A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
Single-Animal, Single-Tube RNA Extraction for Comparison of Relative Transcript Levels via qRT-PCR in the Tardigrade Hypsibius exemplaris
In This Article
Summary
This work presents a rapid RNA extraction and transcript level comparison method for analyzing gene expression in the tardigrade Hypsibius exemplaris. Using physical lysis, this high-throughput method requires a single tardigrade as the starting material and results in robust production of cDNA for quantitative reverse transcription polymerase chain reaction (qRT-PCR).
Abstract
The tardigrade Hypsibius exemplaris is an emerging model organism renowned for its ability to survive environmental extremes. To explore the molecular mechanisms and genetic basis of such extremotolerance, many studies rely on RNA-sequencing (RNA-seq), which can be performed on populations ranging from large cohorts to individual animals. Reverse transcription polymerase chain reaction (RT-PCR) and RNA interference (RNAi) are subsequently used to confirm RNA-seq findings and assess the genetic requirements for candidate genes, respectively. Such studies require an efficient, accurate, and affordable method for RNA extraction and measurement of relative transcript levels by quantitative RT-PCR (qRT-PCR). This work presents an efficient single-tardigrade, single-tube RNA extraction method (STST) that not only reliably isolates RNA from individual tardigrades but also reduces the required time and cost for each extraction. This RNA extraction method yields quantities of cDNA that can be used to amplify and detect multiple transcripts by quantitative PCR (qRT-PCR). The method is validated by analyzing dynamic changes in the expression of genes encoding two heat-shock-regulated proteins, Heat-Shock Protein 70 β2 (HSP70 β2) and Heat-Shock Protein 90α (HSP90α), making it possible to assess their relative expression levels in heat-exposed individuals using qRT-PCR. STST effectively complements existing bulk and single tardigrade RNA extraction methods, permitting rapid and affordable examination of individual tardigrade transcriptional levels by qRT-PCR.
Introduction
Tardigrades are small multicellular animals renowned for their ability to survive extreme conditions that are lethal to most other forms of life1. For example, these animals can survive nearly 1000 times the dose of ionizing radiation that is lethal to humans2,3,4,5,6,7,8,9,10, nearly complete desiccation11,12,13,14,15, freezing in the absence of added cryoprotectants16,17,18, and, in their desiccated state, even the vacuum of space19,20. Owing to their unique capacity for survival in extreme environments, these animals have become foundational models for understanding extremotolerance in complex, multicellular organisms1,21,22,23.
Stable genetic manipulation of these remarkable animals, including transgenesis and germline gene modification, has remained elusive until recently24,25. As such, most experiments to reveal molecular mechanisms of extremotolerance are performed through transcriptional profiling via RNA sequencing. Many valuable and informative RNA sequencing data sets exist for tardigrades under various extreme conditions, ranging from radiation8,9,26,27,28, heat stress29, freezing stress12, and desiccation27,30,31,32,33. Some of these studies have utilized bulk RNA extraction and purification methods to illuminate our molecular understanding of extremotolerance. However, bulk extraction of RNA transcripts from many animals prevents analysis of variation in gene expression between individuals, thus missing the potential richness of more refined data sets. Importantly, these studies often analyze heterogeneous populations of animals that include both animals that survive environmental stressors and those that do not. As such, these studies are confounded by averaging expression data from multiple and potentially dramatically different response states. To address this issue, Arakawa et al., 201634 developed an elegant low-input RNA-seq pipeline that applies an RNA extraction kit followed by a linear PCR amplification step using single34,35,36 or multiple30,37,38animals as input. These studies have been foundational to our understanding of tardigrade extremotolerance22. Interestingly, this protocol has also been applied to qRT-PCR using seven animals as starting material24.
In most model organisms, having identified potential targets via RNA-seq, qRT-PCR is then performed to confirm transcriptional changes identified by RNA-seq and assess the expression time course of candidate genes in a high-resolution manner. To test the function of identified genes, such studies are often followed by RNAi-mediated knockdown of molecular targets39,40 and analysis of extremotolerant capacity12,41. The efficacy of each RNAi knockdown is typically confirmed by qRT-PCR by directly monitoring the decrease in transcript abundance. However, RNAi is a labor-intensive process in tardigrades as each dsRNA must be delivered via manual microinjection of individuals39,40. Owing to the low throughput nature of this strategy, a rapid, low-cost RNA extraction method adapted for qRT-PCR from single animals would be highly valuable for tardigrade research. Although previous methods have been developed to extract RNA from single tardigrades, these protocols have not combined their extraction with qRT-PCR, instead relying on optical density-based methods12,40,41. Motivated by these challenges, we sought to develop a protocol that reliably yields RNA in quantity and quality that can be used for qRT-PCR from single H. exemplaris.
Adapted from a single-animal RNA extraction protocol developed for Caenorhabditis elegans42, STST is optimized for H. exemplaris. The extraction method consists of six rapid freeze-thaw steps, physically disrupting the cuticle, allowing RNA extraction and subsequent cDNA synthesis. The STST method decreases extraction time by more than 24-fold compared to bulk RNA extraction methods, as described by Boothby, 201843, and by 30% compared to single tardigrade RNA extraction kits, as described by Arakawa et al., 201634. Further, the number of sample-experimenter interactions is decreased from 5 to only 1 compared to RNA extraction kit preparations, thus reducing the risk of contamination by exogenous ribonucleases. When querying for highly expressed genes, the STST method produces sufficient cDNA for 25 quantitative RT-PCR reactions per single tardigrade, requiring only 1 μL of the total 25 μL cDNA volume per reaction. However, template concentrations need to be empirically determined for lower abundance transcripts.
The efficacy of the STST method for analyzing dynamic changes in gene expression was evaluated by investigating the differential expression of the genes encoding heat-shock protein-90α (HSP90α) and heat-shock protein 70β2 (HSP70β2) in response to short-term heat-shock at 35°C for 20 minutes. Both HSP70β2 and HSP90α in most eukaryotic organisms are rapidly upregulated following short-term heat-shock exposure (20 min)42. Analysis in H. exemplaris revealed that both the HSP70β2 and HSP90α-encoding RNAs extracted from single heat-treated tardigrades showed statistically significant increases in expression following short-term heat exposure. These findings demonstrate that the STST protocol can be used to analyze dynamic changes in gene expression in individual animals over time.
The STST extraction method should complement existing experimental methods such as RNA-seq by facilitating rapid and inexpensive RNA extraction and subsequent comparison of transcript levels by qRT-PCR. This method will also be valuable for assessing the efficiency and penetrance of RNAi in manually injected individuals more quantitatively than optical density alone. Finally, owing to their similar cuticular structures and physical characteristics, it is likely that this method will also be effective for analyzing gene expression in other tardigrade species44.
Access restricted. Please log in or start a trial to view this content.
Protocol
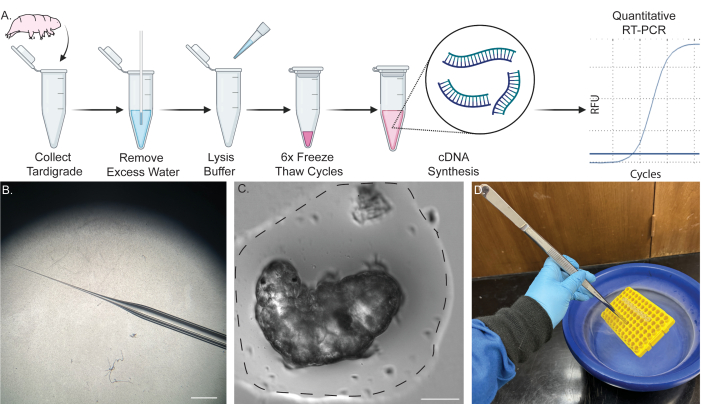
Figure 1: Single-tube pipeline for RNA extraction from a single tardigrade. (A) Scheme showing the protocol for RNA extraction from a single tardigrade, including six freeze-thaw cycles and subsequent cDNA synthesis. Samples may subsequently be used for RT-PCR and qRT-PCR. (B) Image of micropipette taper used for removal of water. Scale bar: 2 mm. (C) Bright field image of a tardigrade in a small volume of water (dotted line). Removal of most water to the extent shown is required for successful extraction and prevents dilution of lysis buffer. Scale bar: 50 μm. (D) Image showing immersion of samples in liquid nitrogen using long forceps to rapidly freeze-thaw the samples safely. Some of the content was created in BioRender. Kirk, M. (2022) BioRender.com/d93s511 Please click here to view a larger version of this figure.
NOTE: Figure 1A shows a schematic of the procedure. For detailed tardigrade and algal culturing procedures, refer to previously published reports45,46,47.
1. Sterilization of spring water
- Pour 2 L of spring water from a 5-gallon water jug (see Table of Materials for Specifics) into a 2 L autoclave-safe glass bottle.
- Place the cap on the autoclave-safe bottle and seal with a small amount of autoclave tape. Do not tighten the bottle; place the cap on top.
- Autoclave the spring water for 50 min on a wet cycle with no drying step.
- Allow the water to come to room temperature (RT), and seal the cap firmly before storing it at RT.
2. Glass micropipette pulling (with a pipette puller)
- Secure a glass micropipette (O.D.: 1 mm, I.D.: 0.58 mm, Length: 10 cm) on a micropipette puller. Avoid contact with the heating filament, as this will alter the pipette shape and damage the filament.
- Determine the pulling of the pipette empirically for each filament and pipette puller. However, to serve as a starting point for optimization, use 78 °C and a single pull step of pull weight of 182.2 g.
- Allow the filament to heat and gravity to separate the glass micropipette into two glass micropipettes with sharp points (Figure 1B).
- Store these pulled glass micropipettes in a closed 100 mm Petri dish with wax or clay to hold them in place and prevent the sharp tips from breaking.
3. Glass micropipette pulling (without a pipette puller)
- Light a Bunsen burner or other controlled flame source on a low setting.
- Take a glass micropipette with one end in each hand.
- Hold the center of the glass micropipette over the flame until the glass begins to melt. Then, rapidly pull the two ends apart. This will create two very delicate sharp tips.
- Lightly break the tip with a pair of sterile fine forceps.
- Store these pulled glass micropipettes in a closed 100 mm Petri dish with wax or clay to hold them in place and prevent the sharp tips from breaking.
4. RNA extraction
- Obtain 0.5 L of liquid nitrogen in a cryo-safe container.
CAUTION: Liquid nitrogen is cryogenic and may cause burns if exposed to skin or eyes. When handling, use protective clothing, splash goggles, nitrile gloves, cryo-gloves, a lab coat, and closed-toed shoes. Ascertain that the container is liquid nitrogen safe before transporting the liquid. Using an ethanol-dry ice bath for this step may also be possible. - Make cDNA synthesis master mix: a 10 µL solution containing 1 µL of random hexamer primer, 2 µL of DNase, 4 µL of 5x RT Buffer, 1 µL Enzyme Mix, 1 µL of H2O, and 1 µL of 10 mM dNTPs. Store this solution on ice.
- Prepare tardigrade lysis buffer (5 mM Tris (pH = 8), 0.5% (v/v) Detergent 1, 0.5% (v/v) Detergent 2, 0.25 mM EDTA in sterile nuclease-free Water).
NOTE: This solution can be stored on the bench top for 6 months. However, maintain sterility and avoid potential RNAse-contaminating sources. - Aliquot enough lysis buffer for extractions (2 μL/ tardigrade).
- Add RNAse inhibitor to the tardigrade lysis buffer solution to a final concentration of 4 U/μL.
- Vortex and spin down the solution at RT on a bench-top centrifuge at a speed of 2000 x g for 5 s before storing the solution on ice.
- Remove as many tardigrades as needed for the experiment from culture using a sterile filter-tipped P1000 pipette and place them in a sterile 35 mm Petri dish.
NOTE: Any number of tardigrades may be processed in this way. Usually, three tardigrades per condition are processed for extraction. - Wash the tardigrades three times, using 1 mL of autoclaved sterile spring water and a sterile filter-tipped P1000 pipette. Slowly pipetting them up and down helps to remove algal contaminants.
- Using a dissecting microscope at 25x to 50x magnification, transfer a single tardigrade from this washed culture to a new sterile 35 mm Petri dish using a sterile filter-tipped P10 pipette.
- Use a sterile filter-tipped P200 pipette to wash the single tardigrade in 100 μL of sterile nuclease-free water.
NOTE: This wash step is used to further remove contaminants, including ribonucleases. - Transfer the washed tardigrade to the bottom of a clean, sterile PCR tube in 1-2 μL of sterile nuclease-free water using a sterile filter-tipped P10 pipette, carefully ensuring the tardigrade does not stick to the side of the tip.
- Visualize the tardigrade under a dissecting microscope at 25x magnification.
- To facilitate water removal of water, break the tip of the pulled glass micropipette lightly outside of the tube. Ensure the bore is big enough to pull up the water but not the tardigrade.
- Using the capillary action of a pulled glass micropipette, remove water until the animal is surrounded by a small bubble of water approximately two tardigrade lengths in diameter.
- Monitor the water removal process via the dissecting scope to ensure the water level is appropriate and the tardigrade remains hydrated.
NOTE: Figure 1C offers an example of how much water to remove. This is a critical step. A small bubble of water will surround the tardigrade to prevent it from drying out, but as much excess water as possible should be removed to prevent dilution of the lysis buffer. For an example of the remaining water levels, please refer to Figure 1C. - Immediately after removing the water, add 2 μL of tardigrade lysis buffer to the bottom of the tube, briefly vortex, and centrifuge the tube at RT for 5 s at 2000 x g on a tabletop centrifuge.
- Immediately place the samples containing the tardigrades into a PCR tube rack and ensure that they are held tight by the rack.
- Grip the rack using a pair of long coarse forceps and gently dip the rack containing the samples into the liquid nitrogen until fully frozen (Figure 1D).
- Remove the rack from the liquid nitrogen and immediately place it on ice. Allow the sample to thaw (takes ~45 s to 1 min). Monitor the sample every 15 s by removing it from the ice and visibly inspecting it. Once the sample is visibly transparent, move on to the next step.
- Repeat steps 4.18-4.19 five more times. A total of six freeze-thaw cycles are required for maximal lysis and extraction (Figure 2A,B).
- Once the freeze-thaw is complete, place samples on ice and immediately progress to the next step. Do not freeze the samples at this point for storage, as this will diminish available RNA for cDNA preparation.
5. cDNA synthesis
- Add 2 μL of cDNA synthesis master mix to the PCR tube containing tardigrade lysate. Briefly flick the tube and spin it at RT at 2000 x g for 5 s with a tabletop centrifuge before replacing the samples on ice.
- Place the samples in a thermocycler and incubate at 25 °C for 10 min to anneal primers, at 55 °C for 30 min to perform reverse transcription, and finally, heat-inactivate enzymes at 85 °C for 5 min.
- After the incubation, immediately place the tube on ice and dilute the sample to a total volume of 25 μL by adding 21 μL of sterile nuclease-free water. For low-copy number transcripts, alter this dilution step as determined empirically.
6. qPCR
- Determine the annealing temperature of the primer set using total RNA prepared from larger amounts of tardigrades, for example, the bulk extraction method presented in Boothby, 201843.
- Run a PCR temperature gradient to determine the optimal annealing temperature before running qRT-PCR (for all PCR settings used in this protocol, refer to Table 1 and Table 2).
- Thaw one tube of indicator dye super mix on ice and isolate from light. Place a 96-well qPCR plate on ice and place 5 μL of super mix, 2 μL of water, 1 μL of each primer (10 μM), and 1 μL of cDNA product in the number of desired wells.
- Seal the PCR plate with plate seal and run the qRT-PCR using an annealing temperature appropriate for the primer set (for all qRT-PCR settings used in this paper, refer to Table 3).
7. Quantification and results interpretation
- Compare the results quantitatively to one or more control housekeeping genes, whose expression is expected to be constant over the imposed conditions. For this study, the actin gene was used.
- Obtain and compare the Ct-values or cycle threshold for each well to the Ct values of the control housekeeping gene reactions. Calculate the fold change in gene expression using the following equation:
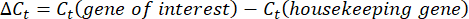
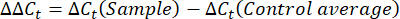
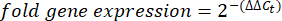
NOTE: Fold gene expression is plotted for each transcript and tardigrade as a 2-(ΔΔCt)48. - To obtain a rough estimate of the transcript number from the Ct-value, use the following equation:
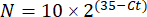
Where N is the number of transcripts, and 2 is the assumed PCR efficiency or the fold increase in fluorescence per cycle of PCR48.
Access restricted. Please log in or start a trial to view this content.
Results
Development and optimization of single-tardigrade RNA extraction
Adapting the protocol from Ly et al., 201542 for RNA extraction in tardigrades, the STST system is optimized to maximize the quantity and quality of the preparation (Figure 1A). RT-PCR was performed for actin transcripts, quantifying transcript yield by amplifying a 527 bp region spanning exons 1 and 2 (sequences for these primers can be found in Table 1). The opti...
Access restricted. Please log in or start a trial to view this content.
Discussion
This study presents an efficient method for the extraction of RNA for single-tardigrade qRT-PCR. Directly comparing the STST methodology to an existing single tardigrade RNA extraction kit revealed that STST RNA extraction yields >200-fold higher amounts of actin RNA transcripts reduces the cost to less than one dollar per sample, and reduces the time required for extraction by 30%. To apply STST to a relevant biological question, we assessed the short-term heat-shock response expression profile. We found that transcr...
Access restricted. Please log in or start a trial to view this content.
Disclosures
The authors declare no conflicts of interest to disclose.
Acknowledgements
We want to acknowledge the NIH Ruth Kirschstein Fellowship # 5F32AG081056-02 and the Errett Fisher Post-Doctoral Fellowship, which supported Dr. Molly J. Kirk, the Crowe Family Fellowship, which supported Chaoming Xu, and a University of California, Santa Barbara Academic Senate Grant, and NIH grants #R01GM143771 and #2R01HD081266, which supported these research efforts. The authors also acknowledge the use of the Biological Nanostructures Laboratory within the California NanoSystems Institute, supported by the University of California, Santa Barbara, and the University of California, Office of the President.
Access restricted. Please log in or start a trial to view this content.
Materials
| Name | Company | Catalog Number | Comments |
| 10 µL Premium Barrier Tips Low Binding, Racked, Sterile | Genesee Scientific | 23-401 | Refered to as Sterile Filter-Tipped P 10 Pipette Tips |
| 1000 µL Premium Pipet Tips, Low Binding, Racked, Sterile | Genesee Scientific | 23-165RS | Refered to as Sterile Filter-Tipped P 1000 Pipette Tips |
| 200 µL Premium Barrier Tips Low Binding, Racked, Sterile | Genesee Scientific | 23-412 | Refered to as Sterile Filter-Tipped P 200 Pipette Tips |
| 4 Star Straight Strong Medium Point Tweezer | Excelta | 00-SA-DC | Refered to as Long forceps |
| 96-Well PCR Rack with Lid Assorted, 5 Racks/Unit | Genesee Scientific | 27-202A | Refered to as PCR Rack |
| Andwin Scientific 3M LEAD FREE AUTOCLAVE TAPE 1" | Thermo Fisher Scientific | NC0802040 | Refered to as Autoclave Tape |
| Autoclave Tape | Thermo Fisher Scientific | AB1170 | Refered to as PCR Plate Seals |
| Benchling v8 | Benchling | N/A | Refered to as Benchling |
| BioRadHard-Shell 96-Well PCR Plate | BioRad | HSS9641 | Refered to as PCR Plate |
| BULWARK FR Lab Coat: | Grainger | 26CF64 | Refered to as Lab Coat |
| C1000 Touch Bio-rad Thermocycler | BioRad | 1851148 | Refered to as Thermocycler |
| C1000 Touch Bio-rad Thermocycler with CFX Optics Module | BioRad | 1845097 | Refered to as qPCR thermocycler |
| Chloroccoccum hypnosporum. | Carolina | 152091 | Refered to as Algae |
| Corning PYREX Reusable Media Storage Bottles | Thermo Fisher Scientific | 06-414-1E | Refered to as 2 L Autoclave-safe Glass Bottle |
| Daigger & Company Vortex-Genie 2 Laboratory Mixer | Thermo Fisher Scientific | 3030A | Refered to as Vortexer |
| Direct-zol Micro Prep | Zymo Research | R2060 | Refered to as RNA extraction kit |
| Dumont 5 Biology Tweezers | Fine Science Tools | 11254-20 | Refered to as Fine Forceps |
| EDTA | Fisher Scientific | S311-500 | Refered to as EDTA |
| FIJI v 2.14.0/1.54f | ImageJ, | N/A | Refered to as FIJI/ImageJ |
| Filament for pippette Puller | Tritech Research | PC-10H | Refered to as Filament |
| Fisherbrand Economy Impact Goggles | Fisher Scientific | 19-181-501 | Refered to as Splash Goggles |
| Glass Micropipette O.D. 1mm ID 0.58, Length 10 cm | TriTech Research | GD-1 | Reffered to as glass micropipette |
| Hypsibius exemplaris Z151 Strain | Carolina | 133960 | Refered to as Tardigrades or H. exemplaris |
| Liquid Nitrogen Dewar 1 L | Agar Scientific | AGB7475 | Refered to as Cryo-safe container |
| Maxima H Minus First Strand cDNA Synthesis Kit | Thermo Fisher Scientific | K1651 | Refered to as cDNA Synthesis Master Mix |
| Narishige Dual-Stage Glass Micropipette Puller | Tritech Research | PC-10 | Refered to as micropipette puller |
| Nitrile Gloves | Fisher Scientific | 17-000-314 | Refered to as Nitrile Gloves |
| PETRI DISH, PS, 35/10 mm, WITH VENTS | Grenier | 627102 | Refered to as 35 mm Petri dish |
| PIPETMAN P10, 1–10 µL, Metal Ejector | Gilson | F144055M | Refered to as P 10 Pipette |
| PIPETMAN P1000, 100–1000 µL, Metal Ejector | Gilson | F144059M | Refered to as P 1000 Pipette |
| PIPETMAN P200, 20–200 µL, Metal Ejector | Gilson | F144058M | Refered to as P 200 Pipette |
| Pound This 4-Color Modeling Clay | American Science Surplus | 96517P001 | Refered to as Clay |
| Prism v10.0 | GraphPad | N/A | Refered to a Prism |
| RNAse-Free, 8 Strip 0.2 mL PCR Tubes with caps | Invitrogen | AM12230 | Refered to as Sterile PCR Tube |
| RNasin Ribonuclease Inhibitor | Promega | N2111 | Refered to as RNAse inhibitor |
| Spring water | Nestle Pure Life | 44221229 | Refered to as Spring Water |
| SsoAdvanced Universal SYBR Green Supermix | BIO RAD | 1725271 | Refered to as Indicator Dye Super mix |
| Stereo-Microscope System w/optics and illumination | TriTech Research | SMT1 | Refered to as Dissecting Microscope |
| Supertek Scientific Tirrill Burners | Thermo Fisher Scientific | S09572B | Refered to as Bunsen Burner |
| Table Top Centrifuge | Qualitron | DW-41-115-NEW | Refered to as Table Top Centrifuge |
| Tempshield Cryo-Gloves | Fisher Scientific | 11-394-305 | Refered to as Cryo Gloves |
| Thermo Scientific Nunc Petri Dishes | Thermo Fisher Scientific | 08-757-099 | Refered to as 100 mm Petri dish |
| Tris base | Fisher Scientific | T395-500 | Refered to as Tris or Tris Base |
| Triton X-100 | Fluka | 93443 | Refered to as Detergent 1 |
| TWEEN 20 | Sigma aldrich | P1379-500 | Refered to as Detergent 2 |
| Water - PCR/RT-PCR certified, nuclease-free | Growcells | PCPW-0500 | Refered to as Sterile Nuclease Free Water |
References
- Møbjerg, N., Neves, R. C. New insights into survival strategies of tardigrades. Comp Biochem Physiol Part A Mol Integr Physiol. 254, 110890(2021).
- Jönsson, K. I., Harms-Ringdahl, M., Torudd, J. Radiation tolerance in the eutardigrade Richtersius coronifer. Int J Radiat Biol. 81 (9), 649-656 (2005).
- Horikawa, D. D., et al. Radiation tolerance in the tardigrade Milnesium tardigradum. Int J Radiat Biol. 82 (12), 843-848 (2006).
- Bruckbauer, S. T., Cox, M. M. Experimental evolution of extremophile resistance to ionizing radiation. Trends Genet. 37 (9), 830-845 (2021).
- Jönsson, K. I., Hygum, T. L., Andersen, K. N., Clausen, L. K. B., Møbjerg, N. Tolerance to gamma radiation in the marine heterotardigrade, Echiniscoides sigismundi. PLoS One. 11 (12), e0168884(2016).
- Jönsson, K. I. Radiation tolerance in tardigrades: Current knowledge and potential applications in medicine. Cancers (Basel). 11 (9), 1333(2019).
- Yoshida, Y., et al. RNA sequencing data for gamma radiation response in the extremotolerant tardigrade Ramazzottius varieornatus. Data Brief. 36, 107111(2021).
- Clark-Hachtel, C. M., et al. The tardigrade Hypsibius exemplaris dramatically upregulates DNA repair pathway genes in response to ionizing radiation. Curr Biol. 34 (9), 1819-1830.e6 (2024).
- Anoud, M., et al. Comparative transcriptomics reveal a novel tardigrade specific DNA binding protein induced in response to ionizing radiation. Elife. 13, RP92621(2024).
- Jönsson, K. I., Schill, R. O. Induction of Hsp70 by desiccation, ionising radiation and heat-shock in the eutardigrade Richtersius coronifer. Comp Biochem Physiol B Biochem Mol Biol. 146 (4), 456-460 (2007).
- Boothby, T. C. Desiccation of Hypsibius exemplaris. Cold Spring Harb Protoc. 2018 (11), 871-873 (2018).
- Boothby, T. C., et al. Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell. 65 (6), 975-984.e5 (2017).
- Horikawa, D. D., Higashi, S. Desiccation tolerance of the tardigrade Milnesium tardigradum collected in Sapporo, Japan, and Bogor, Indonesia. Zoolog Sci. 21 (8), 813-816 (2004).
- Halberg, K. A., Jørgensen, A., Møbjerg, N. Desiccation tolerance in the tardigrade Richtersius coronifer relies on muscle mediated structural reorganization. PLoS One. 8 (12), e3330(2013).
- Sørensen-Hygum, T. L., Stuart, R. M., Jørgensen, A., Møbjerg, N. Modelling extreme desiccation tolerance in a marine tardigrade. Sci Rep. 8 (1), 11495(2018).
- Lyons, A. M., Roberts, K. T., Williams, C. M. Survival of tardigrades (Hypsibius exemplaris) to subzero temperatures depends on exposure intensity, duration, and ice-nucleation - as shown by large-scale mortality dye-based assays. bioRxiv. , (2024).
- Møbjerg, A., et al. Extreme freeze-tolerance in cryophilic tardigrades relies on controlled ice formation but does not involve significant change in transcription. Comp Biochem Physiol Part A Mol Integr Physiol. 271, 111245(2022).
- Tsujimoto, M., Imura, S., Kanda, H. Recovery and reproduction of an Antarctic tardigrade retrieved from a moss sample frozen for over 30 years. Cryobiology. 72 (1), 78-81 (2016).
- Jönsson, K. I. Tardigrades as a potential model organism in space research. Astrobiology. 7 (5), 757-766 (2007).
- Jönsson, K. I., Rabbow, E., Schill, R. O., Harms-Ringdahl, M., Rettberg, P. Tardigrades survive exposure to space in low Earth orbit. Curr Biol. 18 (17), R729-R731 (2008).
- Kasianchuk, N., Rzymski, P., Kaczmarek, Ł The biomedical potential of tardigrade proteins: A review. Biomed Pharmacother. 158, 113983(2023).
- Arakawa, K. Examples of extreme survival: Tardigrade genomics and molecular anhydrobiology. Annu Rev Anim Biosci. 10 (1), 519-542 (2022).
- Hvidepil, L. K. B., Møbjerg, N. New insights into osmobiosis and chemobiosis in tardigrades. Front Physiol. 14, 1274522(2023).
- Tanaka, S., Aoki, K., Arakawa, K. In vivo expression vector derived from anhydrobiotic tardigrade genome enables live imaging in Eutardigrada. Proc Natl Acad Sci U S A. 120 (5), e2216739120(2023).
- Kondo, K., Tanaka, A., Kunieda, T. Single-step generation of homozygous knockout/knock-in individuals in an extremotolerant parthenogenetic tardigrade using DIPA-CRISPR. PloS Genet. 20 (6), e1011298(2024).
- Yoshida, Y., Hirayama, A., Arakawa, K. Transcriptome analysis of the tardigrade Hypsibius exemplaris exposed to the DNA-damaging agent bleomycin. bioRxiv. , (2024).
- Yoshida, Y., et al. Time-series transcriptomic screening of factors contributing to the cross-tolerance to UV radiation and anhydrobiosis in tardigrades. BMC Genomics. 23 (1), 405(2022).
- Yoshida, Y., et al. RNA sequencing data for gamma radiation response in the extremotolerant tardigrade Ramazzottius varieornatus. Data Brief. 36, 107111(2021).
- Neves, R. C., et al. Differential expression profiling of heat stressed tardigrades reveals major shift in the transcriptome. Comp Biochem Physiol Part A Mol Integr Physiol. 267, 111143(2022).
- Yoshida, Y., et al. Comparative genomics of the tardigrades Hypsibius dujardini and Ramazzottius varieornatus. PLoS Biol. 15 (7), e2002266(2017).
- Wang, C., Grohme, M. A., Mali, B., Schill, R. O., Frohme, M. Towards decrypting cryptobiosis - analyzing anhydrobiosis in the tardigrade Milnesium tardigradum using transcriptome sequencing. PLoS One. 9 (3), e92663(2014).
- Mali, B., et al. Transcriptome survey of the anhydrobiotic tardigrade Milnesium tardigradum in comparison with Hypsibius dujardini and Richtersius coronifer. BMC Genomics. 11 (1), 168(2010).
- Förster, F., et al. Transcriptome analysis in tardigrade species reveals specific molecular pathways for stress adaptations. Bioinform Biol Insights. 6, 69-96 (2012).
- Arakawa, K., Yoshida, Y., Tomita, M. Genome sequencing of a single tardigrade Hypsibius dujardini individual. Sci Data. 3 (1), 160063(2016).
- Arakawa, K. Transcriptome assembly of Richtersius coronifer with annotated BLAST result against Ramazzottius varieornatus. Figshare. Dataset. , (2019).
- Yoshida, Y., Konno, S., Nishino, R., Murai, Y., Tomita, M., Arakawa, K. Ultralow input genome sequencing library preparation from a single tardigrade specimen. J Vis Exp. (137), (2018).
- Murai, Y., et al. Multiomics study of a heterotardigrade, Echiniscus testudo, suggests convergent evolution of anhydrobiosis-related proteins in Tardigrada. bioRxiv. , (2020).
- Yoshida, Y., Sugiura, K., Tomita, M., Matsumoto, M., Arakawa, K. Comparison of the transcriptomes of two tardigrades with different hatching coordination. BMC Dev Biol. 19 (1), 24(2019).
- Tenlen, J. R. Microinjection of dsRNA in tardigrades. Cold Spring Harb Protoc. 2018 (11), (2018).
- Tenlen, J. R., McCaskill, S., Goldstein, B. RNA interference can be used to disrupt gene function in tardigrades. Dev Genes Evol. 223 (3), 171-181 (2013).
- Giovannini, I., et al. Production of reactive oxygen species and involvement of bioprotectants during anhydrobiosis in the tardigrade Paramacrobiotus spatialis. Sci Rep. 12 (1), 15888(2022).
- Ly, K., Reid, S. J., Snell, R. G. Rapid RNA analysis of individual Caenorhabditis elegans. MethodsX. 2, 59-63 (2015).
- Boothby, T. C. Total RNA extraction from tardigrades. Cold Spring Harb Protoc. 2018 (11), 905-907 (2018).
- Czerneková, M., Vinopal, S. The tardigrade cuticle. Limnol Rev. 21 (3), 127-146 (2021).
- Goldstein, B. Hypsibius dujardini. collection notes and culture protocol from Bob McNuff. , At http://tardigrades.bio.unc.edu/protocols/CollectionCulture.pdf (2007).
- McNuff, R. Laboratory culture of Hypsibius exemplaris. Cold Spring Harb Protoc. 2018 (11), 867-870 (2018).
- Gabriel, W. N., et al. The tardigrade Hypsibius dujardini, a new model for studying the evolution of development. Dev Biol. 312 (2), 545-559 (2007).
- Ruiz-Villalba, A., Ruijter, J. M., van den Hoff, M. J. B. Use and misuse of cq in qPCR data analysis and reporting. Life (Basel). 11 (6), 508(2021).
- Antonov, J., et al. Reliable gene expression measurements from degraded RNA by quantitative real-time PCR depend on short amplicons and a proper normalization. Lab Invest. 85 (8), 1040-1050 (2005).
- Toussaint, J., et al. Improvement of the clinical applicability of the genomic grade index through a qRT-PCR test performed on frozen and formalin-fixed paraffin-embedded tissues. BMC Genomics. 10, 424(2009).
Access restricted. Please log in or start a trial to view this content.
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved