commercially available cytotoxicity detection kit. LDH signal in basal medium was often below the limit of detection and often less reproducible than LDH quantified in ASL samples from HCoV-infected cultures.
- Prepare additional controls necessary for LDH assay.
- Background control: use PBS (use PBS containing calcium and magnesium if ASL samples were collected in this way).
- Positive control (ceiling value): treat ALIs apically with Triton X-100. Collect Triton control wells (typically use three ALI cultures for this at each time point).
- Add 200 μL of 2% Triton X-100 in PBS directly to the apical compartment of the transwell.
- Incubate for 10-15 min to allow cells to completely lyse.
- Collect the entire volume as a Triton ceiling sample.
- Negative control (low control/baseline LDH release): collect ASL from mock-infected ALI cultures derived from the same donor as infected cultures.
- Collect negative control mock ASL following ASL collection procedure above.
NOTE: The same transwells can be used for this negative control for all time points, whereas fresh transwells will be necessary for each time point for the Triton control.
- Collect negative control mock ASL following ASL collection procedure above.
- To allow for quantification of shed virus in addition to LDH readings from each ASL sample, load an optically clear flat-bottom black 96-well plate as follows:
- Dilute all samples in PBS, using 45 μL of sample and 55 μL of PBS (100 μL total volume).
- Treat Triton positive control and mock background control samples in the same manner.
NOTE: This dilution allows for triplicate loading of each experimental sample (45 μL x 3) and enough residual ASL volume for titering via plaque assay. - Load LDH plate in triplicate: Triton ceiling, mock background control, PBS control, and experimental samples.
- Prepare the reaction mixture as indicated by the manufacturer (dye solution + catalyst).
- Add 100 μL of reaction mixture to each well, and incubate the plate at room temperature for 20 min protected from light.
- After 20 min, measure absorbance at 492 nm.
- Calculate percentage cytotoxicity relative to the Triton ceiling value using equation (2).
Cytotoxicity (%)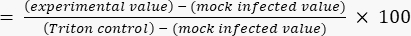 (2)
(2)
6. Preparation of nasal ALI cultures for immunofluorescence (IF) imaging
- Move the transwell into a fresh 24-well plate, and wash 3x apically with PBS (collect the first PBS wash as ASL if titering; then, perform two additional PBS washes to remove excess shed virus that might hinder IF quality)
- After the final PBS wash, cover the transwell apically and basally in 4% PFA.
- Incubate for 30 min to fix in 4% PFA, and then remove and wash 3x with PBS.
- Excise the transwell support containing the cells using a sharpened razor blade or scissors.
- To increase the number of IF targets for each transwell, cut each membrane in half, and stain each half with different antibody combinations.
- Permeabilize with 0.2% Triton X-100 in PBS for 10 min.
- Block with 10% normal donkey serum and 1% BSA in PBST (PBS + 0.2% Triton X-100) for 60 min at room temperature.
NOTE: Protect samples from light exposure from this step forward. - Incubate in primary antibody solution overnight at 4 °C. Dilute all antibodies 1:1,000 in blocking buffer.
NOTE: Representative antibodies for HCoV nucleocapsid staining, as well as epithelial cell type markers and cytoskeletal stains, are listed below (see the Table of Materials for manufacturer information and catalog numbers).- For HCoV antigen staining, use antibodies targeting SARS-CoV-2 nucleocapsid, MERS-CoV nucleocapsid, and HCoV-NL63 nucleocapsid.
- To identify epithelial cell types, use antibodies targeting goblet cell marker MUC5AC and ciliated cell marker type IV β-tubulin.
- For cytoskeletal markers, use antibodies directed against phalloidin (binds F-actin) and epithelial cell adhesion marker: EpCAM (CD326).
- Incubate in secondary antibody solution for 60 min at room temperature; use secondary antibody dyes diluted 1:1,000 in blocking buffer.
- After staining is complete, transfer the membrane onto a glass slide with a spatula, orient the transwell with the apical side toward the slide, and add mounting solution. Allow to settle for 15-30 min prior to applying clear nail polish around edges.
- Acquire images using a confocal microscope (Z-axis step: 0.5 μm; sequential scanning)1,16,17.
NOTE: After fixation of cultures in 4% PFA and washing with PBS, fixed cultures can be stored at 4 °C for weeks to months prior to staining and preparation for imaging. After staining and mounting of membranes, samples can be stored long-term (>2 years) at 4 °C in the dark.
7. Collection of intracellular protein for western immunoblotting or RNA for RT-qPCR analysis
- Collect ASL as described above if quantifying viral titers (protocol section 2).
- Move the transwell to a clean 24-well plate, as scraping the membrane can lead to breaking the insert.
- For western blot analysis, collect total protein lysate in 125 μL of RIPA buffer (50 mM Tris, pH 8, 150 mM NaCl, 0.5% deoxycholate, 0.1% SDS, 1% NP40) supplemented with protease inhibitors and phosphatase inhibitors.
- Add 125 μL of RIPA buffer to the apical compartment, and incubate for 5-10 min.
- Scrape the membrane using a P200 pipette tip to remove any remaining attached cells, and collect the entire volume (scrape vigorously across the entire surface of the membrane, and then pipette up and down multiple times to collect the entire sample).
- Incubate protein lysate samples on ice for 10 min, and then centrifuge at maximum speed (20,000 × g) for 10 min at 4 °C.
- Mix the supernatant with 4x Laemmli sample buffer with β-mercaptoethanol (reducing agent) according to manufacturer protocols.
- Boil protein samples at 95 °C for 5 min, and then run using traditional western blotting protocols1,16,17.
- For collection of total RNA, use a commercially available RNA extraction kit of choice.
NOTE: The apical compartment of 24-well transwell inserts has a maximal volume of ~200 μL; thus, we perform two sequential washes on each culture to reach total recommended volume. Details below correspond to recommended collection of RNA samples in a 350 μL total volume. - Add 200 μL of lysis buffer to the apical compartment of the infected transwell.
- Let sit for 5-10 min, scrape any remaining cells from the membrane using a pipette tip, and collect the entire volume into a labeled microcentrifuge tube.
- Add 150 μL of additional lysis buffer to the apical compartment of the transwell, and pipette up and down before collecting into same microcentrifuge tube.
- Extract RNA according to the manufacturer's protocol.
Access restricted. Please log in or start a trial to view this content.
Results
The representative figures are partially adapted from data that can be found in the manuscript Otter et al.1. Nasal ALI cultures derived from four or six donors were infected with one of four HCoVs (SARS-CoV-2, MERS-CoV, HCoV-NL63, and HCoV-229E) according to the protocols described above, and the average apically shed viral titers for each virus are depicted in Figure 1A. While all four of these HCoVs replicate productively in nasal ALI cultures, SARS-CoV-2 and HCoV-...
Access restricted. Please log in or start a trial to view this content.
Discussion
The methods detailed here describe a primary epithelial culture system in which patient-derived nasal epithelial cells are grown at an air-liquid interface and applied to the study of HCoV-host interactions. Once differentiated, these nasal ALI cultures recapitulate many features of the in vivo nasal epithelium, including a heterogeneous cellular population with ciliated, goblet, and basal cells represented, as well as intact mucociliary function with robustly beating cilia and mucus secretion. This heterogeneou...
Access restricted. Please log in or start a trial to view this content.
Disclosures
Susan Weiss is on the Scientific Advisory Boards for Ocugen. Noam A. Cohen consults for GSK, AstraZeneca, Novartis, Sanofi/Regeron, and Oyster Point Pharmaceuticals and has a US Patent, "Therapy and Diagnostics for Respiratory Infection" (10,881,698 B2, WO20913112865), and a licensing agreement with GeneOne Life Sciences.
Acknowledgements
This study has the following funding sources: National Institutes of Health (NIH) R01AI 169537 (S.R.W. and N.A.C.), NIH R01AI 140442 (S.R.W.), VA Merit Review CX001717 (N.A.C.), VA Merit Review BX005432 (S.R.W. and N.A.C.), Penn Center for Research on Coronaviruses and other Emerging Pathogens (S.R.W.), Laffey-McHugh Foundation (S.R.W. and N.A.C.), T32 AI055400 (CJO), T32 AI007324 (AF).
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| Alexa Fluor secondary antibodies (488, 594, 647) | Invitrogen | Various | |
| BSA (bovine serum albumin) | Sigma-Aldrich | A7906 | |
| cOmplete mini EDTA-free protease inhibitor | Roche | 11836170001 | |
| Cytotoxicity detection kit | Roche | 11644793001 | |
| DMEM (Dulbecco's Modified Eagle Media) | Gibco | 11965-084 | |
| DPBS (Dulbecco's Phosphate Buffered Saline) | Gibco | 14190136 | |
| DPBS + calcium + magnesium | Gibco | 14040-117 | |
| Endohm-6G measurement chamber | World Precision Instruments | ENDOHM-6G | |
| Epithelial cell adhesion marker (EpCAM; CD326) | eBiosciences | 14-9326-82 | |
| Epithelial Volt/Ohm (TEER) Meter (EVOM) | World Precision Instruments | 300523 | |
| FBS (Fetal Bovine Serum) | HyClone | SH30071.03 | |
| FV10-ASW software for imaging | Olympus | Version 4.02 | |
| HCoV-NL63 (Human coronavirus, NL63) | BEI Resources | NR-470 | |
| HCoV-NL63 nucleocapsid antibody | Sino Biological | 40641-V07E | |
| Hoescht stain | Thermo Fisher | H3570 | |
| Laemmli sample buffer (4x) | BIO-RAD | 1610747 | |
| LLC-MK2 cells | ATCC | CCL-7 | To titrate HCoV-NL63 |
| MERS-CoV (Human coronavirus, Middle East Respiratory Syndrome Coronavirus (MERS-CoV), EMC/2012) | BEI Resources | NR-44260 | |
| MERS-CoV nucleocapsid antibody | Sino Biological | 40068-MM10 | |
| MUC5AC antibody | Sigma-Aldrich | AMAB91539 | |
| Olympus Fluoview confocal microscope | Olympus | FV1000 | |
| Phalloidin-iFluor 647 stain | Abcam | ab176759 | |
| PhosStop easy pack (phosphatase inhibitors) | Roche | PHOSS-RO | |
| Plate reader | Perkin Elmer | HH34000000 | Any plate reader or ELISA reader is sufficient; must be able to read absorbance at 492 nm |
| RIPA buffer (50 mM Tris pH 8; 150 mM NaCl; 0.5% deoxycholate; 0.1% SDS; 1% NP40) | Thermo Fisher | 89990 | Can prep in-house or purchase |
| RNeasy Plus Kit | Qiagen | 74134 | |
| SARS-CoV-2 (SARS-Related Coronavirus 2, Isolate USA-WA1/2020) | BEI Resources | NR-52281 | |
| SARS-CoV-2 nucleocapsid antibody | Genetex | GTX135357 | |
| Triton-X 100 | Fisher Scientific | BP151100 | |
| Type IV β- tubulin antibody | Abcam | ab11315 | |
| VeroCCL81 cells | ATCC | CCL-81 | To titrate MERS-CoV |
| VeroE6 cells | ATCC | CRL-1586 | To titrate SARS-CoV-2 |
References
- Otter, C. J., et al. Infection of primary nasal epithelial cells differentiates among lethal and seasonal human coronaviruses. Proceedings of the National Academy of Sciences of the United States of America. 120 (15), 2218083120(2023).
- Fehr, A., Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods in Molecular Biology. 1282, 1-23 (2015).
- Gaunt, E. R., Hardie, A., Claas, E. C. J., Simmonds, P., Templeton, K. E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. Journal of Clinical Microbiology. 48 (8), 2940-2947 (2010).
- Kesheh, M. M., Hosseini, P., Soltani, S., Zandi, M. An overview on the seven pathogenic human coronaviruses. Reviews in Medical Virology. 32 (2), 2282(2022).
- MERS-CoV Worldwide Overview. European Centre for Disease Prevention and Control. , Available from: https://www.ecdc.europa.eu/en/middle-east-respiratory-syndrome-coronavirus-mers-cov-situation-update (2022).
- MERS Situation Update. World Health Organization Regional Office for the Eastern Mediterranean. , Available from: http://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html (2022).
- Cao, Y., Liu, X., Xiong, L., Cai, K. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2: A systematic review and meta-analysis. Journal of Medical Virology. 92 (9), 1449-1459 (2020).
- Vareille, M., Kieninger, E., Edwards, M. R., Regamey, N. The airway epithelium: Soldier in the fight against respiratory viruses. Clinical Microbiology Reviews. 24 (1), 210-229 (2011).
- Farzal, Z., et al. Comparative study of simulated nebulized and spray particle deposition in chronic rhinosinusitis patients. International Forum of Allergy and Rhinology. 9 (7), 746-758 (2019).
- Gaeckle, N. T., Pragman, A. A., Pendleton, K. M., Baldomero, A. K., Criner, G. J. The oral-lung axis: The impact of oral health on lung health. Respiratory Care. 65 (8), 1211-1220 (2020).
- Hou, Y., et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 182, 429-446 (2020).
- Hariri, B. M., Cohen, N. A. New insights into upper airway innate immunity. American Journal of Rhinology and Allergy. 30 (5), 319-323 (2016).
- Hiemstra, P. S., McCray, P. B., Bals, R. The innate immune function of airway epithelial cells in inflammatory lung disease. European Respiratory Journal. 45 (4), 1150-1162 (2015).
- Hatton, C. F., et al. Delayed induction of type I and III interferons mediates nasal epithelial cell permissiveness to SARS-CoV-2. Nature Communications. 12 (1), 7092(2021).
- Sungnak, W., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nature Medicine. 26 (5), 681-687 (2020).
- Li, Y., et al. SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proceedings of the National Academy of Sciences of the United States of America. 118 (16), 2022643118(2021).
- Comar, C. E., et al. MERS-CoV endoribonuclease and accessory proteins jointly evade host innate immunity during infection of lung and nasal epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 119 (21), 2123208119(2022).
- Lee, R. J., et al. Bacterial D-amino acids suppress sinonasal innate immunity through sweet taste receptors in solitary chemosensory cells. Science Signaling. 10 (495), (2017).
- Brewington, J. J., et al. Brushed nasal epithelial cells are a surrogate for bronchial epithelial CFTR studies. JCI Insight. 3 (13), 99385(2018).
- Comer, D. M., Elborn, J. S., Ennis, M. Comparison of nasal and bronchial epithelial cells obtained from patients with COPD. PLoS One. 7 (3), e32924(2012).
- Vanders, R. L., Hsu, A., Gibson, P. G., Murphy, V. E., Wark, P. A. B. Nasal epithelial cells to assess in vitro immune responses to respiratory virus infection in pregnant women with asthma. Respiratory Research. 20 (1), 259(2019).
- Lee, R. J., et al. Fungal aflatoxins reduce respiratory mucosal ciliary function. Scientific Reports. 6, 33221(2016).
- Patel, N. N., et al. Fungal extracts stimulate solitary chemosensory cell expansion in noninvasive fungal rhinosinusitis. International Forum of Allergy and Rhinology. 9 (7), 730-737 (2019).
- Baer, A., Kehn-Hall, K. Viral concentration determination through plaque assays: Using traditional and novel overlay systems. Journal of Visualized Experiments. 93 (93), e52065(2014).
- Robinot, R., et al. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nature Communications. 12 (1), 4354(2021).
- Whitsett, J. A. Airway epithelial differentiation and mucociliary clearance. Annals of the American Thoracic Society. 15, S143-S148 (2018).
- Gao, N., Raduka, A., Rezaee, F. Respiratory syncytial virus disrupts the airway epithelial barrier by decreasing cortactin and destabilizing F-actin. Journal of Cell Science. 135 (16), 259871(2022).
- Schmidt, H., et al. IL-13 impairs tight junctions in airway epithelia. International Journal of Molecular Sciences. 20 (13), 3222(2019).
- Huang, Z. Q., et al. Interleukin-13 alters tight junction proteins expression thereby compromising barrier function and dampens rhinovirus induced immune responses in nasal epithelium. Frontiers in Cell and Developmental Biology. 8, 572749(2020).
- Saatian, B., et al. Interleukin-4 and interleukin-13 cause barrier dysfunction in human airway epithelial cells. Tissue Barriers. 1 (2), e24333(2013).
- Coles, J. L., et al. A revised protocol for culture of airway epithelial cells as a diagnostic tool for primary ciliary dyskinesia. Journal of Clinical Medicine. 9 (11), 3753(2020).
- Baldassi, D., Gabold, B., Merkel, O. M. Air−liquid interface cultures of the healthy and diseased human respiratory tract: Promises, challenges, and future directions. Advanced NanoBiomed Research. 1 (6), 2000111(2021).
- Seibold, M. A. Interleukin-13 stimulation reveals the cellular and functional plasticity of the airway epithelium. Annals of the American Thoracic Society. 15, S98-S106 (2018).
- Morrison, C. B., et al. SARS-CoV-2 infection of airway cells causes intense viral and cell shedding, two spreading mechanisms affected by IL-13. Proceedings of the National Academy of Sciences of the United States of America. 119 (16), 2119680119(2022).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved