A Practical Guide to Phage- and Robotics-Assisted Near-Continuous Evolution
In This Article
Summary
Phage- and Robotics-assisted Near-continuous Evolution (PRANCE) is a technique for rapid, robust protein evolution. Robotics allows the parallelization of experiments, real-time monitoring, and feedback control.
Abstract
Robotics-accelerated Evolution techniques improve the reliability and speed of evolution using feedback control, improving the outcomes of protein and organism evolution experiments. In this article, we present a guide to setting up the hardware and software necessary to implement Phage- and Robotics-assisted Near-continuous Evolution (PRANCE). PRANCE combines fast phage-based molecular evolution with the ability to run hundreds of independent, feedback-controlled evolution experiments simultaneously. This paper will describe the hardware requirements and setup for PRANCE, including a liquid-handling instrument, a plate reader, auxiliary pumps, heaters, and 3D-printed containers. We describe how to configure the liquid handling robot to be compatible with Python-based open-source software. Finally, we provide suggestions for the first two experiments that can be conducted with a newly constructed PRANCE system that exercises its capabilities and validates that the system is ready to conduct multiplexed evolution. This guide is intended to serve as a handbook for navigating the considerable equipment setup associated with conducting robotics-accelerated evolution.
Introduction
PRANCE is a combination of two powerful directed evolution techniques. First is PACE1, a molecular technique that couples rounds of gene diversification and selection to the fast life cycle of the M13 bacteriophage, enabling rapid rounds of evolution to occur continuously in liquid phage culture. This selection is driven by the use of a plasmid-encoded gene circuit that couples the function of the evolving protein to the expression of pIII, M13's tail coat protein, which is needed for phage propagation, this is illustrated in Figure 1. At the experimental level, continuous dilution of the liquid phage culture allows for continuous selection. Selection stringency can thus be modulated both at the level of the gene circuit as well as at the experimental level by controlling the phage culture dilution rate. PACE can therefore be applied to any biomolecule engineering challenge for which there is a molecular sensor that can detect the desired activity in E. coli bacteria to induce pIII expression. Applications include the evolution of protein-protein binding2,3,4, protein-DNA binding5, protein solubility6, and numerous specific enzymatic functions7. Second is Robotics-accelerated Evolution8,9, which uses a feedback controller to eliminate two common failure modes of directed evolution: extinction, which occurs when the environment is too stringent, and lack of evolution, which occurs when the environment is too lenient. Unlike serial passaging of phage as done in PANCE (Phage-assisted Non-continuous Evolution)7,10, Robotics-accelerated "near-continuous" evolution involves rapid pipetting that maintains cultures at mid-log phase, allowing populations to experience continuous cycles of infection and propagation. When these two technologies are used together, they are referred to as PRANCE, for Phage and Robotics-assisted Near-continuous Evolution8, which enables robust, multiplexed, and rapid continuous evolution. PRANCE has been used to evolve polymerases, tRNAs, and amino-acyl tRNA synthetases and to do feedback control during those evolutions to improve their speed and reliability8.
There are several details of the hardware and software setup for PRANCE that enable the use of bacteriophage on a liquid-handling robot. Instead of using default software provided by the robot manufacturer, we use a python-based open-source software package11, which enables fast, concurrent execution and thus, the ability to keep the semi-continuous bioreactors at mid-log phase. Researcher hands-off time can be extended to several days by having several on-deck components routinely self-sterilize, and this is achieved with automatic control of pumps that can bleach and rinse these components. Phage cross-contamination can be eliminated by the use of a liquid handling robot that does not use force-fit tips and careful adjustment of liquid handling settings.
Protocol
1. Hardware setup
NOTE: See Figure 2 for an overview of the hardware components of a PRANCE system and Figure 3 for photos of these components physically assembled.
- Obtain the primary hardware for the PRANCE system, including a liquid handling instrument, a plate reader, and auxiliary pumps.
NOTE: All PRANCE systems to date have been implemented on Medium to Large liquid handling instruments equipped with an 8-channel, individually addressable pipetting arms, a single-piston 96-tip pipetting arm, a robotic gripper for moving plates, an integrated wash station for tip sterilization, and an integrated plate reader capable of absorbance and luminescence measurements. - Configure the heating strategies depending on the model and features of the liquid-handling robot. Use a heated plate carrier or heater-mediated robot climate control.
- Establish a tip-wash station to allow for tip reuse.
NOTE: To date, PRANCE systems have used off-the-shelf wash stations, although, in principle, this component could easily be constructed out of low-cost components. - Establish a source of bacterial culture maintained at log-phase by setting up a real-time bioreactor running at 37 °C as a chemostat/turbidostat. Alternatively, arrest a log-phase bacterial culture of at least 1 L volume pre-grown at 37 °C in log-phase (OD600 between 0.25 and 0.45) at 4 °C in a nearby refrigerator. Ensure that the culture, whether chilled or warm, is stirred regularly using a shaker plate or stir plate to prevent sedimentation.
- Configure the preferred pumps for robotic integration with the necessary software and drivers. Implement the software to enable the pumps to deliver defined quantities of liquid on the order of 10-100 mL.
NOTE: See the Table of Materials for pumps used in this implementation and the manufacturer's website for software used to operate these pumps and documentation on how to configure them. Such software for the pumps used in the PRANCE setup illustrated in this manuscript is provided open source in the following GitHub repository https://github.com/dgretton/std-96-pace PRANCE requires at least a three-pump manifold capable of pumping three separate channels (deliver bacteria to bacterial reservoir, deliver bleach to bacterial reservoir, and drain bacterial reservoir to waste), with the speed of each calibrated and controlled independently. In the past, people have used fish tank pumps and hydroponics pump arrays, although, in principle, any python-controllable peristaltic pump can be used. Essential functions include the ability to use a robot gripper to transfer plates in or out of the reader, to initiate a plate reader measurement, and to access measurements. - 3D-Print the required custom deck components for the PRANCE system, including, at minimum, the bacterial reservoir/distribution manifold ("waffle"), as found in Supplemental File 1 (https://drive.google.com/file/d/16ELcvfFPzBzNSto0xUrBe-shi23J9Na7/view?usp=share_link). Secure these containers onto the deck and calibrate their positions using standard Liquid Handling Robot software. Connect the reservoir to the pump array.
NOTE: Consult the robot manufacturer's documentation for details of how to perform the calibration as it will be robot-dependent. Resin-based 3D printers are most appropriate; an example of the printer type used is given in the Table of Materials; standard clear resin was used with the default printer settings. - Equip the system with a drain compatible with the local biosafety recommendations.
- Place labware on the deck of the Liquid Handling Robot as exemplified in Figure 4.
- Follow standard safety procedures, including the use of standard laboratory personal protective equipment (i.e., lab coat, gloves, and eye protection).
2. Software preparation
- Install open-source software used for controlling Liquid Handling robots with python11, available from the open-source PyHamilton repository. https://github.com/dgretton/pyhamilton
- Modify and calibrate the deck layout file for the Liquid Handling robot software to accurately reflect the labware positions on the robot deck, as shown in Figure 4.
NOTE: The setup used here uses the software provided by the manufacturer of the liquid handling robot, according to the provided documentation. - Run the PRANCE robot method program in simulation mode.
- Open the Command Line with the following commands (in the Windows operating system), as shown in Figure 5.
Windows key + R
Enter: cmd - Change the parent directory to the directory of the robot method program. Enter a command as below with the correct path, as shown in Figure 5.
CD c:\Robot_methods_directory\PRANCE - Call the robot method program with Python with the simulation mode flag, as shown in Figure 5.
py robot_method.py --simulate - Select the PLAY button in the upper left of the Robot Run Control window that will open when the program is executed (Figure 5).
NOTE: Ensure that the PRANCE method can run without errors in simulation before moving forward. It becomes obvious if the script is able to operate in simulation mode without errors, as it will complete multiple loops of the main program without the system's error handling being called, which terminates the main program loop.
- Open the Command Line with the following commands (in the Windows operating system), as shown in Figure 5.
- Run the PRANCE robot method program with the simulation mode disabled.
- Open the Command Line in the appropriate directory (Figure 5).
Windows key + R
Enter: cmd
CD c:\Robot_methods_directory\PRANCE - Call the robot method program with Python without flags:
py robot_method.py - Select the PLAY button in the upper left of the Robot Run Control window that will open when the program is executed.
- Confirm that PyHamilton can control the instrument and cause it to initialize.
- Open the Command Line in the appropriate directory (Figure 5).
- Establish real-time data synchronization.
NOTE: To date, PRANCE systems have used networked computers that allow users to monitor the log files and real-time plate reader measurement graphs via remote file-sharing software, or via a remote desktop. - Turn off automatic updates.
3. Pre-run preparation
- Ensure that log-phase bacterial culture sources are available for all cultures required for the planned run and that they are being actively stirred to prevent sedimentation. Use an active chemostat/turbidostat or a growth-arrested refrigerated pregrown culture.
- Update the controller manifest file with the details of what volume (range 0-500 µL) of which bacterial culture is to be pumped into each well of the 96-well lagoon per program cycle. This allows precise control of the effective lagoon dilution rate. This can be seen in Figure 6.
- Calculate the Dilution rate of the lagoon using the DilutionCalculator.xlsx spreadsheet (provided as Supplemental File 2), as seen in Figure 7.
- Update the robot_method.py file with the intended lagoon height. To follow this protocol, use 14 (in millimeter units) as the default value for the variable fixed_lagoon_height in the program. This corresponds to a lagoon volume of 550 µL on the system but may differ depending on the particular 96-deep-well plate used.
- Place clean filtered pipette tips onto the robot deck in their designated positions and tape the tip racks to the tip holders to ensure stability during the run.
- Place clean 96-deep-well plates onto the robot deck in their designated positions.
- Place clean 96-well reader plates onto the robot deck in their designated positions.
- Ensure that the plate reader tray is not occupied by a pre-existing plate.
- Ensure that pumps are connected to the computer and are assigned to the correct address.
- Clean the pump lines by activating the pumps to pump bleach and then water.
- Connect pump lines to the appropriate sources and outputs, paying close attention to ensure the correct lines are connected to the relevant bacterial cultures.
- Refill tanks/buckets containing bleach/water for bacterial reservoir and pipette tip washing.
- Ensure all on-deck components, particularly mobile elements, are stabilized in their designated positions.
- Activate heaters as per local implementation to target temperature (i.e. 37 °C; Figure 8).
- Run the UV Sterilization Protocol file for 10 min to operate the built-in UV sterilization lamp in the liquid-handling robots as supplied by the manufacturer (Figure 9).
- Select the PLAY button in the upper left of the Robot Run Control window that will open when the program is executed.
- Run the file with the parametrized option for 600 s.
- Ensure that the Robot Run Control software is closed.
NOTE: The robot method program will crash if there are any existing instances of the Run Control software running.
4. Hardware and software integration
- Conduct a 'water run,' where the PRANCE robot method program is run overnight with water substituting for all cultures and wet reagents.
NOTE: This test can be run at room temperature.- Complete the pre-run preparation as detailed above with the controller_manifest and robot_method set up for an effective lagoon dilution rate of 1 volume/h as shown in Figure 5 and Figure 6.
- Connect the 'bacteria in' line to a container of water to replace log-phase bacteria for the water run.
NOTE: Food coloring can be added to the water sources to track liquid movement through the experiment. - Open the Command Line in the appropriate directory.
- Call the robot method program with Python with the new run flag (py robot_method.py --new) and input the requested arguments, including log file name (TestRun), number of lagoon wells (16), cycle duration (30), number of cycles per reader plate measurement (4), and inducer volume (inducer volume is 0 µL for this test run, during an evolution where mutagenesis is induced with arabinose, this value may be 10 µL), as shown in Figure 5.
- Select the PLAY button in the upper left of the Robot Run Control window that will open when the program is executed once arguments have been provided.
NOTE: The PRANCE method can be started using an empty lagoon plate, and the liquid volume of the lagoons will equilibrate to the final volume over the first six cycles.
- Conduct a 'bacteria-only run', where the PRANCE protocol is run overnight only with bacterial culture at target temperature but with no bacteriophage.
- Complete the pre-run preparation as detailed above with the controller_manifest and robot_method set up for an effective lagoon dilution rate of 1 volume/h, as shown in Figure 5 and Figure 6. Ensure that heaters are switched on for a target temperature of 37 °C.
- Connect the 'bacteria in' line to the selected source of log-phase bacteria.
- Open the Command Line in the appropriate directory.
- Call the robot method program with Python with the new run flag (py robot_method.py --new) and input the requested arguments, as detailed previously in section 4.1.4.
- Select the PLAY button in the upper left of the Robot Run Control window that will open when the program is executed once the arguments have been provided.
- Run an 'infection test,' where phages bearing an evolved protein are challenged to propagate on bacteria requiring that protein.
NOTE: Decide in advance which lagoons will be inoculated with phage and which lagoons will not be inoculated and thus serve as no-phage control lagoons to detect cross-contamination.- Complete the pre-run preparation as detailed above with the controller_manifest and robot_method set up for an effective dilution rate of 1 volume/h, as shown in Figure 5 and Figure 6. Ensure that heaters are switched on for a target temperature of 37 °C.
- Connect the 'bacteria in' line to the selected source of log-phase bacteria.
- Open the Command Line in the appropriate directory.
- Call the robot method program with Python with the new run flag (py robot_method.py --new) and input the requested arguments as detailed previously in section 4.1.4.
- Select the PLAY button in the upper left of the Robot Run Control window that will open when the program is executed once arguments have been provided.
- Before adding bacteriophage, run the method for 2-3 h to equilibrate the volume and bacteria OD in the lagoon plates.
- Inoculate the with-phage lagoons with 106 pfu/mL of bacteriophage at the end of a run cycle when the program is sleeping (e.g., 5.5 µL of phage aliquot at 108 pfu/mL, as determined by plaque assay or qPCR), into a 550 µL lagoon.
- Run the program overnight and then check the phage titer in the lagoon wells by plaque assay or qPCR.
Representative Results
Infection test results
This test will reveal problems with bacterial culture, phage cloning and titer, temperature stability of the equipment, liquid handling settings, and plate reader integration. A successful phage infection test will reveal clear and rapid phage infection in lagoons inoculated with phage, and no signal in no-phage lagoons. Figure 10 shows some representative results of a phage infection test. Experimental results can also be compared to Figures 1d and 1c of this PRANCE paper8, depending on whether a "hot PRANCE" (fed by a live bacterial turbidostat) or "cool PRANCE" (fed by chilled mid-log phase culture) configuration is being implemented. This test may reveal several common issues. Issues with bacterial culture preparation can often result in weak or absent infection. Bacteria can only be optimally infected by M13 phage when they are in mid-log phase and at 37 °C. At other temperatures and growth stages, they exhibit weaker pilus expression and thus are less susceptible to phage infection12. Inoculating with low-titer phage, or phage with backbone mutations can result in delayed or absent signal. Issues with plate reader gain settings for fluorescence or luminescence will be revealed by this test.
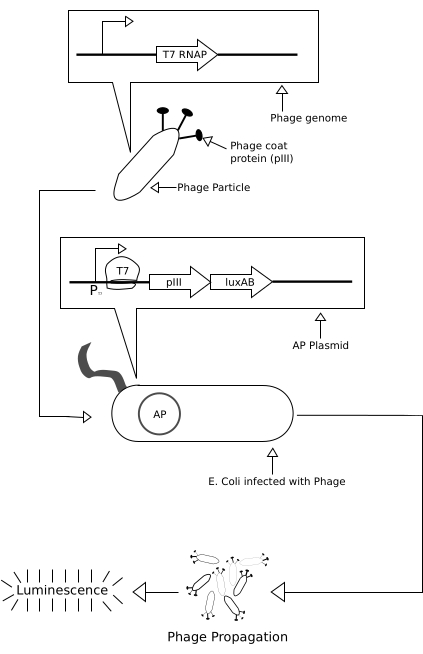
Figure 1: Schematic of the genetic circuit operating during the infection test run of the PRANCE apparatus. When T7 RNA polymerase, encoded on the phage genome, infects the Escherichia coli host, it is transcribed and binds on the AP at the T7 promoter, which leads to transcription of the pIII phage protein and luxAB protein, which, in turn, facilitates phage propagation and production of luminescence. Abbreviations: PRANCE = Phage- and Robotics-assisted Near-continuous Evolution; AP = accessory plasmid. Please click here to view a larger version of this figure.
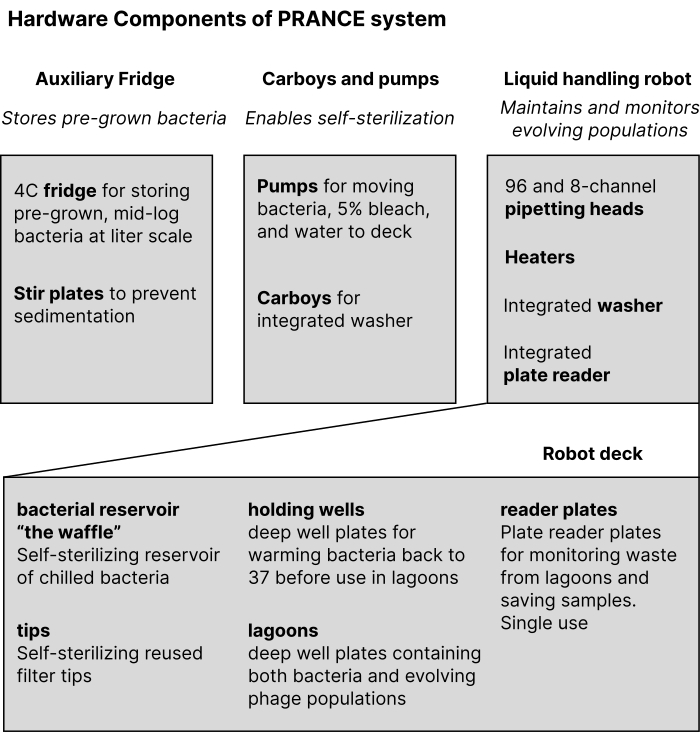
Figure 2: A schematic of the physical components of the PRANCE system. A fridge stores stirred cultures, which are then moved onto the robot deck by an array of pumps, to the bacterial reservoir, "the waffle." The liquid-handling robot is used to move bacterial cultures from "the waffle" using the pipetting head to the holding wells to warm up to incubation temperature, and then to the lagoons where the main incubation occurs. Both the holding wells and the lagoons are standard 2 mL deep-well plates. The robot takes samples into single-use reader plates, which are in turn moved to a plate reader for measurement. Abbreviation: PRANCE = Phage- and Robotics-assisted Near-continuous Evolution. Please click here to view a larger version of this figure.

Figure 3: The PRANCE robotic apparatus. (A) PRANCE setup. (I) HEPA filter and external heater. (II) Culture refrigerator. (III) Main robot enclosure. (IV) Plate reader. (V) Pumps and tanks. (B) Robot enclosure. (VI) Main culture pumps. (VII) Water, waste, and bleach tanks. (VIII) Washer pumps. (C) Robot enclosure. (IX) Robot pipetting arm and gripper. (X) Pipette tips. (XI) 3D-Printed component to allow culture distribution onto the robot ("the waffle"). (XII) Plates for sampling in the plate reader. (XIII) Buckets for tip washing. (XIV) "Lagoons": culture vessels where evolutionary culturing takes place. Abbreviations: PRANCE = Phage- and Robotics-assisted Near-continuous Evolution; HEPA = high-efficiency particulate air. Please click here to view a larger version of this figure.
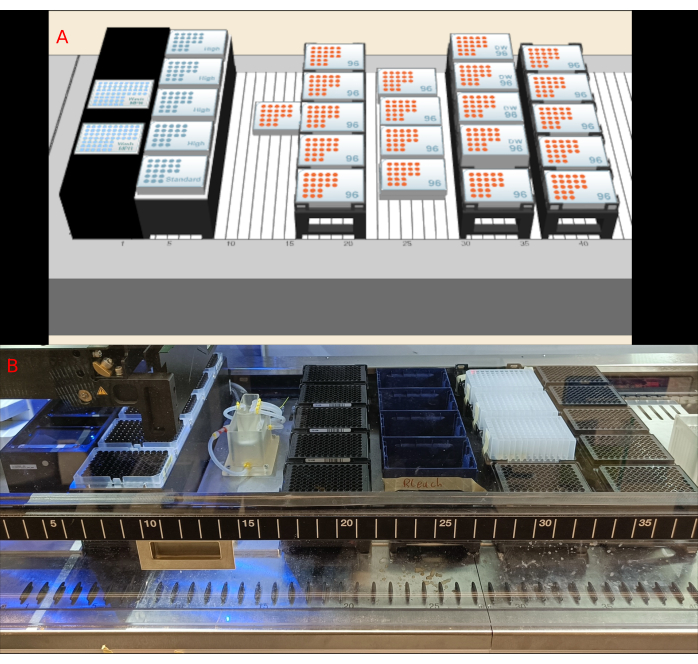
Figure 4: Deck Layout. (A) 3D representation of the deck layout in the robot control software. (B) Photograph of the deck components. Please click here to view a larger version of this figure.
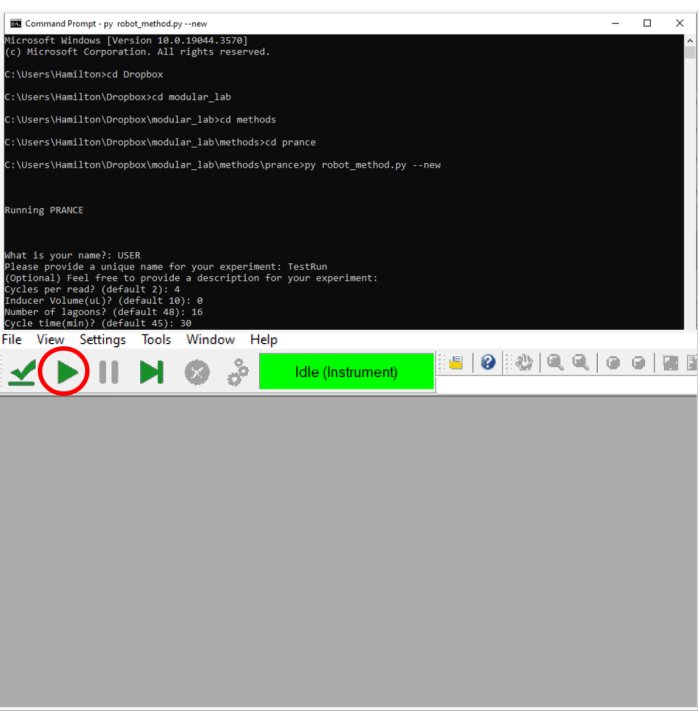
Figure 5: Screenshot of the command line with example parameters (above) and run control software (below). The play button is located at the top left and can be clicked with a mouse or actuated with a touchscreen depending on local implementation. Please click here to view a larger version of this figure.
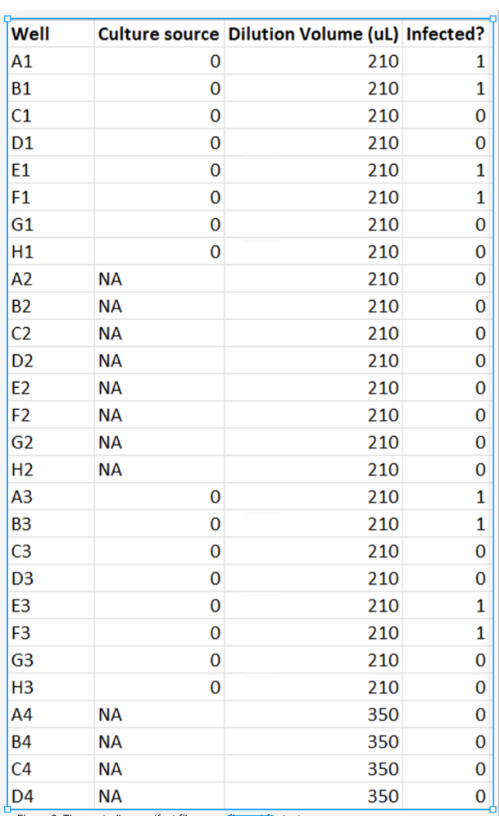
Figure 6: The controller manifest file as configured for test runs. Lagoons containing culture #0 would be in columns 1 and 3 of the 96-deep-well plate. Remaining columns would be empty. Rows A, B, D, and E of the 96-deep-well-plate are marked on the right column for infection by phage (1), the other rows (0) are no-phage controls.This instance of the controller manifest would result in the program diluting the lagoon with 210 µL of culture every cycle. Please click here to view a larger version of this figure.
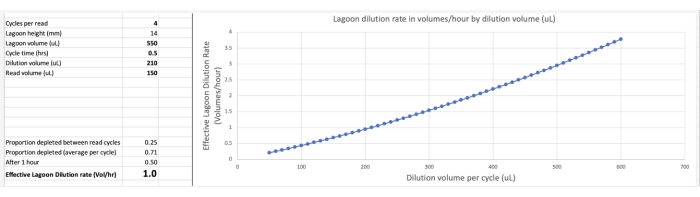
Figure 7: Calculation of the effective lagoon dilution rate using the DilutionCalculator Spreadsheet. See Supplemental File 2 for the DilutionCalculator Spreadsheet. As seen in this figure, a 550 µL lagoon that is diluted by 210 µL of fresh culture every 30 min cycle, with 150 µL samples for reader plate measurement being taken every four cycles will correspond to an effective dilution rate of 1.0 lagoon volumes/h (after every 1 h, 50% of the original lagoon liquid at the start of the hour will remain) Please click here to view a larger version of this figure.

Figure 8: Robot heater system. The heater is activated by plugging in the power supply as indicated by the red circle. Please click here to view a larger version of this figure.
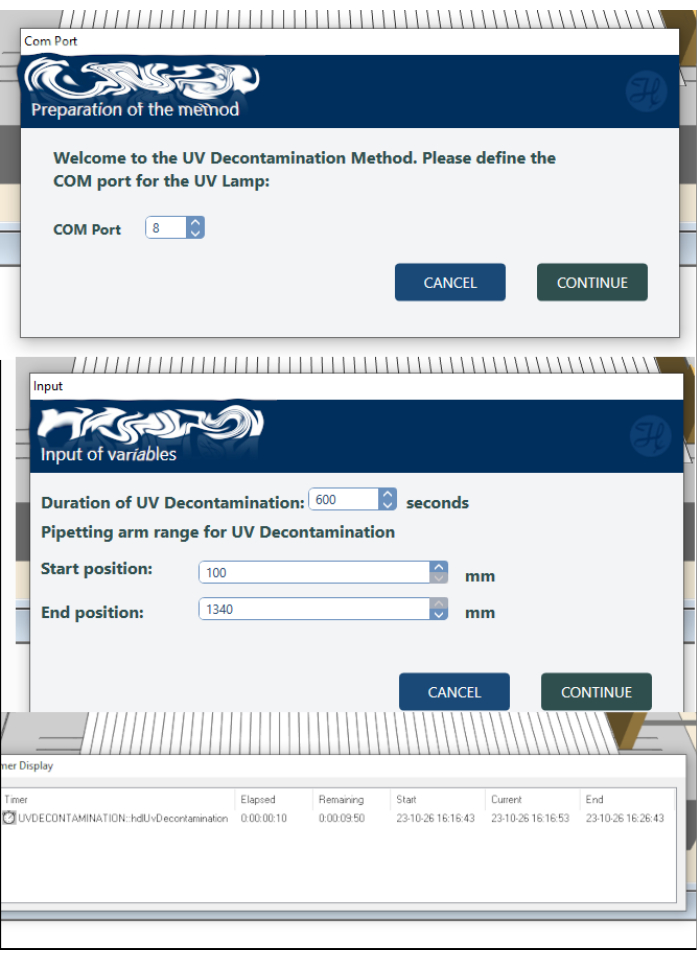
Figure 9: Settings of the UV decontamination protocol. Please click here to view a larger version of this figure.
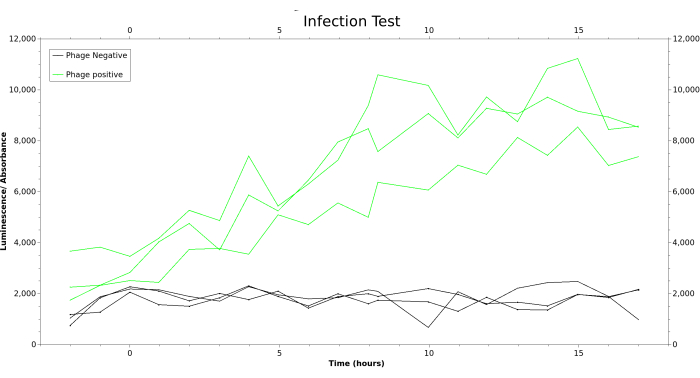
Figure 10: A measurement of an infection test run on the PRANCE system. Samples are taken during the run and measurements of the luminescence and absorbance are made. For each lagoon, the luminescence measurements are divided by the corresponding absorbance measurement and plotted as a function of time. The lagoons that have been infected with Phage are colored in green, whereas the uninfected control lagoons are colored in black. Abbreviation: PRANCE = Phage- and Robotics-assisted Near-continuous Evolution. Please click here to view a larger version of this figure.
Supplemental File 1: STL file for 3D-printing the required custom deck components for the PRANCE system, including, at minimum, the bacterial reservoir/distribution manifold ("waffle"). Please click here to download this File.
Supplemental File 2: DilutionCalculator Spreadsheet. Please click here to download this File.
Discussion
Despite efforts to standardize equipment, practically speaking, every PRANCE setup will be different due to changes in equipment supply, hardware, and software versioning. As a result, each PRANCE setup manifests unique setup challenges, demanding a comprehensive understanding of the purpose of each component for effective modular troubleshooting.
This method delineates a step-by-step protocol for the setup and testing of an established PRANCE system. We first focus on the critical elements of the hardware and software and then detail the essential steps to prepare for and conduct a series of test runs, which establish that the system is ready for PRANCE.
An essential feature of the hardware is optimization to reduce the risk of sample cross-contamination during multiplexed experiments using bacteriophage. It is recommended to use exclusively filtered tips with robot tip technology that is compatible with tip reuse and is thought to minimize aerosols produced during tip ejection by avoiding force-fit tips. Robust tip washing as per this protocol allows for tip reuse although the adequacy of this must be validated as part of the infection test on each system. Self-sterilization is also dependent on a consistent supply of water and bleach for the system. These are stored in tanks/buckets and if depleted will result in impaired self-sterilization and rapid cross-contamination. Photographs can be taken of the tanks/buckets taken before and after the program runs to benchmark the rate at which the washing equipment consumes water and bleach given a particular pump setup.
Another key element of the system is the maintenance of the bacterial growth phase and temperature. PRANCE experiments are conducted using the S2060 E. coli bacterial strain (Addgene: #105064). This is a K12-derived F-plasmid-containing strain optimized to reduce biofilms7. In addition, the F-plasmid in this strain has been edited with the addition of a tetracycline resistance cassette for plasmid maintenance, luxCDE and luxR to complement luxAB-mediated luminescence monitoring, as well as lacZ under the phage shock promoter to allow for colorimetric visualization of plaques. The F-plasmid-encoded F-pilus is necessary for M13 phage infection. Bacteria used in PACE must therefore be cultured at 37 °C and at mid-log phase when the F-pilus12 is expressed and M13 phage infection, propagation, and evolution are possible. For static temperature regulation, an off-the-shelf heated plate carrier can be employed. An alternative is simply heating the air going into the HEPA filter using inexpensive heaters, though this is not recommended as it may lead to accelerated wear and tear on the hardware. In addition, this accelerates the evaporation of auxiliary on-deck fluids, such as the bleach/water buckets and inducer, when used.
Calibration of the software packages is also essential for proper system function. Divergences between the software deck layout and the actual robot deck are the most common cause of system failure during operation. Regular calibration of the auxiliary pumps that supply bacterial culture, bleach, and drain the system is vital as peristaltic pump usage can lead to tubing wear and fluid volume alterations.
The water run test will rapidly reveal a number of common setup problems, including incorrect liquid handling settings, fluidics leaks/faulty connections, and software instability. A successful water run will exhibit no unexpected liquid leaks and run stably without errors overnight. There are a number of common issues that may arise during a water run such as failure to execute certain liquid-handling steps, dripping from pipettes, and the protocol stopping mid-run. In case of failure to execute certain liquid handling steps, confirm that all liquid classes have been installed. These list the appropriate viscosity and pipetting speeds and are adjusted in the robot control software provided by the manufacturer. If there is dripping from pipettes, it is important for robot pipetting arm settings to be correct to enable clean pipetting and eliminate phage cross-contamination. Successful robotic pipetting requires, in addition to correct liquid classes, correct deck layout heights of all labware, and appropriate pipetting height offsets specified in the PRANCE robot method program. These height offsets may require direct adjustment. If the protocol stops mid-run, often this will be generated by a wide array of errors that indicate that the deck layout file may not match the actual deck configuration.
The bacteria-only run test will reveal issues with plate reader settings and real-time data visualization, problems with excessive bleach concentration or insufficient rinsing, and temperature stability. A successful bacteria-only run will exhibit equilibration of lagoon absorbance over the first three cycles, followed by stable absorbance for the duration of the run. In addition, it may reveal several common issues. This is the first step where the data generated by the plate reader are plotted. Data in the plate reader database may not be saved properly or plotted properly. If bacteria fail to equilibrate in their absorbance, this may indicate that the bleach concentration is too high. Excessive bleach or insufficient washing can sterilize the entire experiment, rather than just the piece of labware. If this is suspected, bleach-detecting strips can be used to test the lagoon. The stability of the temperature of the culture can be checked with a thermometer gun.
A successful infection test indicates that the system is ready for PRANCE runs. An infection test can be performed by inoculating a subset of lagoons containing bacterial culture. These bacteria will express pIII when infected by the appropriate phage that lacks the gene for pIII (ΔgIII), allowing phage propagation. One possible combination for testing is to use S2060 bacteria transformed with a plasmid expressing pIII under the phage shock promoter with any ΔgIII phage. We recommend using ΔgIII phage bearing the wild-type T7 RNA Polymerase with S2060 bacteria transformed with an accessory plasmid, in which pIII and luxAB are driven by the T7 promoter (Plasmid pJC173b13), as illustrated in Figure 1. This also allows plate-reader-mediated monitoring of infection during the test run. Definitive evidence of the success of the infection test and lack of cross-contamination will come from phage titering of test and control lagoons. Where a luciferase reporter is used, an increase in luminescence in test wells only, as seen in Figure 3, is also an indicator of successful phage infection and propagation. The gold standard for phage titer quantification is the plaque assay7. There is also a protocol for M13 quantification by qPCR7 that may be faster, although this does not discriminate between infectious and non-infectious phage particles and thus may overestimate titers.
The main program references a manifest file, this is a plain text database file, which dictates the dilution volume per cycle of each propagating culture as well as the selection of any number of potential bacterial culture feedstocks, which may differ in selection stringency. In this manner, the manifest file defines many of the parameters of the PRANCE run. It should be noted that this file can be edited during the run by either the operator or the system, meaning that manual or automatic feedback control can be effected.
The utility of a fully functioning PRANCE setup lies in its capacity to rapidly evolve large populations in a carefully monitored and controlled environment. The plate-based format distinguishes PRANCE from other techniques, like using smaller off-the-shelf turbidostat-based systems14,15. The plate-based setup not only facilitates easy integration with additional robotic processing steps but also compatibility with other laboratory instruments such as centrifuges. Moreover, the ability to conduct accelerated evolution concurrently across multiple instances introduces an additional dimension to the experiment, enhancing the prospect of achieving diverse and robust results. The granular control and feedback system integral to PRANCE further bolsters the predictability and reliability of the experiment, marking a significant advancement in the field of directed evolution techniques. However, this technique is limited in the number of parallel experiments it can conduct. Depending on the configuration, PRANCE setups are usually limited either by robot pipetting speed or by available deck space.
The same hardware and software used for PRANCE can also be applied to evolution methods that do not involve bacteriophage. As demonstrated in the many-turbidostats method11, this same instrument can be employed exclusively with bacteria, enabling whole-genome adaptive evolution experiments. This adaptability widens the scope of this instrument, paving the way for new forms of Robotics-accelerated Evolution.
Acknowledgements
We thank Emma Chory and Kevin Esvelt for their help and advice with hardware and software setup. Samir Aoudjane, Osaid Ather, and Erika DeBenedictis are supported by the Steel Perlot Early Investigator Grant. This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (CC2239), the UK Medical Research Council (CC2239), and the Wellcome Trust (CC2239).
Materials
| Name | Company | Catalog Number | Comments |
| 3D printed bacterial reservoir "waffle" | - | - | https://drive.google.com/file/d/16ELcvfFPzBzNSto0xUrBe-shi23J9Na7/view; For Robot deck |
| 3D printer | FormLabs | Form 3B+ | 3D printer components |
| 3D printer resin (clear) | FormLabs | RS-F2-GPCL-04 | consumable for 3D printer |
| 8-1,000 µL head | Hamilton | 10140943 | For Liquid handling robot |
| 96-1,000 µL pipetting head | Hamilton | 10120001 | For Liquid handling robot |
| Black polystyrene plate reader microplates | Millipore Sigma | CLS3603 | For Robot deck |
| BMG Labtech Spectrostar FLuorstar Omega | BMG Labtech | 10086700 | For Liquid handling robot |
| Cleaning solution | Fluorochem Limited | F545154-1L | used to clean the liquid handling parts of the robot |
| Deep Well plates | Appleton Woods | ACP006 | these are used to contain evolving bacteria on the deck of the robot |
| encolsure heater | Stego | 13060.0-01 | heats inside robot enclosure |
| Hamilton STAR | Hamilton | 870101 | For Liquid handling robot |
| Heater | Erbauer | BGP2108-25 | For Liquid handling robot |
| HIG Bionex centrifuge | Hamilton | 10086700 | For Liquid handling robot |
| iSWAP plate gripper | Hamilton | 190220 | For Liquid handling robot |
| laboratory tubing | Merck | Z280356 | to construct liquid handling manifold |
| luer to barb connector | AIEX | B13193/B13246 | for connectorizing tubing |
| Magnetic stir plate | Camlab | SKU - 1189930 | For Auxiliary Fridge |
| Molcular pipetting arm | Hamilton | 173051 | For Liquid handling robot |
| Omega | BMG labtech | 5.7 | plate reader control software |
| One way Check Valves | Masterflex | MFLX30505-91 | to one way sections of liquid handling manifold |
| pyhamilton | MIT/Open source | https://github.com/dgretton/std-96-pace%20PRANCE | open source python robot control software |
| pymodbus | opensource | 3.5.2 | python pump software interface |
| Refrigetator | Tefcold | FSC175H | allows cooled bacteria to be used instead of turbidostat |
| S2060 Bacterial strain | Addgene | Addgene: #105064 | E. coli |
| temperature controller | Digiten | DTC102UK | Used to control heaters thermostatically |
| Thermostat switch controller | WILLHI | WH1436A | WILLHI WH1436A 10 A Temperature Controller 110 V Digital Thermostat Switch Sous Vide Controller NTC 10K Sensor Improved Version; for Liquid handling robot |
| Venus | Hamilton | 4.6 | proprietary robot control software |
| Wash Station for MPH 96/384 | Hamilton | 190248 | For Liquid handling robot |
| Suggested pump manufacturers | |||
| Company | Catalog number | Notes | Documentation |
| Agrowtek | AD6i Hexa Pump | https://www.agrowtek.com/doc/im/IM_ADi.pdf | |
| Amazon | INTLLAB 12V DC | ||
| Cole-Parmer | EW-07522-3 | Masterflex L/S Digital Drive, 100 RPM, 115/230 VAC | https://pim-resources.coleparmer.com/instruction-manual/a-1299-1127b-en.pdf |
| Cole-Parmer | EW-07554-80 | Masterflex L/S Economy variable-speed drive, 7 to 200 rpm, 115 VAC | https://pim-resources.coleparmer.com/instruction-manual/a-1299-1127b-en.pdf |
References
- Esvelt, K. M., Carlson, J. C., Liu, D. R. A system for the continuous directed evolution of biomolecules. Nature. 472, 499-503 (2011).
- Pu, J., Zinkus-Boltz, J., Dickinson, B. C. Evolution of a split RNA polymerase as a versatile biosensor platform. Nat Chem Biol. 13 (4), 432-438 (2017).
- Pu, J., Disare, M., Dickinson, B. C. Evolution of C-terminal modification tolerance in full-length and split T7 RNA polymerase biosensors. Chembiochem. 20 (12), 1547-1553 (2019).
- Xie, V. C., Styles, M. J., Dickinson, B. C. Methods for the directed evolution of biomolecular interactions. Trends Biochem Sci. 47 (5), 403-416 (2022).
- Popa, S. C., Inamoto, I., Thuronyi, B. W., Shin, J. A. Phage-assisted continuous evolution (PACE): A guide focused on evolving protein-DNA interactions. ACS Omega. 5 (42), 26957-26966 (2020).
- Wang, T., Badran, A. H., Huang, T. P., Liu, D. R. Continuous directed evolution of proteins with improved soluble expression. Nat Chem Biol. 14 (10), 972-980 (2018).
- Miller, S. M., Wang, T., Liu, D. R. Phage-assisted continuous and non-continuous evolution. Nat Protoc. 15 (12), 4101-4127 (2020).
- DeBenedictis, E. A., et al. Systematic molecular evolution enables robust biomolecule discovery. Nat Methods. 19 (1), 55-64 (2022).
- Zhong, Z., et al. Automated continuous evolution of proteins in vivo. ACS Synth Biol. 9 (6), 1270-1276 (2020).
- Roth, T. B., Woolston, B. M., Stephanopoulos, G., Liu, D. R. Phage-assisted evolution of Bacillus methanolicus methanol dehydrogenase 2. ACS Synth Biol. 8 (4), 796-806 (2019).
- Chory, E. J., Gretton, D. W., DeBenedictis, E. A. Enabling high-throughput biology with flexible open-source automation. Mol Syst Biol. 17 (3), 9942 (2021).
- Novotny, C. P., Lavin, K. Some effects of temperature on the growth of F pili. J Bacteriol. 107 (3), 671-682 (1971).
- Carlson, J. C., Badran, A. H., Guggiana-Nilo, D. A., Liu, D. R. Negative selection and stringency modulation in phage-assisted continuous evolution. Nat Chem Biol. 10 (3), 216-222 (2014).
- Steel, H., Habgood, R., Kelly, C., Papachristodoulou, A. In situ characterization and manipulation of biological systems with Chi.Bio. PLOS Biology. 18 (7), e3000794 (2020).
- Wong, B. G., Mancuso, C. P., Kiriakov, S., Bashor, C. J., Khalil, A. S. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nat Biotechnol. 36 (7), 614-623 (2018).
This article has been published
Video Coming Soon
ABOUT JoVE
Copyright © 2024 MyJoVE Corporation. All rights reserved