Method Article
A Knowledge Graph Approach to Elucidate the Role of Organellar Pathways in Disease via Biomedical Reports
In This Article
Summary
A computational protocol, CaseOLAP LIFT, and a use case are presented for investigating mitochondrial proteins and their associations with cardiovascular disease as described in biomedical reports. This protocol can be easily adapted to study user-selected cellular components and diseases.
Abstract
The rapidly increasing and vast quantities of biomedical reports, each containing numerous entities and rich information, represent a rich resource for biomedical text-mining applications. These tools enable investigators to integrate, conceptualize, and translate these discoveries to uncover new insights into disease pathology and therapeutics. In this protocol, we present CaseOLAP LIFT, a new computational pipeline to investigate cellular components and their disease associations by extracting user-selected information from text datasets (e.g., biomedical literature). The software identifies sub-cellular proteins and their functional partners within disease-relevant documents. Additional disease-relevant documents are identified via the software's label imputation method. To contextualize the resulting protein-disease associations and to integrate information from multiple relevant biomedical resources, a knowledge graph is automatically constructed for further analyses. We present one use case with a corpus of ~34 million text documents downloaded online to provide an example of elucidating the role of mitochondrial proteins in distinct cardiovascular disease phenotypes using this method. Furthermore, a deep learning model was applied to the resulting knowledge graph to predict previously unreported relationships between proteins and disease, resulting in 1,583 associations with predicted probabilities >0.90 and with an area under the receiver operating characteristic curve (AUROC) of 0.91 on the test set. This software features a highly customizable and automated workflow, with a broad scope of raw data available for analysis; therefore, using this method, protein-disease associations can be identified with enhanced reliability within a text corpus.
Introduction
Studying disease-related proteins enhances the scientific knowledge of pathogenesis and helps to identify potential therapeutics. Several large text corpora of biomedical publications, such as PubMed's 34 million articles containing publication titles, abstracts, and full-text documents, report novel findings that link proteins with diseases. However, these findings are fragmented across various sources and must be integrated to generate new biomedical insights. Several biomedical resources exist to integrate protein-disease associations1,2,3,4,5,6,7. However, these curated resources are often incomplete and may not encompass the latest research findings. Text-mining approaches are essential to extract and synthesize protein-disease associations in large text corpora, which would result in a more comprehensive understanding of these biomedical concepts in the scientific literature.
Multiple biomedical text-mining approaches exist to uncover protein-disease relationships8,9,10,11,12,13,14, and others contribute in part to determining these relationships by identifying the proteins, diseases, or other biomedical entities mentioned in text13,15,16,17,18,19. However, many of these tools lack access to the most up-to-date literature, with the exception of a few that are periodically updated8,11,13,15. Similarly, many tools also have a limited scope of study, as they are confined to broad predefined diseases or proteins9,13. Several approaches are also prone to the identification of false positives within the text; others have addressed these issues with an interpretable and global blacklist of protein names9,11 or less interpretable name entity recognition techniques15,20. While most resources present only pre-computed results, some tools offer interactivity via web apps or accessible software code8,9,11.
To address the above limitations, we present the following protocol, CaseOLAP with label imputation and full text (CaseOLAP LIFT), as a flexible and customizable platform to investigate associations between proteins (e.g., proteins associated with a cellular component) and diseases from text datasets. This platform features automated curation of gene ontology (GO) term-specific proteins (e.g., organelle-specific proteins), imputation of missing document topic labels, analysis of full-text documents, as well as analysis tools and predictive tools (Figure 1, Figure 2, and Table 1). CaseOLAP LIFT curates organelle-specific proteins by using user-provided GO terms (e.g., organelle compartment) and functionally related proteins by using STRING21, Reactome22, and GRNdb23. Disease-studying documents are identified by their PubMed-annotated medical subject header (MeSH) labels. For the ~15.1% of unlabeled documents, labels are imputed if at least one MeSH term synonym is found in the title or at least two are found in the abstract. This enables previously uncategorized publications to be considered in the text-mining analysis. CaseOLAP LIFT also allows the user to select sections of publications (e.g., titles and abstracts only, full text, or full text excluding methods) within a specified timeframe (e.g., 2012-2022). The software also semi-automatically curates a use case-specific blacklist of protein names, vitally reducing the false-positive protein-disease associations present in other approaches. Overall, these improvements enable greater customizability and automation, expand the quantity of data available for analysis, and yield more confident protein-disease associations from large biomedical text corpora.
CaseOLAP LIFT incorporates biomedical knowledge and represents the relationship of various biomedical concepts using a knowledge graph, which is leveraged to predict hidden relationships in the graph. Recently, graph-based computation methods have been applied to biological settings, including integrating and organizing biomedical concepts24,25, drug repurposing and development26,27,28, and for clinical decision-making from proteomics data29.
To demonstrate the utilities of CaseOLAP LIFT in the setting of constructing a knowledge graph, we highlight a use case on the investigation of the associations between mitochondrial proteins and eight categories of cardiovascular disease. Evidence from ~362,000 disease-relevant documents was analyzed to identify the top mitochondrial proteins and pathways associated with the diseases. Next, these proteins, their functionally related proteins, and their text-mining results were incorporated into a knowledge graph. This graph was leveraged in a deep learning-based link prediction analysis to predict protein-disease associations so far unreported within biomedical publications.
The introduction section describes the background information and objectives of our protocol. The following section describes the steps of the computational protocol. Subsequently, the representative results of this protocol are described. Finally, we briefly discuss the computational protocol use cases, advantages, drawbacks, and future applications.
Protocol
1. Running the docker container
- Download the CaseOLAP LIFT docker container by using the terminal window and typing in docker pull caseolap/caseolap_lift:latest.
- Create a directory that will store all the program data and output (e.g. mkdir caseolap_lift_shared_folder).
- Start the docker container with the command docker run --name caseolap_lift -it -v PATH_TO_FOLDER:/caseolap_lift_shared_folder caseolap/caseolap_lift:latest bash with PATH_TO_FOLDER as the full file path for the folder (e.g., /Users/caseolap/caseolap_lift_shared_folder). Future commands from section 2 will be issued on this terminal window.
- Start the elastic search within the container. In a new terminal window, type docker exec -it --user elastic caseolap_lift bash /workspace/start_elastic_search.sh.
NOTE: In this protocol, CaseOLAP LIFT is run interactively, with every step performed sequentially. This analysis can also be executed end-to-end by passing it in as a parameters.txt file. The parameters.txt used in this study are in /workspace/caseolap_lift/parameters.txt. To access more details on each step, run the command with the --help flag, or visit the documentation on the GitHub repository (https://github.com/CaseOLAP/caseolap_lift).
2. Preparing the diseases and proteins
- Navigate to the caseolap_lift folder with cd /workspace/caseolap_lift
- Make sure that the download links in config/knowledge_base_links.json are up-to-date and accurate for the latest version of each knowledge base resource. By default, the files are only downloaded once; to update these files and re-download, run the preprocessing step with -r in step 2.4.
- Determine the GO term and disease categories to use for this study. Find the identifiers for all GO terms and MeSH identifiers at http://geneontology.org/ and https://meshb.nlm.nih.gov/, respectively.
- Execute the pre-processing module using command-line options. This preprocessing step assembles specified diseases, lists proteins to study, and gathers protein synonyms for text-mining. Indicate the user-defined studied GO terms using the -c flag and the disease MeSH tree numbers using the -d flag, and specify abbreviations with -a.
Example command:
python caseolap_lift.py preprocessing -a "CM ARR CHD VD IHD CCD VOO OTH" -d "C14.280.238,C14.280.434 C14.280.067,C23.550.073 C14.280.400 C14.280.484 C14.280.647 C14.280.123 C14.280.955 C14.280.195,C14.280.282,C14.280.383,C14.280.470,
C14.280.945,C14.280.459,C14.280.720" -c "GO:0005739" --include-synonyms --include-ppi -k 1 -s 0.99 --include-pw -n 4 -r 0.5 --include-tfd - Examine the categories.txt, core_proteins.txt, and proteins_of_interest.txt files from the previous step in the output folder. Ensure that all the disease categories in categories.txt are correct and that a reasonable amount of proteins are identified within core_proteins.txt and proteins_of_interest.txt. If necessary, repeat step 2.4, and modify the parameters to include a greater or fewer number of proteins.
NOTE: The number of proteins included in the study is determined by --include-ppi, --include-pw, and --include-tfd flags to include protein-protein interactions, proteins with shared reactome pathways, and proteins with transcription factor dependence, respectively. Their specific functionality is specified with additional flags such as -k, -s, -n, and -r (see documentation).
3. Text-mining
- Make sure the categories.txt, core_proteins.txt, and proteins_of_interest.txt files from the previous step are found in the output folder. Use these files as the input for the text-mining. Optionally, adjust the configurations pertaining to the document parsing and indexing in the config folder. See a previous version of the CaseOLAP protocol for more details on configuration and troubleshooting8.
- Execute the text-mining module with python caseolap_lift.py text_mining. Add the -l flag to impute the topics of uncategorized documents and the -t flag to download the full text of disease-relevant documents. Other optional flags specify a date range of publications to download (-d) and provide options to screen the protein names (described in step 3.3). A sample of a parsed document is shown in Figure 3.
Example command: python caseolap_lift.py text_mining -d "2012-10-01,2022-10-01" -l -t
NOTE: A bulk of the computational protocol time is spent on step 3.2, which can potentially span over 24 h. The runtime will depend on the size of the text corpus to be downloaded, which will also depend on the date range and whether label imputation and full-text functionality are enabled. - (Recommended) Screen the protein names. The protein names identified in disease-relevant publications contribute to protein disease associations but are prone to false positives (i.e., homonyms with other words). To address this, enumerate possible homonyms in a blacklist (config/remove_these_synonyms.txt) so that they are excluded from the downstream steps.
- Find names to inspect: Under the result folder, find the protein names with the highest frequency under all_proteins or core_proteins (ranked_synonyms/ranked_synonyms_TOTAL.txt) and protein names with the highest scores under the folders in ranked_proteins depending on the score(s) of interest. If there are many names, prioritize the inspection of the top-scoring names.
- Inspect the names: Type python caseolap_lift.py text_mining -c followed by a protein name to display up to 10 name-containing publications. Then, for each name, check if the name is protein-specific.
- Recalculate the scores: Type python caseolap_lift.py text_mining -s. Repeat step 3.1, step 3.2, and step 3.3 until the names in step 3.1 appear correct.
4. Analyzing the results
- Make sure the text-mining results are in the result folder (e.g., result/all_proteins and result/core_proteins directories and associated files), which will be used as input for the analysis step. Specifically, a score indicating the strength of each protein-disease association is reported in the caseolap.csv results from the text-mining. Indicate which set of text-mining results to use for the analysis by specifying either --analyze_core_proteins to include only the GO-term related proteins or --analyze_all_proteins to include all the functionally related proteins.
- Identify the top proteins and pathways for each disease. Significant protein-disease associations are defined as those with scores exceeding a specified threshold. Z-score transform the CaseOLAP scores within each disease category, and consider the proteins with scores above a specified threshold (indicated by the -z flag) as significant.
NOTE: Biological pathways significant to each disease are identified automatically using significant proteins as input for the reactome pathway analysis. All such proteins are reported in the resulting result_table.csv in the analysis_results folder, and relevant figures and pathway analysis results are automatically generated in the analysis_results folder.
Example command: python caseolap_lift.py analyze_results -z 3.0 --analyze_core_proteins - Review the analysis results, and adjust as necessary. The number of proteins and, therefore, the enriched reactome pathways significant to each disease category depend on the z-score threshold used in the analysis. A z-score table, generated at output/analysis_results/zscore_cutoff_table.csv, indicates the number of proteins significant to each disease category to aid in the selection of a z-score threshold as high as possible while yielding several proteins significant to each disease category.
5. Predictive analysis
- Construct a knowledge graph.
- Ensure the required files are in the results folder, including the kg folder generated from preprocessing (step 2.4) and the caseolap.csv from the text-mining results under the all_proteins or core_proteins folders (step 3.2).
- Design the knowledge graph. Depending on the downstream task, include or exclude components of the complete knowledge graph. The knowledge graph consists of protein-disease scores from the text-mining and connections to the knowledge base resources used in step 2.4 (Figure 4). Include the MeSH disease tree with the --include_mesh flag, the protein-protein interactions from STRING with --include_ppi, the shared reactome pathways with --include_pw, and the transcription factor dependence from GRNdb/GTEx with --include_tfd.
- Run the knowledge graph construction module. Indicate which set of text-mining results to use for the analysis by specifying --analyze_core_proteins to only include the GO-term related proteins or --analyze_all_proteins to include all the functionally related proteins. By default, raw CaseOLAP scores are loaded as the edge weights between the protein and disease nodes; to scale the edge weights, indicate --use_z_score, or non-negative z-scores with --scale_z_score.
Example command: python caseolap_lift.py prepare_knowledge_graph --scale_z_score
- Predict novel protein-disease associations.
- Make sure the knowledge graph files, merged_edges.tsv and merged_nodes.tsv, are output from the previous step (step 5.1.3).
- Run the knowledge graph prediction script to predict protein-disease associations so far unreported within the scientific literature by typing python kg_analysis/run_kg_analysis.py. This is implemented with GraPE30 and uses DistMult31 to produce knowledge graph embeddings, which a multi-layer perceptron uses to predict the protein-disease associations. In the output/kg_analysis folder, predictions with a predicted probability >0.90 (predictions.csv) and model evaluation metrics (eval_results.csv) are saved.
NOTE: In this work, the chosen model parameters (e.g., embedding method, link prediction model, hyperparameters) were tailored for the representative study. This code serves as an example and a starting point for other analyses. To explore model parameters, refer to GraPE's documentation (https://github.com/AnacletoLAB/grape).
Results
Representative results were produced following this protocol to study the associations between mitochondrial proteins (Table 2) and eight cardiovascular disease categories (Table 3). In these categories, we found 363,567 publications published from 2012 to October 2022 (362,878 categorized by MeSH metadata, 6,923 categorized by label imputation). All the publications had titles, 276,524 had abstracts, and 51,065 had the full text available. Overall, 584 of the 1,687 queried mitochondrial proteins were identified within the publications, while 3,284 of their 8,026 queried functionally related proteins were identified. In total, 14 unique proteins were identified with significant scores across all the disease categories, with a z-score threshold of 3.0 (Figure 5). The Reactome pathway analysis of these proteins revealed 12 pathways significant to all the diseases (Figure 6). All the proteins, pathways, diseases, and scores were integrated into a knowledge graph (Table 4). This knowledge graph was leveraged to predict 12,688 novel protein-disease associations and filtered with a probability score of 0.90 to yield 1,583 high-confidence predictions. A highlighted example of two protein-disease associations is shown in Figure 7, illustrated in the context of other relevant biological entities functionally related to the proteins. The model evaluation metrics are reported in Table 5.
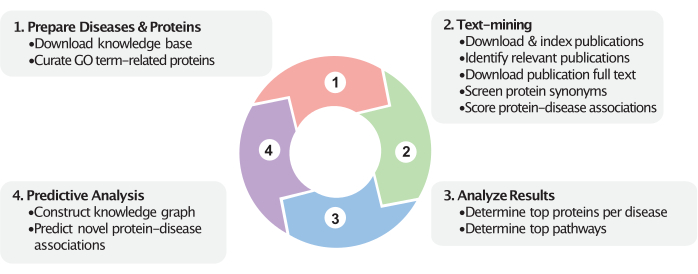
Figure 1: Dynamic view of the workflow. This figure represents the four major steps in this workflow. First, relevant proteins are curated based on the user-provided GO terms (e.g., cellular components), and disease categories are prepared based on the user-provided disease MeSH identifiers. Second, associations between proteins and diseases are calculated in the text-mining step. Publications within a certain date range are downloaded and indexed. Disease-studying publications are identified (via MeSH labels and optionally via imputed labels), and their full texts are downloaded and indexed. Protein names are queried within the publications and used to calculate the protein-disease association scores. Next, following text-mining, these scores help identify the top protein and pathway associations. Finally, a knowledge graph is constructed encompassing these proteins, diseases, and their relationships within the biomedical knowledge base. Novel protein-disease associations are predicted based on the constructed knowledge graph. These steps use the most recently available data from the biomedical knowledge bases and PubMed. Please click here to view a larger version of this figure.
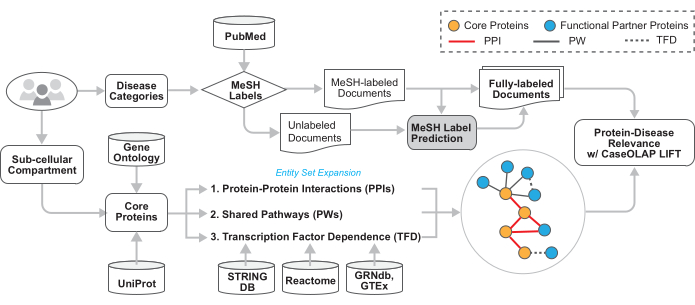
Figure 2: Technical architecture of the workflow. The technical details of this workflow are illustrated in this figure. The user provides the MeSH tree numbers of the disease categories and GO term(s). Text documents are downloaded from PubMed, disease-relevant documents are identified based on the provided MeSH labels, and documents without topic-indicating MeSH labels receive imputed category labels. The proteins associated with the provided GO term(s) are acquired. This protein set is expanded to include proteins that are functionally related via protein-protein interactions, shared biological pathways, and transcription factor dependence. These proteins are queried within disease-relevant documents and scored by CaseOLAP. Please click here to view a larger version of this figure.

Figure 3: An example of a processed document. An example of a parsed, indexed text document is presented here. In order, relevant fields indicate the index name (_index, _type), the PubMed ID (_id, pmid), the document subsections (title, abstract, full_text, introduction, methods, results, discussion), and other metadata (year, MeSH, location, journal). For display purposes only, the document subsections are truncated with ellipses. The MeSH field contains the document topics, which may sometimes be provided by our label-imputation step. Please click here to view a larger version of this figure.
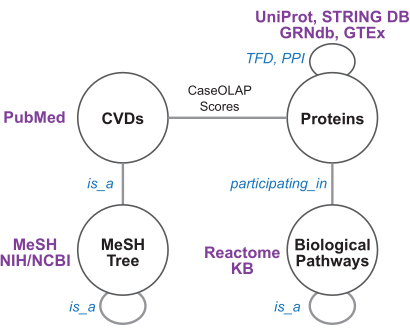
Figure 4: Knowledge graph schema and biomedical resources. This figure depicts the knowledge graph schema. Each node and edge represents a node or edge type, respectively. The edges between cardiovascular diseases (CVDs) and proteins are weighted by CaseOLAP scores. The protein-protein interaction (PPI) edges are weighted by STRING confidence scores. The GRNdb/GTEx-derived transcription factor dependence (TFD) edges, MeSH-derived disease tree edges, and reactome-derived pathway edges are unweighted. Please click here to view a larger version of this figure.
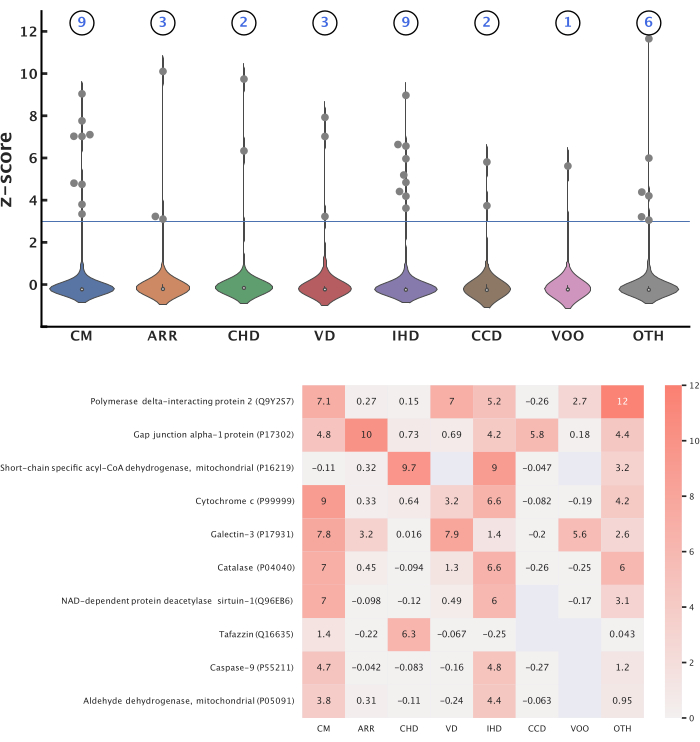
Figure 5: Top protein-disease associations. This figure presents mitochondrial proteins significant to each disease category. Z-score transformation was applied to the CaseOLAP scores within each category to identify significant proteins using a threshold of 3.0. (Top) Number of mitochondrial proteins significant to each disease: These violin plots depict the distribution of z-scores for proteins in each disease category. The total number of proteins significant to each disease category is shown above each violin plot. A total of 14 unique proteins were identified as significant across all the diseases, and some proteins were significant to multiple diseases. (Bottom) Top-scoring proteins: The heatmap displays the top 10 proteins that obtained the highest average z-scores across all the diseases. The blank values represent no obtained score between the protein and disease. Please click here to view a larger version of this figure.
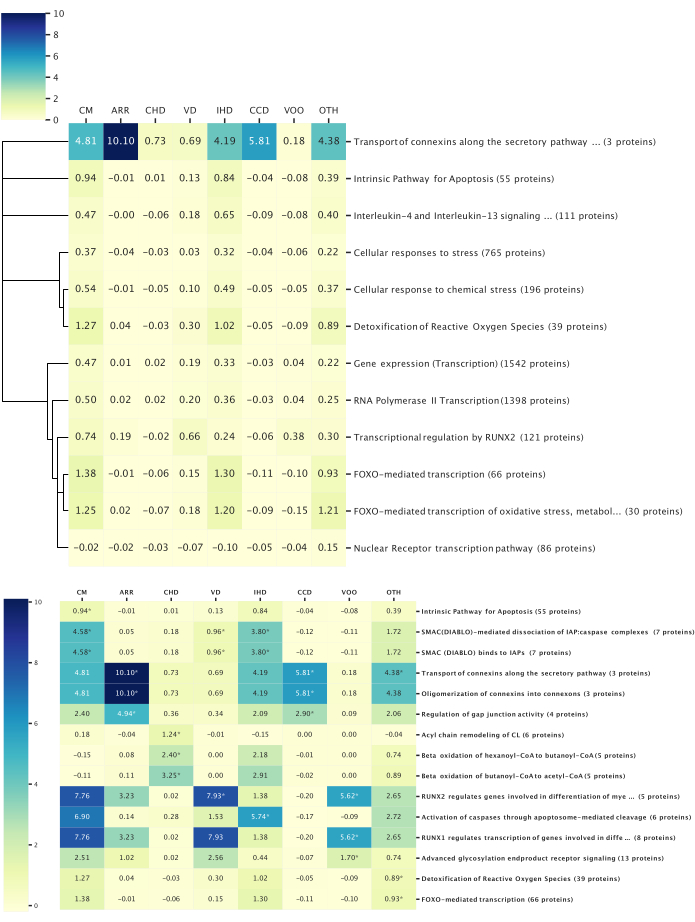
Figure 6: Top pathway-disease associations. This figure illustrates the top biological pathways associated with the studied disease categories, as determined via reactome pathway analysis. All the pathway analyses were filtered with p < 0.05. The heatmap values represent the average z-score of all the proteins within the pathway. (Top) Pathways conserved among all the diseases: Overall, 14 proteins were identified with relevance to all the disease categories, and 12 conserved pathways among all the disease categories were revealed. A dendrogram was constructed based on the pathway hierarchical structure to link the pathways with similar biological functions. The dendrogram height represents the relative depth within the pathway hierarchy; broad biological functions have longer limbs, and more specific pathways have shorter limbs. (Bottom) Pathways distinct to a disease category: Pathway analysis was performed using proteins achieving a significant z-score in each disease. The top three pathways with the lowest p-values associated with each disease are shown and indicated by asterisks. The pathways could be within the top three in multiple diseases. Please click here to view a larger version of this figure.
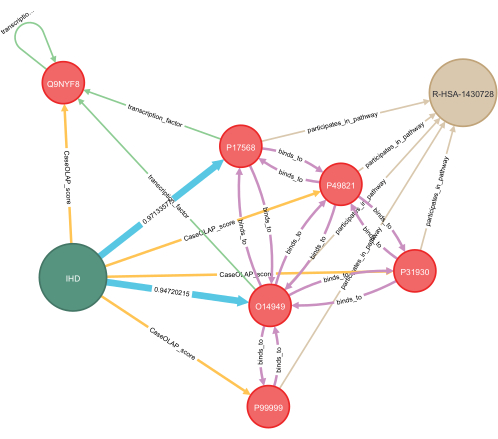
Figure 7: Application of deep learning for knowledge graph completion. An example of applying deep learning to a disease-specific knowledge graph is presented in this figure. Hidden relationships between proteins and disease are predicted, and these are indicated in blue. Computed probabilities for both predictions are displayed, with values ranging from 0.0 to 1.0 and with 1.0 indicating a strong prediction. Several proteins with known interactions are included, representing protein-protein interactions, transcription factor dependence, and shared biological pathways. For visualization, a subgraph of a few nodes with relevance to the highlighted example is shown. Key: IHD = ischemic heart disease; R-HSA-1430728 = metabolism; O14949 = cytochrome b-c1 complex subunit 8; P17568 = NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 7; Q9NYF8 Bcl-2-associated transcription factor 1, score: 7.24 x 10−7; P49821 = NADH dehydrogenase (ubiquinone) flavoprotein 1, mitochondrial, score: 1.06 x 10−5; P31930 = cytochrome b-c1 complex subunit 1, mitochondrial, score: 4.98 x 10−5; P99999 = cytochrome c, score: 0.399. Please click here to view a larger version of this figure.
Table 1: Workflow and rate-limiting steps. This table presents rough estimates of the computational time for each stage of the workflow. Options to include components of the pipeline will change the total runtime needed to complete the analysis. The total time estimate varies depending on the computational resources available, including the hardware specifications and software settings. As a rough estimate, the protocol took 36 h of active runtime to execute on our computational server, with six cores, 32 Gb of RAM, and 2 Tb of storage, but this may be faster or slower on other devices. Please click here to download this Table.
Table 2: Automatic assembly of the cellular component proteins. This table shows the number of proteins associated with a given cellular component (i.e., GO term), proteins functionally related to them via protein-protein interactions (PPI), shared pathways (PW), and transcription factor dependence (TFD). The number of total proteins is the number of proteins from all the prior categories combined. All the functionally related proteins were obtained using CaseOLAP LIFT's default parameters. Please click here to download this Table.
Table 3: MeSH label-imputation statistics. This table displays the disease categories, the MeSH tree numbers used as the parent term of all the diseases included in the category, the number of PubMed articles found in each category from 2012-2022, and the number of additional articles included based on the label-imputation step. Please click here to download this Table.
Table 4: Knowledge graph construction statistics. This table describes the statistics for the size of the constructed knowledge graph, including the various nodes and edge types. The CaseOLAP scores represent the relationship between a protein and a cardiovascular disease (CVD) category. Please click here to download this Table.
Table 5: Knowledge graph prediction statistics and validations. This table reports the evaluation metrics for the knowledge graph link prediction of novel/hidden protein-disease associations. The knowledge graph edges were partitioned into 70/30 training and test datasets, and graph connectivity of the edges was preserved in both datasets. The accuracy indicates the proportion of predictions correctly classified, while the balanced accuracy corrects for class imbalance. The specificity indicates the proportion of negative predictions correctly classified. The precision indicates the proportion of correct positive predictions out of all the positive predictions, while the recall indicates the proportion of correct positive predictions out of all the positive edges (i.e., protein-disease associations identified via text-mining). The F1 score is the harmonic mean of the precision and recall. The area under the receiver operating characteristic curve (AUROC) describes how well the model distinguishes between positive and negative predictions, with 1.0 indicating a perfect classifier. The area under the precision-recall curve (AUPRC) measures the trade-off between precision and recall at varying probability thresholds, with higher values indicating better performance. Please click here to download this Table.
Discussion
CaseOLAP LIFT empowers researchers to investigate associations between functional proteins (e.g., proteins associated with a cellular component, biological process, or molecular function) and biological categories (e.g., diseases). The described protocol should be executed in the specified sequence, with protocol section 2 and protocol section 3 being the most critical steps, as protocol section 4 and protocol section 5 depend on their results. As an alternative to protocol section 1, the CaseOLAP LIFT code can be cloned and accessed from the GitHub repository (https://github.com/CaseOLAP/caseolap_lift). It should be noted that despite testing during the software development, bugs may occur. If so, the failed step should be repeated. If the issue persists, it is recommended to repeat protocol section 1 to ensure that the latest version of the docker container is used. Further assistance is available by creating an issue on the GitHub repository for additional support.
This method supports hypothesis generation by enabling investigators to identify entities of interest and reveal the potential associations between them, which may not be readily accessible in existing biomedical resources. The resulting protein-disease associations allow researchers to gain new insights via the scores' interpretable metrics: the popularity scores indicate the most studied proteins in relation to a disease, the distinctiveness scores indicate diseases most unique to a protein, and the combined CaseOLAP score is a combination of the two. To prevent false-positive identifications (e.g., due to homonyms), some text-mining tools utilize a blacklist of terms to avoid9,11. Likewise, CaseOLAP LIFT also utilizes a blacklist but allows the user to tailor the blacklist to their use case. For example, when studying coronary artery disease (CAD), "CAD" should not be considered a name for the protein "caspase-activated deoxyribonuclease". However, when studying other topics, "CAD" might usually refer to the protein.
CaseOLAP LIFT adapts to the quantity of data available for text mining. The date range functionality alleviates the computational burden and creates flexibility for hypothesis generation (e.g., studying how the scientific knowledge on a protein-disease association has changed over time). Meanwhile, the label imputation and full-text components enhance the scope of data available for text-mining. Both components are disabled by default to reduce the computational costs, but the user may decide to include either component. The label imputation is conservative, and it categorizes most publications correctly (87% precision) but misses other category labels (2% recall). This method currently relies on a rule-based heuristic that matches disease keywords, and there are plans to enhance the performance through the use of document topic modeling techniques. Since many uncategorized reports tend to be recent publications, studies investigating a recent date range (e.g., all publications within the last 3 years) are better served by disabling label imputation. The full-text component increases the runtime and storage requirements. Notably, only a minority of documents have the full text available (~14% of documents in our study). Assuming that the protein names mentioned within the publications' methods section are less likely to be related to the disease topics, querying full-text articles excluding the methods section is recommended.
The resulting protein-disease association scores are useful for traditional analyses such as clustering, dimensionality reduction, or enrichment analyses (e.g., GO, pathways), with some implementation included in this software package. To contextualize these scores within existing biomedical knowledge, a knowledge graph is automatically constructed and can be explored using graph visualization tools (e.g., Neo4j32, Cytoscape33). The knowledge graph can also be used for predictive analyses (e.g., link prediction of unreported protein-disease relationships, community detection of protein networks, prize-collecting path-walking methods).
We have examined the model evaluation metrics for the predicted protein-disease associations (Table 5). The model assigns a probability score between 0.0 and 1.0 to each protein-disease association, with scores closer to 1.0 indicating a higher level of confidence in the prediction. The internal evaluation of the model performance, which was based on various metrics including the AUROC, accuracy, balanced accuracy, specificity, and recall, indicated excellent overall performance int his work. However, the evaluation also highlighted a rather poor score for the precision (0.15) of the model, resulting in both a lower AUPRC and F1 score. Future studies to improve this metric will help to elevate the overall performance of the model. We envision this could be achieved by implementing more sophisticated knowledge graph embedding and graph prediction models. Based on the model's precision of 0.15, investigators should anticipate approximately 15% positive identifications; in particular, out of all the 12,688 protein-disease associations predicted by the model, approximately 15% are true-positive associations. This can be mitigated by considering only protein-disease associations with a high probability score (e.g., >0.90); in our use case, filtering with a probability threshold of 0.90 led to high-confidence predictions of 1,583 associations. Investigators may find it helpful to also manually inspect these predictions to ensure high validity (see Figure 7 as an example). An external evaluation of our predictions determined that of the 310 protein-disease associations from an extensive curated database DisGeNet19, 103 were identified in our text-mining study, and 88 additional associations were predicted by our knowledge graph analysis with a probability score >0.90.
Overall, CaseOLAP LIFT features improved flexibility and usability in designing custom analyses of the associations between functional protein groups and multiple categories of disease in large text corpora. This package is streamlined in a new user-friendly command line interface and is released as a docker container, thus reducing the issues associated with configuring the programming environments and software dependencies. The CaseOLAP LIFT pipeline to study mitochondrial proteins in cardiovascular diseases can be easily adapted; for example, future applications of this technique could involve investigating the associations between any proteins associated with any GO terms and any biomedical category. Furthermore, the ranked protein-disease associations identified by this text-mining platform are important in the preparation of the dataset for the use of advanced natural language techniques. The resulting knowledge graph enables investigators to convert these findings into biologically informative knowledge and lays the foundation for follow-up graph-based analyses.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by National Institutes of Health (NIH) R35 HL135772 to P.P., NIH T32 HL13945 to A.R.P. and D.S., NIH T32 EB016640 to A.R.P., National Science Foundation Research Traineeship (NRT) 1829071 to A.R.P. and D.S., NIH R01 HL146739 for I.A., J.R., A.V., K.B., and the TC Laubisch Endowment to P.P. at UCLA.
Materials
| Name | Company | Catalog Number | Comments |
| Software - Docker | Docker | N/A | docker.com |
References
- The UniProt Consortium et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Research. 49, D480-D489 (2021).
- Davis, A. P., et al. Comparative toxicogenomics database (CTD): Update 2023. Nucleic Acids Research. 51, D1257-D1262 (2023).
- Mohtashamian, M., Abeysinghe, R., Hao, X., Cui, L. Identifying missing IS-A relations in orphanet rare disease ontology. Proceedings. IEEE International Conference on Bioinformatics and Biomedicine. 2022, 3274-3279 (2022).
- Rehm, H. L., et al. ClinGen - The clinical genome resource. New England Journal of Medicine. 372 (23), 2235-2242 (2015).
- Caulfield, M., et al. The National Genomics Research and Healthcare Knowledgebase. , (2019).
- Ma, X., Lee, H., Wang, L., Sun, F. CGI: A new approach for prioritizing genes by combining gene expression and protein-protein interaction data. Bioinformatics. 23 (2), 215-221 (2007).
- Gutiérrez-Sacristán, A., et al. Text mining and expert curation to develop a database on psychiatric diseases and their genes. Database. 2017, 043(2017).
- Sigdel, D., et al. Cloud-based phrase mining and analysis of user-defined phrase-category association in biomedical publications. Journal of Visualized Experiments. (144), e59108(2019).
- Yu, K. -H., et al. Systematic protein prioritization for targeted proteomics studies through literature mining. Journal of Proteome Research. 17 (4), 1383-1396 (2018).
- Lau, E., et al. Identifying high-priority proteins across the human diseasome using semantic similarity. Journal of Proteome Research. 17 (12), 4267-4278 (2018).
- Pletscher-Frankild, S., Pallejà, A., Tsafou, K., Binder, J. X., Jensen, L. J. DISEASES: Text mining and data integration of disease-gene associations. Methods. 74, 83-89 (2015).
- Liu, Y., Liang, Y., Wishart, D. PolySearch2: A significantly improved text-mining system for discovering associations between human diseases, genes, drugs, metabolites, toxins and more. Nucleic Acids Research. 43, W535-W542 (2015).
- Minot, S. S., Barry, K. C., Kasman, C., Golob, J. L., Willis, A. D. geneshot: Gene-level metagenomics identifies genome islands associated with immunotherapy response. Genome Biology. 22 (1), 135(2021).
- Lee, S., et al. BEST: Next-generation biomedical entity search tool for knowledge discovery from biomedical literature. PloS One. 11 (10), 0164680(2016).
- Wei, C. -H., Allot, A., Leaman, R., Lu, Z. PubTator central: Automated concept annotation for biomedical full text articles. Nucleic Acids Research. 47 (W1), W587-W593 (2019).
- Jimeno-Yepes, A. J., Sticco, J. C., Mork, J. G., Aronson, A. R. GeneRIF indexing: Sentence selection based on machine learning. BMC Bioinformatics. 14 (1), 171(2013).
- Wei, C. -H., et al. tmVar 2.0: Integrating genomic variant information from literature with dbSNP and ClinVar for precision medicine. Bioinformatics. 34 (1), 80-87 (2018).
- Maglott, D., Ostell, J., Pruitt, K. D., Tatusova, T. Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Research. 33, D54-D58 (2005).
- Piñero, J., et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Research. 48, D845-D855 (2019).
- Lee, J., et al. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 36 (4), 1234-1240 (2020).
- Szklarczyk, D., et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 47, D607-D613 (2019).
- Gillespie, M., et al. The reactome pathway knowledgebase 2022. Nucleic Acids Research. 50, D687-D692 (2022).
- Fang, L., et al. GRNdb: Decoding the gene regulatory networks in diverse human and mouse conditions. Nucleic Acids Research. 49, D97-D103 (2021).
- Doğan, T., et al. CROssBAR: Comprehensive resource of biomedical relations with knowledge graph representations. Nucleic Acids Research. 49 (16), 96(2021).
- Fernández-Torras, A., Duran-Frigola, M., Bertoni, M., Locatelli, M., Aloy, P. Integrating and formatting biomedical data as pre-calculated knowledge graph embeddings in the Bioteque. Nature Communications. 13 (1), 5304(2022).
- Himmelstein, D. S., et al. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. eLife. 6, e26726(2017).
- Zheng, S., et al. PharmKG: A dedicated knowledge graph benchmark for biomedical data mining. Briefings in Bioinformatics. 22 (4), (2021).
- Morselli Gysi, D., et al. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proceedings of the National Academy of Sciences of the United States of America. 118 (19), 2025581118(2021).
- Santos, A., et al. A knowledge graph to interpret clinical proteomics data. Nature Biotechnology. 40 (5), 692-702 (2022).
- Cappelletti, L., et al. GraPE: Fast and scalable graph processing and embedding. arXiv. , (2021).
- Yang, B., Yih, W., He, X., Gao, J., Deng, L. Embedding entities and relations for learning and inference in knowledge bases. arXiv. , (2014).
- Neo4j Graph Data Platform. , Available from: https://neo4j.com/ (2022).
- Shannon, P., et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 13 (11), 2498-2504 (2003).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved